Abstract
The global knowledge of microbial diversity and function in Sundarbans ecosystem is still scarce, despite global advancement in understanding the microbial diversity. In the present study, we have analyzed the diversity and distribution of bacteria in the tropical mangrove sediments of Sundarbans using 16S rRNA gene amplicon sequencing. Metagenome is comprised of 1,53,926 sequences with 108.8 Mbp data and with 55 ± 2% G + C content. Metagenome sequence data are available at NCBI under the Bioproject database with accession no. PRJNA245459. Bacterial community metagenome sequences were analyzed by MG-RAST software representing the presence of 56,547 species belonging to 44 different phyla. The taxonomic analysis revealed the dominance of phyla Proteobacteria within our dataset. Further taxonomic analysis revealed abundance of Bacteroidetes, Acidobactreia, Firmicutes, Actinobacteria, Nitrospirae, Cyanobacteria, Planctomycetes and Fusobacteria group as the predominant bacterial assemblages in this largely pristine mangrove habitat. The distribution of different community datasets obtained from four sediment samples originated from one sampling station at two different depths providing better understanding of the sediment bacterial diversity and its relationship to the ecosystem dynamics of this pristine mangrove sediment of Dhulibhashani in, Sundarbans.
Keywords: Mangrove sediment, Dhulibhashani, Metagenome, Pyrosequencing, Bacterial diversity
| Specifications | |
|---|---|
| Organism/cell line/tissue | Environmental metagenomes |
| Sex | Not applicable |
| Sequencer or array type | 454 GS junior platform |
| Data format raw data: | Sff file |
| Experimental factors | Environmental samples |
| Experimental features | Bar-coded pyrosequencing and community structure diversity analysis using MG-RAST pipeline |
| Consent | Not applicable |
| Sample source location | Mangrove soil sediment, Sundarbans, India |
1. Direct link to deposited data
http://www.ncbi.nlm.nih.gov/sra/SRX883521
Random mutagenic events followed by natural selective pressure, inflames the evolutionary mechanism to give rise new lineages of microorganisms in the microbial world at an alarming high frequency. This new groups of microorganisms are so nascent to the environment that currently available bacterial culture procedures are completely inept at confining them. Due to the uniqueness in physicochemical characteristics and exclusivity in nutrient composition of a pristine ecosystem, the unculturable microbial community prevails on such part of the earth. The studies of complex microbial communities have considerably advanced in recent years, mainly facilitated by metagenomics and other “omics” coupled with high-throughput DNA sequencing technologies and bioinformatics [1], [2], [3]. Studies using high throughput sequencing conducted in mangrove sediments in Brazil, China and India have enlarged our knowledge on the microbial taxonomic diversity and functional potential [4], [5], [6], [7], however there are still challenges to be overcome regarding the factors involved in shaping the microbial biogeography in mangroves.
Sundarbans is the world's largest tidal halophytic mangrove ecosystem situated in the delta of Ganges, Meghna, and Brahmaputra rivers on the Bay of Bengal, and has been recognized as a UNESCO World Heritage site, plays critical roles on the sea–continent interface providing environmental conditions for the development of important marine and estuarine species. Microbial communities play an important role in the generation of detritus in mangrove areas and involved in biogeochemical cycling of the nutrients [8], [9]. However, no data are yet available regarding the abundance and diversity of bacterial in Dhulibhashani, Sundarbans despite global advancement in understanding the microbial diversity and role of microbes in different mangrove environments, little has been performed in Sundarbans [6]. In order to gain new insight into the bacterial community, we examined temporal changes in the microbial diversity of the sampling station during the monsoon (July 2013) and post-monsoon (December 2013) seasons mainly to understand the role of climate and other parameters in shaping the sediment microbial communities in Sundarbans to build foundational information for future research employing 454-pyrosequencing.
This study comprised of Dhulibhashani (21°37′40.837″N 88°33′47.762″E) mangrove, which is located on the North Eastern coast of India. Sediments were collected from inside of the mangroves during the monsoon (July 13) and post-monsoon season (Dec 2013, aiming to cover the typical habitats in the ecosystems. Samples were collected in triplicate in a sealed sterile container from surface (2 cm) and subsurface (32 cm) sediments of the Sundarbans mangrove wetland. Metagenomic DNA was extracted from soil subsamples using Mo-Bio DNA Power Soil kit (MoBio Laboratories, Carlsbad, CA). To analyze bacterial diversity, the V1–V3 regions of bacterial 16S rRNA gene were amplified by PCR. Pyrosequencing was performed for 200 cycles on a Roche 454 GS-Junior sequencing instrument according to the manufacturer's protocol (454 Life Sciences, USA). The raw data were subjected to initial quality trimming using the MOTHUR software and chimeric sequences were removed by using UCHIME algorithm. The sequences were analyzed with MG-RAST NGS analysis pipeline. The output was 1,53,926 sequences with size 108.8 Mbp and 55 ± 2% G + C.
