Abstract
Serofluid dish (or Jiangshui, in Chinese), a traditional food in the Chinese culture, is made from vegetables by fermentation. In this study, bacterial community of the fermented serofluid dish was assessed by Illumina amplicon sequencing. The metagenome comprised of 49,589 average raw reads with an average 11,497,917 bp and G + C content is 52.46%. This is the first report on V4 hyper-variable region of the 16S rRNA metagenome sequence employing Illumina platform to profile the microbial community of this little known fermented food from Gansu Province, China. The metagenome sequence can be accessed at NCBI, SRA database accession no. SRP065370.
Keywords: Serofluid dish, Jiangshui, 16S rRNA, Cultivation-independent, Microbial diversity
| Specifications | |
|---|---|
| Organism/cell line/tissue | Metagenome of serofluid dish |
| Sex | Not applicable |
| Sequencer or array type | Illumina Hiseq2500 |
| Data format | Raw data: FASTQ file |
| Experimental factors | Environmental sample |
| Experimental features | V4 hypervariable region of 16S rRNA was sequenced using paired end Illumina Hi-Seq technology, sequence analyzed by QIIME data analysis package. |
| Consent | Not applicable |
| Sample source location | A total of 12 serofluid dish samples were collected from 10 handmade areas from Gansu Province, China. |
1. Direct link to deposited data
2. Experimental design, materials and methods
Serofluid dish is a naturally fermented dish from vegetables such as Chinese cabbage and celery that are also called Jiangshui, in Chinese due to its white serofluid [1]. It has been found that serofluid dish making techniques have a history of over two thousand years. In recent years, serofluid dish has been confirmed to be clinically effective for promoting digestion, lowers cholesterol levels, and fall blood pressure in vitro and in vivo [2]. In addition, microbes involved in serofluid dish fermentation have been studied previously [3], [4]. Taxonomically diverse groups of lactic acid bacteria (LAB) have been identified during the fermentation of serofluid dish [5]. However, most of the diversity studies in the last two decades have shown that less than 1% of the microbes of a community are possibly cultured in vitro with known cultivation approach, but 99% still remain unexplored [6]. Culture-based approaches have known limitations in terms of reproducibility and challenges in the culturability of some microbes [7]. Therefore, the set of cultured isolates may not reflect the true microbial composition of fermented serofluid dish. With the advancement of culture independent technique using next generation sequencing or clone library construction, it is time to analyze the entire population in the community as well as their functional potentiality in fermented food [8], [9]. Therefore, the present research was designed to investigate the bacterial community composition by using Illumina based metagenomic approach in serofluid dish, which may represent novel species within this genus which was reported for the first time in association with serofluid dish.
A total of 26 serofluid dish samples were collected from 10 handmade areas across Gansu Province, China (35.22°–36.33°N, 103.59°–105.10°E). The V4 hypervariable region of the 16S rRNA gene was amplified using 515 F and 806R primer combination (5′-GTGCCAGCMGCCGCGGTAA-3′ and 5′-GGACTACHVGGGTWTCTAAT-3′, respectively) [10]. A 6-bp error-correcting tag was added to the forward primer [11]. A total of 12 serofluid dish samples were sent to Novogene Bioinformatics Technology (Beijing, China) to perform amplicon sequencing according to Hiseq protocols. Amplicon pyrosequencing was performed on the Illumina HiSeq PE250 platforms at Novogene Bioinformatics Technology (Beijing, China). Bioinformatic analysis was performed by QIIME data analysis package [12]. Complete data sets are submitted to the NCBI Short Read Archive under accession numbers SRR2919186, SRR2919188, SRR2919190-SRR2919199.
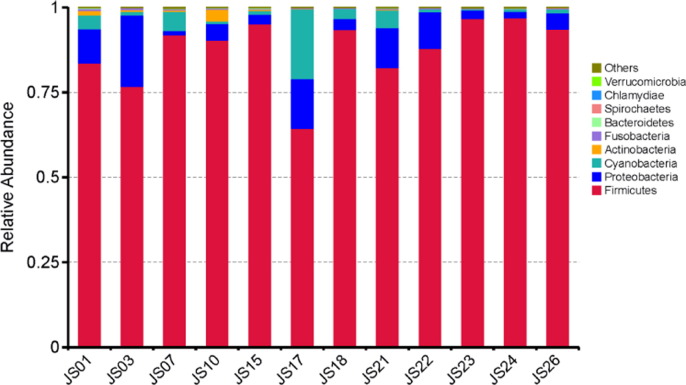
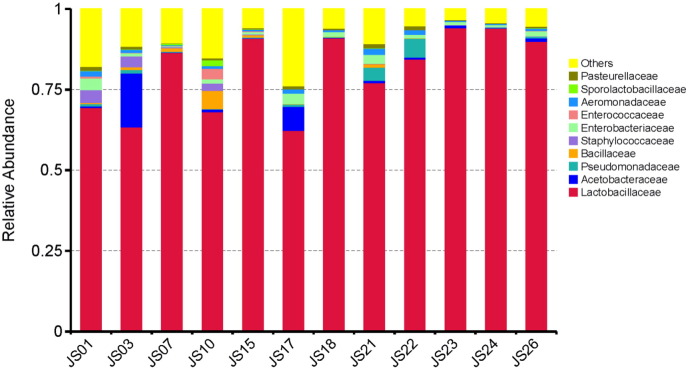
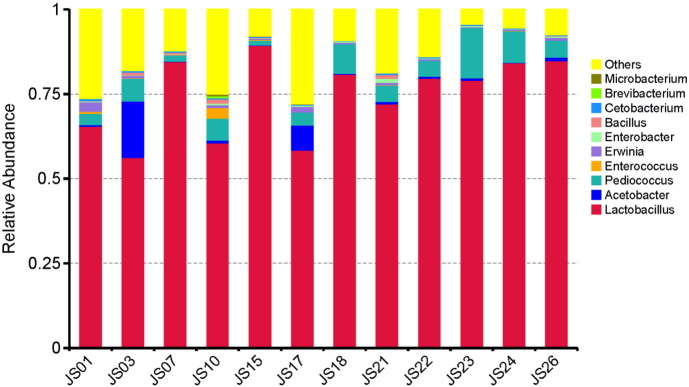
