Abstract
The rock bream (Oplegnathus fasciatus) is considerably one of the most economically important marine fish in East Asia and has a unique neo-Y chromosome system that is a good model to study the sex determination and differentiation in fish. In the present study, we used Illumina sequencing technology (HiSeq2000) to sequence, assemble and annotate the transcriptome of the testis and ovary tissues of rock bream. A total of 40,004,378 (NCBI SRA database SRX1406649) and 53,108,992 (NCBI SRA database SRX1406648) high quality reads were obtained from testis and ovary RNA sequencing, respectively, and 60,421 contigs (with average length of 1301 bp) were obtained after de novo assembling with Trinity software. Digital gene expression analysis reveals 14,036 contigs that show gender-enriched expressional profile with either testis-enriched (237 contigs) or ovary-enriched (581 contigs) with RPKM > 100. There are 237 male- and 582 female-abundant expressed genes that show sex dimorphic expression. We hope that the gonad transcriptome and those gender-enriched transcripts of rock bream can provide some insight into the understanding of genome-wide transcriptome profile of teleost gonad tissue and give useful information in fish gonad development.
Keywords: Gonad transcriptome, Testis, Ovary, Rock bream
| Specifications | |
|---|---|
| Organism/cell line/tissue | Oplegnathus fasciatus/testis and ovary |
| Sex | Male and female |
| Sequencer or array type | Illumina Hiseq2000 |
| Data format | Raw and processed |
| Experimental factors | Transcriptome profiling of testis and ovary at mature stages |
| Experimental features | Testis and Ovary at mature stages were dissected from the rock bream and the total RNAs were extracted by using TRIZOL reagent. Prepared cDNA libraries were paired-end sequenced by HiSeq2000 system. The obtained data was subjected for de novo transcriptome assembly using Trinity, and coding regions were predicted by BLAST. We performed BLASTx against the NR database with an e-value cut off of 1e-6 for unigene annotation. Gene ontology (GO) analysis was conducted by using Blast2GO. |
| Consent | N/A |
| Sample source location | Key Lab of Mariculture and Enhancement of Zhejiang Province, Marine Fisheries Research Institute of Zhejiang Province, Zhoushan, Zhejiang, China |
1. Direct link to deposited data
http://www.ncbi.nlm.nih.gov/sra/SRX1406648 for male gonad.
http://www.ncbi.nlm.nih.gov/sra/SRX1406649 for female gonad.
2. Introduction
Teleost fishes are an outstanding model to study the evolution of sex chromosome since they present a broad range of sex chromosome systems, as well as the absence of differentiated sex chromosomes in most species [3]. Although the diversity of sex determination system in fish, it is hypothesized that genes involved in sex determination are probably conserved throughout evolution. To date, master sex-determination genes have been identified for fish species, including dmy in medaka [11], [12], sdy in rainbow trout [20], amhr2 in fugu [6] and amhy in Patagonian pejerrey [5]. Besides the sex determining genes, some conserved genes shown to play important roles in mammal sex determination and differentiation were analyzed in fish, including cyp19, dax1, dmrt, foxl2, mis, sf1, sox9 and wt1 [13]. These genes act together to constitute complicated network whereby sex phenotype is established in mammals. However, studies on the function and connections of the above genes in fish are in their infancy. More or novel sex-related genes are required to be clarified to further explain the complex mechanism of fish sex determination.
Over the past decade, significant progress has been made in genome-wide gene expression profiling by the development and application of large scale sequencing technique, which can easily show more differential expressional genes in different traits, such as gender. Transcriptome profiling associated with sex determination and differentiation using RNA-seq were reported in several fish species, including platyfish [21], Nile tilapia [15], catfish [14], and Yellow catfish [9]. These data provided transcriptomic information expressed in gonads at particular condition and time and identified sex differentially expressed genes, which are crucial to providing valuable insight in studying fish sex-determination.
Rock bream (Oplegnathus fasciatus), a subtropical and carnivorous species, is an economically important marine fish in East Asia. It has recently been targeted as a promising species for commercial aquaculture and stock enhancement in China. This species possess a differentiated neo-Y chromosome, and is characterized by having a multiple X1X1X2X2/X1X2Y sex chromosome system [16], [19]. Therefore, rock bream fish is an excellent model with which to understand the molecular mechanism of sex determination and sex chromosome evolution in fishes. To gain a global view of the multiple interrelated molecular changes that relate to the sexual dimorphism in rock bream and provide a database for future studies, we initiated a transcriptome project to obtain deep coverage of cDNAs from adult fish of different gender. Our findings may provide a valuable genomic resource for gene annotation and discovery of genes for sex determination as well as development of molecular makers in rock bream.