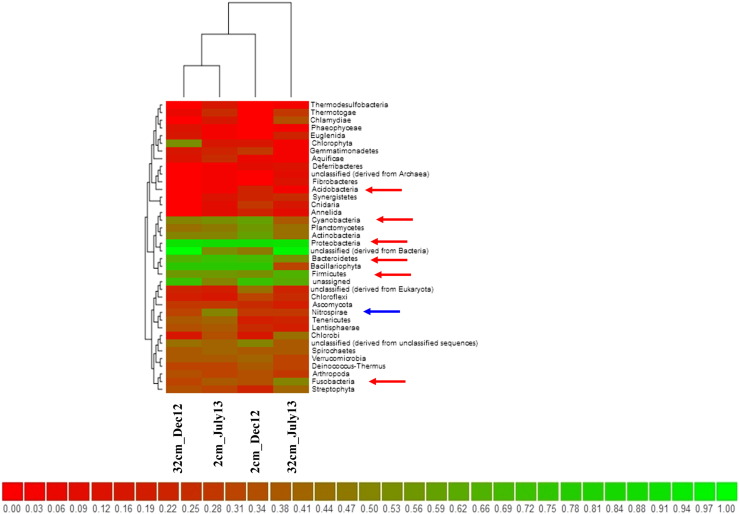
After the analysis of metagenome sequences, a total of 44 phyla were present in two seasons (July 13 and Dec 13) of Dhulibhashani, Sundarbans. In the monsoon sample (July 13) 42 different phyla were recorded whereas 39 phyla were observed at post-monsoon (Dec 13) respectively (Fig. 1). Interestingly a less number of phyla were observed in the soil sample of post-monsoon, possibly due to change in the physico-chemical parameters (salinity, conductivity, and water level). Proteobacteria is the most dominating phyla in both sampling seasons. Out of this experimental data, the abundance of Proteobacteria was found to be 53% at 2 cm_Dec 2013 and 31% at 32 cm_Dec 2013 (post-monsoon) and 44% at 2 cm_ July 13 and 38% at 32 cm_July 13 (monsoon) respectively. Besides Proteobacteria, Bacteroidetes formed 15% and 5% bacterial population in post-monsoon (Dec 13) and 19% and 2% in monsoon (July 13) respectively (Fig. 2). The third abundant phyla were Firmicutes in bothseasons. Insignificant abundance was detected in both sampling seasons for the phyla Nitrospirae. Most interesting phyla, Actinobacteria accounted 7% for the post-monsoon at two different depth samples (2 cm_Dec 13 and 32 cm_Dec 13) and only 5% for monsoon sample only in surface sample (2 cm_July 13). Planctomycetes were found to be the less abundant phyla for both seasons (Dec: 6% and July: 5%). In summary, the use of 454-pyrosequencing platform allowed us to elucidate the bacterial diversity of this pristine mangrove environment of Dhulibhashani, Sundarbans, and indicates the potentiality to explore new bioactive compounds [10] from the mangrove environment leading immense benefit human kind. Our results showed that the microbiomes from the North Eastern coast of India, shaped by the enormous characteristics of the bacterial community suggesting potential abundance of microbial products. Despite the shifts in bacterial composition, mainly represented by the enrichment of Proteobacteria, the potential metabolic functions remain unchanged, and emphasize the importance of the functional redundancy for the resilience of these environments.
Fig. 1.
Taxonomic distribution of the obtained metagenome sequences. Bacterial communities are dominant within the pristine sediment of Dhulibhashani, Sundarbans. The figure was prepared by the Krona interactive visualization program built within the MG-RAST portal. Shown are A (32 cm_Dec 12), B (2 cm_July 13), C (2 cm_Dec 12) and D (32 cm_July 13), respectively.
Fig. 2.
Hierarchical ward clustering heat map generated from taxonomic abundance profile reflecting distribution of Bacterial phyla in the sediment Dhulibhashani, Sundarbans.
