Abstract
Deulajhari hot spring has diverse temperature and pH range varying from 43 °C to 65 °C and 7.83 to 8.10 respectively. Dense foliage around Deulajhari hot spring contributes to the high total organic carbon content (TOC). In our experiment we took sediment samples from the two Deulajhari hot springs (S1 and S2) out of the cluster having temperature of 43 °C and 55 °C and pH of 7.83 and 7.14 respectively. Sediment samples were analysed using 16S rRNA of V3‐V4 region by amplicon metagenome sequencing. Over 34 phyla were detected in cluster S1 and 32 phyla in cluster S2 at the existing physiochemical parameters temperature 43 °C, pH 7.83, electroconductivity 0.019 dSm− 1, and total organic carbon (TOC) 3.80% for S1 and temperature 55 °C, pH 7.14, electroconductivity 0.019 dSm− 1, and total organic carbon (TOC) 0.97% for S2. Existence of a vast number of unresolved sequences 179 out of 292 in S1 and 186 out of 314 in S2 at the genus level emphasizes the significance of our study. Metagenome sequence information for the both clusters S1 and S2 of Deulajhari is available at NCBI, SRA database with accession number SRX1459732 and SRX1459733 respectively.
Direct link to the deposited data:
www.ncbi.nlm.nih.gov/sra/SRX1459732
www.ncbi.nlm.nih.gov/sra/SRX1459733
Keywords: Deulajhari, 16S rRNA, Metagenome, TOC, Illumina platform
1. Experimental design, material and methods
Odisha being bestowed with a range of hot springs located at different geographical locations, is a hub for diverse microbial abundance. Eight hot springs present in different districts of Odisha are Tarabalo in Nayagarh, Deulajhari and Magarmuhan in Angul, Atri and Badaberena in Khurda, Taptapani in Ganjam, Boden in Nuapada, and Bankhol in Cuttack [1]. Our study is the first attempt to explore the microbial diversity of Deulajhari hotspring cluster S1(43 °C) and S2(55 °C) using the Illumina sequencing platform.
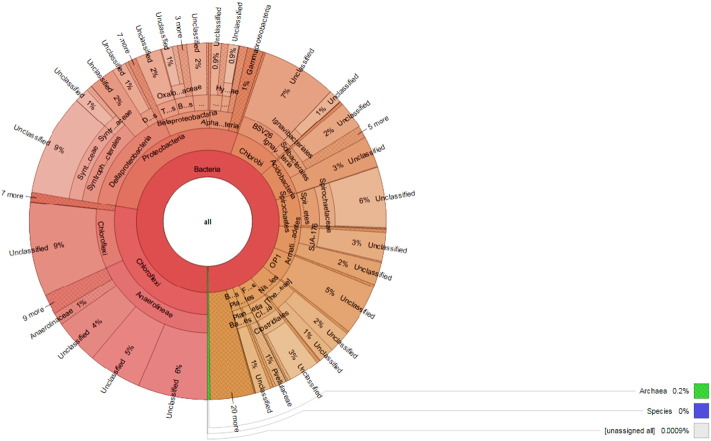
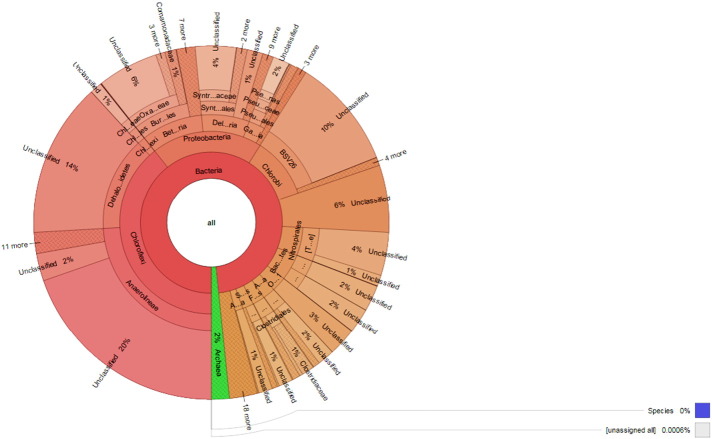
Metagenomic DNA isolation was done from the sediment sample of two clusters S1 and S2 collected from the Deulajhari hot spring (Latitude 20.74199 N, Longitude 84.49206°E) as per Kumar and Khanna, 2014 [2]. Primers 341F, 5′CCTACGGGAGGCAGCAG-3′ and 518R, 5′-ATTACCGCGGCTGCTGG-3′ were used for the amplification of V3–V4 region of 16S rRNA using 50 ng of metagenomic DNA from each cluster S1 and S2. Minelute column (QIAGE, India) was used for the purification of amplified PCR product and Illumina GAIIX sequencer (Genotypic Technology, Pvt. Ltd. Bangalore, India) was used for 150 nucleotide paired and multiplex sequencing. Bioinformatic analysis was done through QIIME pipeline. Krona tool (Fig. 1, Fig. 2) [3] was used for plotting krona graph for each cluster.
Fig. 1.
Abundance of bacterial community in S1 spring of Deulajhari metagenome.
Fig. 2.
Abundance of bacterial community in S2 spring of Deulajhari metagenome.
333,524 and 611,391 high quality reads were obtained from the cluster S1 and S2 respectively and cluster S1 showed the abundance of Chloroflexi (27.65%), Proteobacteria (27.60%), Chlorobi (8.32%), Acidobacteria (6.49%) and Spirochaetes (5.94%) at the phylum level and cluster S2 showed the abundance of Chloroflexi (39.39%), Proteobacteria (19.53%), Chlorobi (10.83%), OP1(6.16%) and Nitrospirae (4.96%) at the phylum level. Total 292 genera were reported in cluster S1 out of which 113 genera were identified while 179 genera still remain unidentified and in cluster S2 total 314 genera were reported out of which 128 were identified and 186 genera remain unidentified.
Thus through the diverse knowledge of microbiota in the two clusters S1and S2 we can further suggest the application of thermophilic bacteria in the diverse field of research.
Acknowledgement
We are thankful to the Siksha O Anushandhan University for providing the infrastructure for carrying out the research work and to the Genotypic Technology Pvt. Ltd. (Bangalore, India) for providing the sequencing platform. And we are also thankful to our colleagues for providing the needful support during sampling and other technical assistance.
Contributor Information
Archana Singh, Email: singh.archana7373@gmail.com.
Enketeswara Subudhi, Email: enketeswarasubudhi@soauniversity.ac.in, esubudhi.omics@gmail.com.
References
- 1.Sahoo R.K., Subudhi E., Kumar M. Investigation of bacterial diversity of hot springs of Odisha, India. Genomics Data. 2015;6:188–190. doi: 10.1016/j.gdata.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar M., Khanna S. Shift in microbial population in response to crystalline cellulose degradation during enrichment with a semi-desert soil. Int. Biodeterior. Biodegrad. 2014;88:134–141. [Google Scholar]
- 3.Ondov B.D., Bergman N.H., Phillippy A.M. Interactive metagenomics visualization in a web browser. BMC Bioinforma. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]