3. Experimental design, materials and methods
3.1. RNA extraction
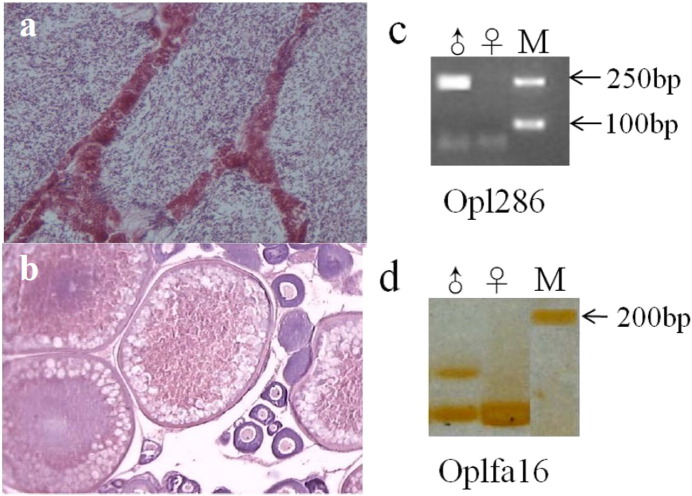
The fully matured testis and ovary were dissected from wild captured rock bream and immediately stored in RNAlater (Qiagen, Hilden, Germany) and then stored at − 80 °C prior to RNA extraction. At the same time, the gonad maturation of the rock bream was checked by histological section. Sperm can be easily striped from the male individual, and the lobular lumen and sperm duct were filled with spermatozoa under microscope (Fig. 1a), which suggest the male is mature at stage V [10]. For the female individual, postovulatory follicles were observed together with oocytes at yolk stage in histological sections of ovaries, indicating the female at maturity stages V (Fig. 1b). Two male specific markers Opl286 and Opfla16 [17], [18] for rock bream sex identification were also used to detect the genetic sex of the fish individuals for RNA-seq (Fig. 1c and d). Total RNAs were extracted by using the TRIZOL Kit (Invitrogen, Carlsbad, CA, USA) following manufacturer's instructions. Total RNA samples were then digested by DNase I to remove potential genomic DNA contamination. Integrity and size distribution were checked with Bioanalyzer 2100 (Agilent technologies, Santa Clara, CA, USA).
Fig. 1.
Gender identification in rock bream by histological section and molecular markers genotyping in rock bream. (a) Paraffin section of mature ovary at stage V, (b) paraffin section of mature testis at stage V. The tissue section was stained with H&E to enhance the contrast. The gender of rock bream is sexing by the male-specific markers of Opl286 (c) and Opfl16 (d).
3.2. RNA isolation, library construction and Illumina sequencing
Initially, about 2.5 μg of starting total RNAs were used to synthesize the cDNA libraries by following the standard protocols of the Illumina TruSeq RNA Sample Preparation Kit (Illumina). The final library had an average fragment size of 180 bp and final yields of ~ 400 ng. After KAPA quantitation and dilution, the library was sequenced on an Illumina HiSeq 2000 with 101 bp paired-end reads. A total of 40,004,378 and 53,108,992 paired-end reads was generated with a read length of 101 bp in the female and male transcriptomes, respectively. The raw transcriptome sequences in the present study was deposited in the NCBI SRA database (SRX1406648 and SRX1406649 for male and female gonads, respectively). Adaptor sequences were trimmed and reads with low quality or length less than 70 were further removed by SolexaQA software. After the removal of ambiguous nucleotides, duplicates and low-quality sequences (Phred quality scores < 20), a total of 63,367,046 cleaned reads (68%) were obtained.
3.3. De novo transcriptome assembly and functional annotation of gonad expressed genes in rock bream
Cleaned reads were de novo assembled into contigs by Trinity software [4] with default parameters settings. The transcriptome was assembled into 60,421 contigs, ranging from 201 to 23,525 bp in length. The average length was 1301 bp, the N50 length was 2560 bp. The contig length distribution is shown in Fig. 2. The assembled transcriptomic contigs were subjected to similarity search against NCBI non-redundant (nr) protein database using BlastX with an e-value cut off of 1e-6. Gene names and descriptions were assigned to each contig based on the BLASTx results. Gene ontology (GO) analysis was then conducted on the assembled transcriptome by using Blast2GO [1]. A total of 33,046 contigs has a significant hit, corresponding to 22,702 unique protein accessions in the nr protein database.
Fig. 2.
