Abstract
Unkeshwar hot springs are located at geographical South East Deccan Continental basalt of India. Here, we report the microbial community analysis of this hot spring using whole metagenome shotgun sequencing approach. The analysis revealed a total of 848,096 reads with 212.87 Mbps with 50.87% G + C content. Metagenomic sequences were deposited in SRA database with accession number (SUB1242219). Community analysis revealed 99.98% sequences belonging to bacteria and 0.01% to archaea and 0.01% to Viruses. The data obtained revealed 41 phyla including bacteria and Archaea and including 719 different species. In taxonomic analysis, the dominant phyla were found as, Actinobacteria (56%), Verrucomicrobia (24%), Bacteriodes (13%), Deinococcus-Thermus (3%) and firmicutes (2%) and Viruses (2%). Furthermore, functional annotation using pathway information revealed dynamic potential of hot spring community in terms of metabolism, environmental information processing, cellular processes and other important aspects. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of each contig sequence by assigning KEGG Orthology (KO) numbers revealed contig sequences that were assigned to metabolism, organismal system, Environmental Information Processing, cellular processes and human diseases with some unclassified sequences. The Unkeshwar hot springs offer rich phylogenetic diversity and metabolic potential for biotechnological applications.
Keywords: hot spring, metagenome, shotgun sequencing, microbial diversity, Unkeshwar
| Specifications | |
|---|---|
| Organism/cell line/tissue | Unkeshwar hot spring metagenome |
| Sex | not applicable |
| Sequencer or array type | Illumina Hi seq 2500 |
| Data format | raw data: fastq |
| Experimental factors | environmental sample |
| Experimental features | shotgun metagenome sequencing followed by microbial community and taxonomic analysis using KEGG pathways |
| Consent | not applicable |
| Sample source location | water sample, Unkeshwar hot spring, Maharashtra State, India |
1. Direct link to deposited data
2. Experimental design, materials and methods
Thermopiles have been discovered in geothermal features all over the world, leading to the discovery of many novel environmental microorganisms with important applications in biotechnology, medicine, and bioremediation [1]. Microbes inhabiting hot springs are dictated by environmental physicochemical characteristics such as pH, redox potential, temperature and concentration of trace elements [2], [3]. Among these factors, water temperature is the major factor in controlling microbial distribution within hot springs [4]. Hot springs microorganisms thrive under multiple environmental stresses and to survive under such stresses, microbial communities use mutualistic or communalistic symbiotic relationships [5].
Metagenomic analyses using high throughput sequencing have been extremely a valuable tool for describing microbial community structure and function in extreme ecosystems [6], [7]. Using such high throughput techniques most of the terrestrial hot springs all over the world, hosting diverse thermophilic microorganisms has been investigated. Majorly studied regions such as Yellowstone National Park, USA [8], Great Basin, USA [4], Philippines [9], Canada and New Zealand [10] and China [11] revealed many novel microbial lineages with promising applications in biotechnology. Metagenome sequencing techniques are also useful for identifying novel genes with novel bioactive molecule production capabilities [12], [13].
India has some high as well as low temperature geothermal springs in different geographic regions. In India thermal springs are found scattered throughout the country and occur either as solitarily or in groups up to 3000 M above sea level [14]. From hot springs of basaltic region of Deccan plateau of India, scanty and superficial reports have been documented [15], [16]. Therefore we investigated microbial diversity of Unkeshwar hot spring located in such region in Maharashtra state of India. Unkeshwar is located at geographical South East Deccan Continental basalt of India (19°34′–19°40'N and 78°22′–78°34'E) from mean sea level. The hot springs in this area emerge through basalt. The main Unkeshwar hot spring is within Unkeshwar temple in the form of kund. The water has sulphurous smell with feeble gaseous activity and discharge through the jointed Deccan basalts. Water samples were collected in sterile containers from Mukhya kund location of Unkeshwar hot spring during November 2012. The discharged water (10,000 l/h) is used by visitors to take bath for a therapeutic belief. Samples were processed for metagenomic and physioco-chemical analysis.
Temperature and pH were measured for the sample at the site. The physiochemical parameters of the samples were analysed at a certified chemical testing laboratory (Accurate Analytical Laboratory Pvt. Ltd., Pune, India) using standard methods of American Public Health Association [17]. The temperature of water sampled was between 50 °C–60 °C and the pH 7.3. The physiological parameters of the water sample tested shown that the total dissolved solids (0.0507%), total volatile solids (0.0180%), Phosphorus (0.008724%), and Sulphates (SO42 −) (0.004375%) were present in higher amount as compared to other elements. Other elements like Calcium, Cobalt, Iron, and Copper were not detected in the tests. Unlike other hot springs, Unkeshwar hot springs content high Phosphorus and Sulphates content. The chemical stress like availability of high phosphorus, sulphur concentrations and slightly higher organic content enriches the microbial diversity of this hot spring.
