Abstract
Histone modification profiles are predictive of gene expression and most of the knowledge gained is acquired through studies done in higher eukaryotes. However, genome-wide studies involving Plasmodium falciparum, the causative agent of malaria, have been rather few, at lower resolution (mostly using ChIP-on-chip), and covering limited number of histone modifications. In our recent study [1], we have performed extensive genome-wide analyses of multiple histone modifications including the active (H3K4me2, H3K4me3, H3K9ac, H3K14ac, H3K27ac and H4ac), inactive (H3K9me3 and H3K27me3), elongation (H3K79me3) and regulatory element (H3K4me1) in a stage-specific manner. Furthermore, we used a ligation-based method suitable for sequencing homopolymeric stretches as seen in P. falciparum for next-generation sequencing library amplification [2], enabling highly quantitative analysis of the extremely AT-rich P. falciparum genome. Our recently published study suggests that transcription regulation by virtue of poised chromatin and differential histone modifications is unique to P. falciparum [1]. Here we describe the experiments, quality controls and chromatin immunoprecipitation-sequencing data analysis of our associated study published in Epigenetics and Chromatin [1]. Stage-specific ChIP-sequencing data for histone modifications is submitted to Gene Expression Omnibus (GEO) database under the accession number GSE63369.
Keywords: Plasmodium falciparum, Genome-wide mapping, Histone modifications, Chromatin, Transcription
| Organism/cell line/tissue | Plasmodium falciparum |
| Sex | Not applicable |
| Sequencer or array type | AB SOLiD 4 system high-throughput genome sequencer |
| Data format | Raw and analyzed |
| Experimental factors | ChIP-sequencing of histone modification using antibody |
| Experimental features | ChIP-sequencing of Plasmodium falciparum histone modifications in a stage specific manner |
| Consent | Not applicable |
| Sample source location | Not applicable |
Direct link to deposited data [provide URL below]
The direct link for the ChIP-sequencing data is -http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE63369
1. Experimental design, materials and methods
1.1. Plasmodium falciparum culture and synchronization
P. falciparum strain 3D7 was cultured in RPMI 1640 medium supplemented with 25 mM HEPES, 0.5% AlbuMAX I, 1.77 mM sodium bicarbonate, 100 μM hypoxanthine and 12.5 μg ml− 1 gentamicin sulfate, at a pH of 7.2 in 5% CO2. Incomplete medium was prepared weekly without sodium bicarbonate and stored at 4 °C. Complete medium was freshly prepared with sodium bicarbonate. Fresh whole blood from a healthy donor (O+ ve) was used for preparing RBCs and stored at 4 °C for at least 1 day. RBCs were washed thrice with washing medium (complete medium without AlbuMAX I) before use. Subculturing was done every 2 days for 6–8 h before invasion by equally dividing the contents of each flask into two or more flasks and quickly restoring the haematocrit between 1 and 1.5% in the required volume of culture medium [3]. Medium was changed every 24 h. Asynchronous culture with early ring stage (less than 10 h) was synchronized using 5% sorbitol, which was added 10 times the volume of infected RBCs pellet followed by vigorous vortexing for 30 s to rupture mature parasitic forms. Culture was then kept for incubation at 37 °C for 8 min under shaking at 240 rpm. Culture was centrifuged at 250 g for 5 min to get rid of ruptured RBCs. Pellet was washed twice with washing medium and transferred to a flask containing complete medium. Parasitemia was monitored with acridine orange stained thin blood smear. The synchronized culture was harvested at 18, 30 and 40 hpi for chromatin immunoprecipitation.