The output file comprised 113 MB data with a total of 595,067 high qualities read having 52.46% GC content. In the serofluid dish sample studied, the classified OTUs represent distinct phyla dominated by Firmicutes (87.79%), Proteobacteria (7.50%) and Cyanobacteria (3.69%) (Fig. 1). At the family level, Lactobacillaceae (81.01%) was predominant followed by Acetobacteraceae (2.43%), Pseudomonadaceae (1.19%), Enterobacteriaceae (1.53%) and Bacillaceae (0.83%) (Fig. 2). The leading genera were Lactobacillus (74.65%), Pediococcus (5.92%), Acetobacter (2.43%), Bacillus (0.34%) and Cetobacterium (0.33%) (Fig. 3). Our data provide the first scientific report on the composition of the bacterial community, using Illumina sequencing method, from the undeveloped serofluid dish located in northwest China region. The most dominated phylum in this study was Lactobacillus which is known to produce beneficial metabolites useful for gut microbiota and human health. 16S rRNA metagenome analysis of serofluid dish revealed the presence of microorganism key functional characteristics. It may contribute to further studies on critical process for human gut microbiota and probiotics, such as nutrient absorption, cholesterol decomposition and blood-pressure control. Consequently, these results indicate that both serofluid dish formation complex and distinct bacterial structure which were not reported before.
Fig. 1.
Taxonomy classification of reads at phylum level. Only top 10 enriched class categories are shown in the figure.
Fig. 2.
Taxonomy classification of reads at family level. Only top 10 enriched class categories are shown in the figure.
Fig. 3.
Taxonomy classification of reads at genera level. Only top 10 enriched class categories are shown in the figure.
3. Nucleotide sequence accession number
Metagenome sequence data are available on NCBI, SRA database accession no. SRP065370.
Competing Interests
The authors declare that there are no competing interests.
Acknowledgments
This work was supported by Gansu Province Science Foundation for Distinguished Young Scholars (Grant No. 1308RJDA014), Longyuan Support Project for Young Creative Talents (Grant No. GANZUTONGZI [2014] no.4), Technology Program of Lanzhou City (Grant No. 2015-3-142, Grant No. 2015-3-97), The Fundamental Research Funds for the Central Universities of China (Grant No. lzujbky-2015-57).
Contributor Information
Peng Chen, Email: chenpeng@lzu.edu.cn.
Hongyu Li, Email: lihy@lzu.edu.cn.
References
- 1.Liu R., Dang S., Yan H., Wang D., Zhao Y., Li Q., Liu X. Association between dietary protein intake and the risk of hypertension: a cross-sectional study from rural western China. Hypertens. Res. 2013;36(11):972–979. doi: 10.1038/hr.2013.71. [DOI] [PubMed] [Google Scholar]
- 2.Hou Z., Yang J., Jia H., Lu X., Wang L. Research overview and prospect of traditional fermented food — serofluid dish. China Condiment. 2015;40(02):132–136. (in Chinese) [Google Scholar]
- 3.Zhang Y., Wang Y., Chen X., Zhao P. Isolation and initiative identification of microorganism from traditional fermentative food-Jiangshui. Food Sci. 2007;28(01):219–222. (in Chinese) [Google Scholar]
- 4.He L., Li Q. Isolation, identification and variation of dominant population from fermented pickle celery. Food Sci. Technol. 2010;35(05):36–40. (in Chinese) [Google Scholar]
- 5.Li X., Li J., Meng X., Zheng N., Xie J. Isolation and identification of microorganisms from Jiangshui, a traditional Chinese fermented vegetable product. Food Sci. 2014;35(23):204–209. (in Chinese) [Google Scholar]
- 6.Amann R.I., Binder B.J., Olson R.J., Chisholm S.W., Devereux R., Stahl D.A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 1990;56(6):1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang R., Dong Y., Chen F., Chen Q. Bacterial diversity analysis during the fermentation processing of traditional Chinese yellow rice wine revealed by 16S rDNA 454 pyrosequencing. J. Food Sci. 2015;80(10):M2265–M2271. doi: 10.1111/1750-3841.13018. [DOI] [PubMed] [Google Scholar]
- 8.Jung J.Y., Lee S.H., Kim J.M., Park M.S., Bae J.W., Hahn Y., Madsen E.L., Jeon C.O. Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl. Environ. Microbiol. 2011;77(7):2264–2274. doi: 10.1128/AEM.02157-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yousif N.M.K., Huch M., Schuster T., Cho G.-S., Dirar H.A., Holzapfel W.H., Franz C.M.A.P. Diversity of lactic acid bacteria from Hussuwa, a traditional African fermented sorghum food. Food Microbiol. 2010;27(6):757–768. doi: 10.1016/j.fm.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 10.Vandeputte D., Falony G., Vieira-Silva S., Tito R.Y., Joossens M., Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2016;65:57–62. doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamady M., Walker J.J., Harris J.K., Gold N.J., Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods. 2008;5(3):235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]