One litre of water sample was filtered using a 0.45 μm (to remove debris) followed by 0.22 μm filter membrane (MOBIO, USA). The 0.22 μm filter membrane was then sliced and subjected to simultaneous metagenomic DNA and RNA extraction using RNA PowerSoil® Total RNA Isolation Kit (MO BIO), according to the manufacturer's protocol. The quantitative analysis of the DNA was done using a Nanodrop and the integrity of DNA samples was checked on 1% agarose gel using a gel documentation system (Protein Simple). The low abundant metagenomic DNA was enriched by Multiple Annealing and Looping Based Amplification Cycles (MALBAC) amplification protocol as described earlier [18]. Enriched metagenomic DNA was extracted and analysed by whole shotgun metagenome analysis for microbial community structure and functional annotations.
Whole metagenome shotgun sequencing of 12UM sample was performed using the Illumina HiSeq 2500 sequencer (Illumina, USA). Metagenome sample library quantification was done using Bioanalyzer 2000 (Agilent, USA). For sequencing, a dual-indexed Paired-End sequencing (2 × 251base pairs) strategy with a total of 250 cycles, six bp index sequence was used. The entire sequencing run was completed in 39 h. Based on quality report of fastq files sequence reads were trimmed wherever necessary to only retain high quality sequence for further analysis. Assembled contigs size > 150 bp was considered for further analysis. Assembly was performed with default k-mer length (21-size) using de-bruijn graph method. In-house PERL and Python code were used to parse the fastq files for further analysis. Taxonomic profiling was performed using NCBI taxonomy data sets. The taxonomy tree was generated based on neighbour-joining method using MEGAN software [19].
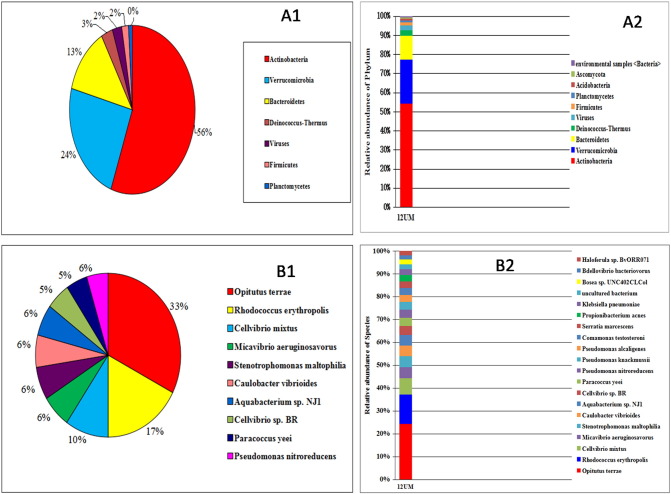
Whole metagenome shotgun sequencing revealed a total of 848,096 reads with 50.87% G + C content. After trimming and assembly, a total 34,123 contigs were obtained. Taxonomic profiling was performed using NCBI taxonomy data sets for 21,424 reads and revealed 41 phyla including bacteria and archaea. The contig sequences presented, Actinobacteria (56%), Verrucomicrobia (24%), Bacteriodes (13%), Deinococcus-Thermus (3%) and firmicutes (2%). At the species level, Opitutus terrae (33%), Rhodococcus erythropolis (17%), Cellovibrio mixtus (10%) were found dominant species (Fig. 1).
Fig. 1.
Taxonomic profiling and microbial community structure in Unkeshwar hot spring using NCBI taxonomy data sets A1: Phylum abundance; A2: Relative abundance phylum of samples; B1: Species abundance; and B2: Relative abundance of species of hot springs (Only top 10 species hits were highlighted).
The prediction of Open reading frames (ORFs) in the assembled contigs was done using a Glimmer-MG tool [20] and the complete functional annotation along with contig ID, gene function and sequences were carried out. For functional annotation the contigs of 12UM were queried to BLASTX programme with optimum e value of 1e− 10. The gene or protein functions of all the contigs from BLASTX output were parsed using in-house PERL (Practical Extraction and Report Language) script. Further, functional annotation was carried out by doing KEGG (Kyoto encyclopedia of genes and genomes) analysis based on taxons. Functional annotation of all the contigs is carried out by SEED Classification [21]. MEGAN software was used to assign the function of each contig. The protein function of each contig having highest alignment score from BLASTX results was considered for functional assignment and revealed functions like protein metabolism (1), cell wall and capsule (1), unassigned (14,281) and not hits (5794).