1.2. Screening of histone modification antibody for chromatin immunoprecipitation (ChIP)
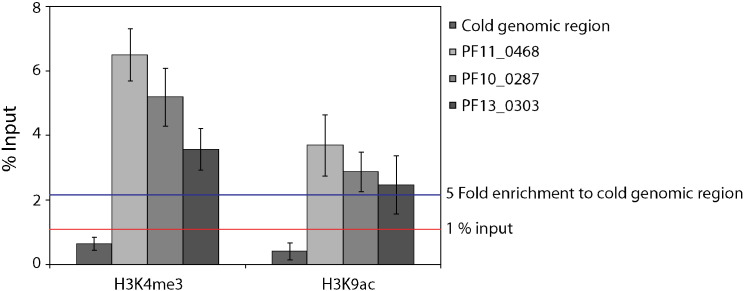
Infected RBCs were cross-linked with 1% formaldehyde (Catalogue number 28908, Thermo Scientific), which was directly added to the culture medium drop-wise in chemical-hood and mixed by rotating for 10 min at room temperature. Formaldehyde fixed cells were quenched with 150 mM glycine for 10 min at room temperature. Infected RBCs were washed twice with 1 × PIC and 1 mM PMSF in cold PBS. Resultant pellet was dissolved in swelling buffer (25 mM Tris pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.1% NP40, 1 mM DTT, 0.5 mM PMSF, 1 × PIC) for nuclei isolation. Nuclei were isolated by dounce homogenization using loose piston (B). Isolated nuclei were lysed and sonicated in sonication buffer (10 mM Tris–HCl pH 7.5, 200 mM NaCl, 1% SDS, 4% NP-40, 1 mM PMSF) to obtain an average chromatin size of 200–400 bp. Chromatin was pre-cleared using 50 μl of a 50% protein A Sepharose (GE healthcare) slurry for 1 h at 4 °C with gentle inverting. Immunoprecipitations were carried out in 1 ml of IP buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton-X 100). Three micrograms of antibody was used per 20 μg purified chromatin. 10% Input chromatin was obtained after preclearing by de-crosslinking and purified using the Qiaquick column (Qiagen) according to the manufacturer's instructions. Immunoprecipitations were carried out with inverting at 4 °C for 14–16 h. The samples were then incubated with 50 μL of a 50% Protein A Sepharose slurry for 3 h at 4 °C with gentle inverting. IP samples were reverse-crosslinked and the DNA was purified using a Qiaquick column (Qiagen). Specificity of ChIP was determined by quantitative PCR for the known histone modification enriched genomic region and an arbitrarily chosen control genomic region. A serial dilution of input sample was performed to calculate the % input enrichment. Samples were processed for ChIP-sequencing if the enrichment was observed more than 1% of input and 5 fold to control genomic region (Fig. 1). Primers used for cold genomic region (Forward 5′-AACGTTAAATTTTGAATCCGAGA-3′, Reverse 5′-AATCTCCGAGACCGGGAAT-3′), Pf11_0468 (Forward 5′-TGTGCACATGGGAATTTCA-3′, Reverse 5′- CTCTTCAATAGCATCCTCTTCATT-3′), PF10_0287(Forward 5′-CCATGAACTGCGACGTCTAC-3′, Reverse 5′-AAAAATCCCTTAAAAAGATGAGTGA-3′), and PF13_0303 (Forward 5′-CAACCATCGTTCCTTGACCT-3′ Reverse 5′-GTAACCGTGCGTGTGCTTTA-3′). We found this method reproducible as the normalization is performed with respect to the control genomic region from the same experimental condition.
Fig. 1.
Assessment of H3K4me3 and H3K9ac ChIP by ChIP-qPCR. Serial dilution of input DNA was performed to make the standard curve to determine the relative concentration for each primer pair. Fold enrichment is calculated over an arbitrarily chosen control genomic region. Error bars represent the standard deviation for three technical replicates. Samples were processed for ChIP-sequencing if the enrichment was observed more than 1% of input and 5 fold to control genomic region.
1.3. RNA extraction and strand-specific RT-PCR
Synchronized culture was harvested at 18 (rings), 30 (trophozoites) and 48 (schizonts) hpi. Parasites were isolated by saponin (8 mg/ml in PBS) lysis at 37 °C for 15 min. Total RNA was extracted from isolated parasites by adding pre-warmed TRIzol to the pellet and incubated at room temperature for 5 min. 0.2 × TRIzol volumes of chloroform was added and vigorous shaking followed by 2–3 min incubation at room temperature. The samples were centrifuged for 30 min to collect upper layer and 0.5 × TRIzol volume of Isopropanol added to precipitate RNA. RNA was treated with DNaseI (Ambion) as described in the manufacturer's protocol followed by phenol–chloroform extraction. RT-PCR was performed using 500 ng total RNA. For each gene, reverse transcription (RT) was performed using a forward and reverse primer. After first strand cDNA synthesis PCR was performed using sense and anti-sense primers.