In summary, the sedimentary bacterial diversity has been studied in the Dhulibhashani island of Sundarbans mangrove ecosystem for the first time by means of 16S rRNA454-pyrosequencing. We found highly diverse bacterial communities in the pristine sediment of Dhulibhashani, Sundarbans. This will be helpful in cataloguing and describing the bacteria diversity of the mangrove sediment of Sundarbans, India that needs to be conserved and simultaneously could be the habitat for large number of economically important microbes. Further research including analyses of expression of functional genes and determination of the active bacterial population within the sediment might unravel the role of bacteria in maintenance of biogeochemical cycle and discovery of bioactive compounds of Sundarbans mangrove ecosystem.
1.1. Nucleotide sequence accession number
All 454-GS Junior sequence data from this study were submitted to the NCBI Sequence Read Archive (SRA) under accession numbers SRR2061029 (Dhulibhashani_2 cm_July 13), SRR2061030 (Dhulibhashani_32 cm_July 13), SRR2061032 (Dhulibhashani_2 cm_Dec 13) and SRR2061033 (Dhulibhashani_32 cm_Dec 13).
Acknowledgments
Funding for this study was provided by the World Bank under the ICZM project grants 54-ICZM/3P/2010 in the Department of Biochemistry, University of Calcutta, India. We acknowledge Mr. Tapas Paul, World Bank, for his continuous support and enthusiasm regarding our study in Sundarbans. The authors express their sincere thanks to Dr. Somnath Bhattacharyya (IESWM) for his support during the study and Mr. Niladri Shekher Majumdar, Roche Diagnostics, for his technical support in pyrosequencing work.
References
- 1.Tringe S.G., Mering C.V., Kobayashi A., Salamov A.A., Chen K., Chang H.W. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 2.Lauber C.L., Hamady M., Knight R., Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards R.A., Rodriguez-Brito B., Wegley L., Haynes M., Breitbart M., Peterson D.M. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics. 2006;7:57. doi: 10.1186/1471-2164-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreote F.D., Jiménez D.J., Chaves D., Dias A.C.F., Luvizotto D.M., Dini-Andreote F. The microbiome of Brazilian mangrove sediments as revealed by metagenomics. PLoS ONE. 2012;7(6) doi: 10.1371/journal.pone.0038600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang X.T., Peng X., Deng G.H., Sheng H.F., Wang Y., Zhou H.W. Illumina sequencing of 16S rRNA tag revealed spatial variations of bacterial communities in a mangrove wetland. Microb. Ecol. 2013;66:96–104. doi: 10.1007/s00248-013-0238-8. [DOI] [PubMed] [Google Scholar]
- 6.Basak P., Majumder N.S., Nag S., Bhattacharyya A., Roy D., Chakraborty A., SenGupta S., Roy A., Mukherjee A., Pattanayak R., Ghosh A., Chattopadhyay D., Bhattacharyya M. Spatiotemporal analysis of bacterial diversity in sediments of Sundarbans using parallel 16S rRNA gene tag sequencing. Microb. Ecol. 2014;69:500–511. doi: 10.1007/s00248-014-0498-y. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharyya A., Majumder N., Basak P., Roy D., Nag S., Haldar A., Mukherjee S., Chattopadhyay D., Mitra S., Bhattacharyya M., Ghosh A. Diversity and distribution of archaea in the mangrove sediment of Sundarbans. Archaea. 2015 doi: 10.1155/2015/968582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh A., Dey N., Bera A. Culture independent molecular analysis of bacterial communities in the mangrove sediment of Sundarbans, India. Saline Syst. 2010;6(1) doi: 10.1186/1746-1448-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakraborty A., Bera A., Mukherjee A., Basak P. Changing bacterial profile of Sundarbans, the world heritage mangrove: impact of anthropogenic interventions. World J. Microbiol. Biotechnol. 2015;31:593–610. doi: 10.1007/s11274-015-1814-5. [DOI] [PubMed] [Google Scholar]
- 10.Sengupta S., Pramanik A., Ghosh A., Bhattacharyya M. Antimicrobial activities of actinomycetes isolated from unexplored regions of Sundarbans mangrove ecosystem. BMC Microbiol. 2015;15(170) doi: 10.1186/s12866-015-0495-4. [DOI] [PMC free article] [PubMed] [Google Scholar]