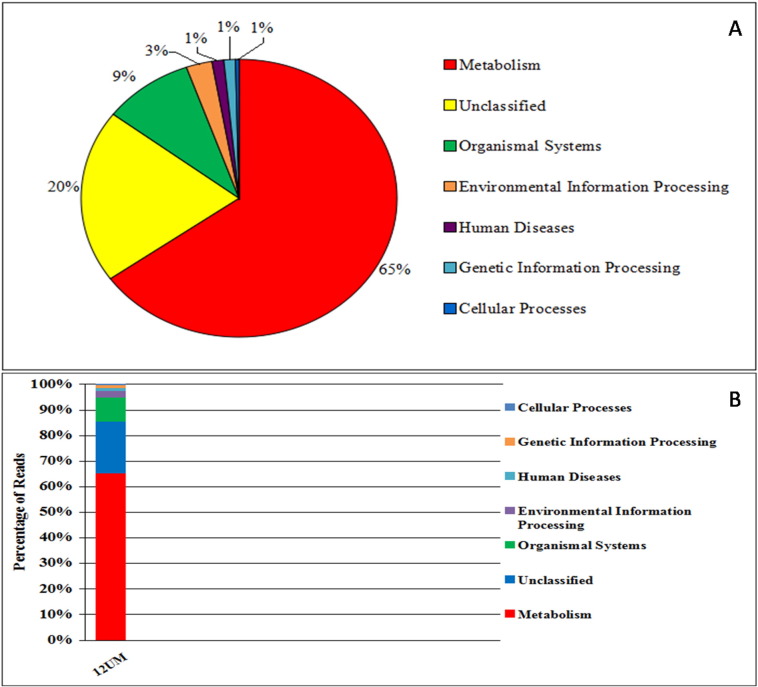
Pathway annotation was done using KEGG pathway analysis performed for each contig sequence by assigning KEGG Orthology (KO) numbers obtained from known reference hits. Around 65% contig sequences were assigned to metabolism, 20% unclassified sequences, 9% organismal system, 3% Environmental Information Processing, 1% cellular processes and 1% human diseases (Fig. 2). Sample read counts of KEGG annotation were metabolism (1990), Organismal systems (283), Environmental information processing (81), Human diseases (37), Genetic information processing (36), cellular processes (11) and unclassified (616).
Fig. 2.
Functional annotation based on KEGG-pathway analysis of Unkeshwar hot spring. A: number of sequence assigned to the pathway; and B: KEGG Relative abundance of Sample.
Taxonomic and functional diversity of a community quantified using whole metagenome shotgun sequencing revealed the dominance of bacterial population and their metabolisms. At the phylum level dominant bacterial phyla were Actinobacteria, Bacteriodes, Deinococcus-Thermus, Firmicutes, and Planctomycetes. Bacterial genera like Rhodococcus, Microbacterium, propionibacterium, Flavobacterium, Deinococcus, Caulobacter, Brevundimonas, Methylobacterium, Paracoccus, Roseomonas, Novosphingobium, Sphingomonas, Achromobacter, Acidovorax, and Aquabacterium were also dominant. KEGG pathway analysis shows that higher number of sequences was contributed to the metabolism as shown in the larger edge size. A number of DNA sequences remained unassigned with respect to taxonomic and functional coherence. To the best of our knowledge, this is the first study that deals with the description of complete profiling of microbial diversity from Unkeshwar hot spring using next generation massively parallel sequencing approach. Metagenome sequencing analysis may significantly provide important breakthroughs in depicting taxonomic structure and functional and/or metabolic pathways of basaltic hot spring of unkeshwar with the promise of novel genes and novel microbes for biotechnological applications.
3. Nucleotide sequence accession number
Metagenome sequence data is available in NCBI SRA under accession number http://www.ncbi.nlm.nih.gov/sra/SRX1499016.
Competing interest
Authors declare that there are no competing interests.
Acknowledgements
GTM is thankful to the Council of Scientific and Industrial Research (CSIR), and Academy of Scientific and Innovative Research (AcSIR), New Delhi, India for providing fellowship and support. This work was funded by Science and Engineering Research Board (SERB), New Delhi, India (EMR/2014/000483). The authors thankfully acknowledge SciGenom Labs, India for the generation of sequencing data generated in the study.