2. Data analysis
2.1. P. falciparum ChIP-seq data processing and quality control measures
Reference genome (PlasmoDB-9.3_P.falciparum 3D7) was converted to color-space using bowtie-build. ChIP-seq data were mapped using the Bowtie allowing two mismatches to generate SAM files. The parameters “–5 5 –3 5 –a –m 1 –C” were used to select for only uniquely mapping reads. Bed files generated using sam2bed tool (https://bedpos.readthedocs.org/en/latest).
2.2. Analysis of ChIP-sequencing data
Our interest was to generate the distribution profile of histone modifications over the gene body in P. falciparum to generate a comprehensive map. However, the transcription start sites (TSSs) and transcription termination sites (TTSs) were not known in P. falciparum. To determine the TSSs and TTSs, we pooled previously reported RNA-seq data covering 8 samples collected at different time points of erythrocytic cycle stage [4]. Pfalciparum3D7_Genome_v9.3 genome index was built by bowtie2-build (version 2.2.3) and all raw reads from RNA-seq results from different stages were aligned using TopHat (v2.0.13) to indexed genome [5] using parameters “–min-intron-length 10 –max-intron-length 10,000”. In the alignment process a total of 89,645,433 (76.6%) reads were mapped out of 117,030,592 reads. Assembly of the transcripts from aligned reads was carried out by cufflinks (v2.2.1) program using PlasmoDB-9.3_Pfalciparum3D7.gff as GTF reference. Cufflinks program was ran with following parameters, minimum intron length set as 10 nt and a maximum intron length as 10,000 nt and assembly was guided by GFF file (PlasmoDB-9.3_Pfalciparum3D7.gff) to annotate the transcripts [5]. GTF file given by Cufflink was merged with reference GTF file (PlasmoDB-9.3_Pfalciparum3D7.gtf) using Cuffmerge function (part of Tuxedo suite). Merged GTF was converted to GFF by ‘gffread’ utility available in Tuxedo suite [5] and a gene co-ordinate GFF file was created using AWK script (awk ‘/gene_name/{print$0}’./merged.gff > ./genes.gff). Genes coding for rRNA and tRNA were removed from this file. This modified file was used to merge overlapping gene features using mergeBed tool from BEDTools. From this Bed files TSSs and TTSs were extracted and used for further analysis.
2.3. Average profile calculations of histone modifications on TSSs and TTSs
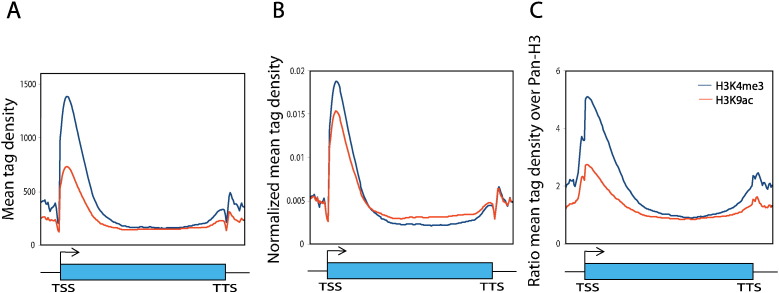
Profile of histone modifications is calculated using the program seqMiner [6]. Histone modification files (.bed) were used to generate average gene profiles. Gene profile was divided into 100 bins (80 inside the gene body and 10 each on 5′ and 3′ ends). ChIP-sequencing tag density values were calculated as described earlier [1], [7] for each dataset and each bin. To normalize the tag density between histone modifications, reads in each bin are divided by total number of reads for a histone modification (Fig. 2A and B). The profiles of histone modification are also normalized by dividing the reads in each bin with the histone H3 ChIP-sequencing (nucleosome) reads to nullify the uneven nucleosome distribution in P. falciparum (Fig. 2C).
Fig. 2.
Profiles of H3K4me3 and H3K9ac over averaged Plasmodium falciparum genes. (A) Mean tag density and (B) Normalized mean tag density of gene profile of H3K4me3 and H3K9ac histone modifications over 500 expressed P. falciparum genes. To normalize the tag density between histone modifications, reads in each bin are divided by total number of reads for a histone modification. (C) Profiles of H3K4me3 and H3K9ac normalized with pan-H3. Gene unit including transcription start site (TSS) and transcription termination site (TTS) is shown below.
2.4. Correlation of histone modifications with gene expression
We categorized 5200 P. falciparum genes into 52 groups based on increasing expression levels. Histone modifications are found to be distributed throughout the gene unit in P. falciparum and therefore enrichment of histone modifications is calculated over the entire gene unit [7], [8]. On each group, histone modification enrichment is summed by adding the mean tag density using R function for the gene unit. Whisker plots were plotted using R boxplot function with default parameters for each category for all the histone modifications. Pearson correlation was calculated between the median of gene expression and histone modification for each category.
3. Discussion
Here we describe information about P. falciparum culturing, chromatin immunoprecipitation controls and data analysis. The dataset comprises ChIP-sequencing of multiple histone modifications in a stage specific manner in P. falciparum. The dataset, which is recently published [1], first time visualized histone modification profiles at the TSSs and TTSs as opposed to translation start sites and translation termination sites [4], [9]. Integrative analysis of datasets we have generated reveals distinct modes of epigenetic and transcriptional regulation in P. falciparum, which can influence parasite virulence and pathogenicity [1]. In addition, the datasets presented here serves as a framework for further investigation of histone modifications to better understand the epigenetic machinery in Plasmodium falciparum and other related parasites.
Acknowledgments
We thank Prof. Sanjeev Galande and Saurabh J. Pradhan for performing ChIP-sequencing, Dr. P.C. Reddy for TSSs and TTSs prediction analysis and critical reading of the manuscript. This work is supported by grants under the DST-INSPIRE Faculty Award (IFA-13 LSBM-53) to KK.
References
- 1.Karmodiya K., Pradhan S.J., Joshi B., Jangid R., Reddy P.C., Galande S. A comprehensive epigenome map of Plasmodium falciparum reveals unique mechanisms of transcriptional regulation and identifies H3K36me2 as a global mark of gene suppression. Epigenetics Chromatin. 2015;8:32. doi: 10.1186/s13072-015-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoeijmakers W.A., Bártfai R., Françoijs K.J., Stunnenberg H.G. Linear amplification for deep sequencing. Nat. Protoc. 2011;6(7):1026–1036. doi: 10.1038/nprot.2011.345. [DOI] [PubMed] [Google Scholar]
- 3.Radfar A., Mendez D., Moneriz C., Linares M., Marín-García P., Puyet A., Diez A., Bautista J.M. Synchronous culture of Plasmodium falciparum at high parasitemia levels. Nat. Protoc. 2009;4:1828–1844. doi: 10.1038/nprot.2009.198. [DOI] [PubMed] [Google Scholar]
- 4.Bártfai R., Hoeijmakers W.A.M., Salcedo-Amaya A.M., Smits A.H., Janssen-Megens E., Kaan A., Treeck M., Gilberger T., Francoijs K., Stunnenberg H.G. H2A.Z demarcates intergenic regions of the Plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye T., Krebs A., Choukrallah M.A., Keime C., Plewniak F., Davidson I., Tora L. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krebs A.R., Karmodiya K., Lindahl-Allen M., Struhl K., Tora L. SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol. Cell. 2011;44:410–423. doi: 10.1016/j.molcel.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karmodiya K., Krebs A., Oulad-Abdelghani M., Kimura H., Tora L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012;13:424. doi: 10.1186/1471-2164-13-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salcedo-Amaya A.M., van Driel M.A., Alako B.T., Trelle M.B., van den Elzen A.M., Cohen A.M., Janssen-Megensa E.M., van de Vegte-Bolmer M., Selzer R.R., Iniguez A.L., Green R.D., Sauerwein R.W., Jensen O.N., Stunnenberg H.G. Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9655–9660. doi: 10.1073/pnas.0902515106. [DOI] [PMC free article] [PubMed] [Google Scholar]