References
- 1.Cardoso A.M., Vieira R.P., Paranhos R., Clementino M.M., Albano R.M., Martins O.B. Hunting for extremophiles in rio de janeiro. Front. Microbiol. 2011;2:100. doi: 10.3389/fmicb.2011.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siering P.L., Clarke J.M., Wilson M.S. Geochemical and biological diversity of acidic, hot springs in Lassen volcanic National Park. Geomicrobiol J. 2006;23:129–141. [Google Scholar]
- 3.Mathur J., Bizzoco R.W., Ellis D.G., Lipson D.A., Poole A.W. Effects of abiotic factors on the phylogenetic diversity of bacterial communities in acidic thermal springs. Appl. Environ. Microbiol. 2007;73:2612–2623. doi: 10.1128/AEM.02567-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole J.K., Peacock J.P., Dodsworth J.A., Williams A.J., Thompson D.B., Dong H., Hedlund B.P. Sediment microbial communities in Great Boiling Spring are controlled by temperature and distinct from water communities. ISME J. 2013;7:718–729. doi: 10.1038/ismej.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan C.S., Chan K.G., Tay Y.L., Chua Y.H., Goh K.M. Diversity of thermophiles in a Malaysian hot spring determined using 16S rRNA and shotgun metagenome sequencing. Front. Microbiol. 2015;6:177. doi: 10.3389/fmicb.2015.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon C., Wiezer A., Strittmatter A.W. Daniel R (2009) Phylogenetic diversity and metabolic potential revealed in a glacier ice metagenome. Appl. Environ. Microbiol. 2009;75:7519–7526. doi: 10.1128/AEM.00946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie W., Wang F., Guo L., Chen Z., Sievert S.M. Comparative metagenomics of microbial communities inhabiting deep-sea hydrothermal vent chimneys with contrasting chemistries. ISME J. 2011;5:414–426. doi: 10.1038/ismej.2010.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer-Dombard D.R., Shock E.L., Amend J.P. Archaeal and bacterial communities in geochemically diverse hot springs of Yellowstone National Park, USA. Geobiology. 2005;3:211–227. [Google Scholar]
- 9.Huang Q., Jiang H., Briggs B.R., Wang S., Hou W., Li G., Wu G., Solis R., Arcilla C.A., Abrajano T., Dong H. Archaeal and bacterial diversity in acidic to circum neutral hot springs in the Philippines. FEMS Microbiol. Ecol. 2013;85:452–464. doi: 10.1111/1574-6941.12134. [DOI] [PubMed] [Google Scholar]
- 10.Sharp C.E., Brady A.L., Sharp G.H., Grasby S.E., Stott M.B., Dunfield P.F. Humboldt's spa: microbial diversity is controlled by temperature in geothermal environments. ISME J. 2014;8:1166–1174. doi: 10.1038/ismej.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yim L.C., Hongmei J., Aitchison J.C., Pointing S.B. Highly diverse community structure in a remote central Tibetan geothermal spring does not display monotonic variation to thermal stress. FEMS Microbiol. Ecol. 2006;57:80–91. doi: 10.1111/j.1574-6941.2006.00104.x. [DOI] [PubMed] [Google Scholar]
- 12.Tirawongsaroj P., Sriprang R., Harnpicharnchai P., Thongaram T., Champreda V. Novel thermophilic and thermostable lipolytic enzymes from a Thailand hot spring metagenomic library. J. Biotechnol. 2008;133:42–49. doi: 10.1016/j.jbiotec.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 13.Jiménez D.J., Andreote F.D., Chaves D., Montaña J.S., Osorio-Forero C., Junca H., Zambrano M.M., Baena S. Structural and functional insights from the metagenome of an acidic hot spring microbial planktonic community in the Colombian Andes. PLoS One. 2012;7 doi: 10.1371/journal.pone.0052069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verma A., Dhiman K., Gupta M., P. S. Bioprospecting of thermotolerant bacteria from Hot Water Springs of Himachal Pradesh for the production of Taq DNA polymerase. Proc. Natl. Acad. Sci. India, Sect. B Biol. Sci. 2015;85:739–749. [Google Scholar]
- 15.Yannawar V.B., Bhosle A.B., Shaikh P.R., Gaikwad S.R. Water quality of Hot Water Unkeshwar Spring of Maharashtra, India. IJIAS. 2013;3:541–551. [Google Scholar]
- 16.Pathak A.P., Rathod M.G. Cultivable bacterial diversity of terrestrial thermal spring of Unkeshwar, India. J. Biochem. Tech. 2014;5:814–818. [Google Scholar]
- 17.Apha A.W.W.A.W.E.F. Standard methods for the examination of water and wastewater. 2012;22 [Google Scholar]
- 18.Zong C., Lu S., Chapman A.R., Xie X.S. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huson D.H., Auch A.F., Qi J., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelley D.R., Liu B., Delcher A.L., Pop M., Salzberg S.L. Gene prediction with Glimmer for metagenomic sequences augmented by classification and clustering. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gkr1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overbeek R., Begley T., Butler R.M., Choudhuri J.V., Chuang H.Y., Cohoon M. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]