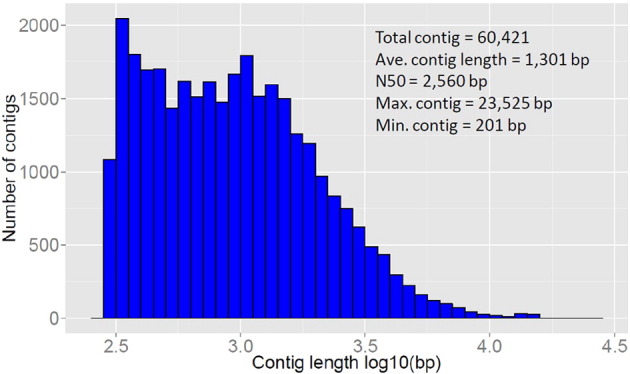
Length distribution of the assembled unigene of rock bream gonad transcriptome.
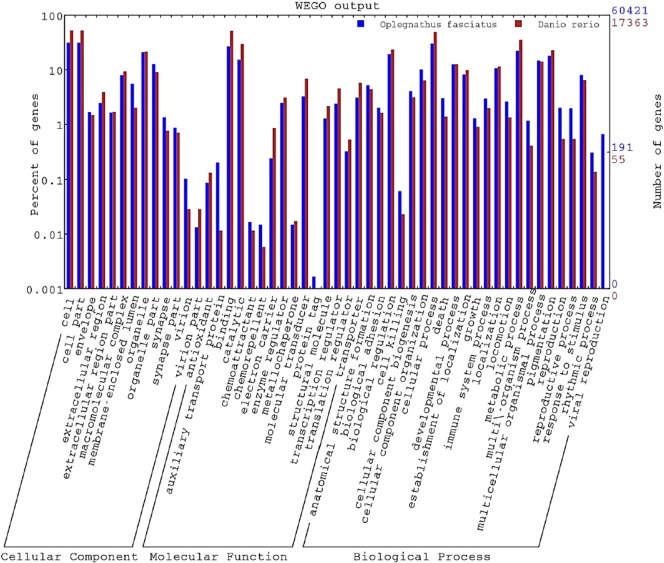
All assembled contigs were analyzed by transcripts_to_best_scoring_ORFs.pl from Trinity package to search for ORFs, which could distinguish between coding and non-coding sequences. KEGG pathways were assigned to assembled contigs using the online KEGG Automatic Annotation Server (KAAS) (http://www.genome.jp/tools/kaas/). The Bi-directional Best Hit (BBH) method was used to obtain KEGG Orthology (KO) assignment. Gene ontology (GO) analysis was conducted on those 22,702 unique proteins by Blast2GO. A total of 17,363 unique proteins were assigned at least one GO term for describing biological processes, molecular functions and cellular components. The Blast2GO output file was input into the BGI WEGO program (http://wego.genomics.org.cn) and GO annotations were plotted. To assess the functional diversity of assembled transcriptome, GO annotation of zebrafish were compared with those of the rock bream transcriptome, reflecting a similar functional distribution on GO categories and indicating the sequence diversity of the transcriptome study (Fig. 3).
Fig. 3.
Comparison of the Gene Ontology (GO) categories of the unigene between rock bream and zebrafish. The GO terms of the unigene of rock bream (blue) and zebrafish (red) were categorized into cellular components, molecular functions, and biological processes.
3.4. Gene expression quantification and differential expression analysis
The high-quality cleaned reads of each RNA-seq library were mapped to the assembled transcripts with Bowtie program [7]. The counting of alignments was done using RSEM [8]. The differential expression statistical analysis was done using the statistical method described in the R package [2]. Differentially expressed gene (fold changes > 2 and adjusted p-value < 0.001) between two samples were identified with the software. Mapping results show 14,036 contigs display significant expression dimorphic between ovary and testis. Among these sex-biased contigs, 5736 male-enriched contigs and 4391 ovary-enriched contigs were annotated with known genes. The contigs with FPKM (fragments per kilobase of exon per million reads mapped) ≥ 100 was defined as abundant expressed genes (AEGs). There are 237 male-AEGs and 582 female-AEGs show sex dimorphic expression (summarized in Table S1). Apparently, the sex-biased AEGs are mostly associated with gonadal differentiation and development, gametogenesis and gametes constitution. On one hand, the male-biased AEGs including piwi-like protein, synaptonemal complex protein, spermatogenesis associated 4, meiosis expressed gene 1 protein homolog, meiotic recombination protein dmc1 lim15 homolog, radial spoke head 1 homolog, sperm flagellar protein, dynein light chain cytoplasmic, dynein intermediate chain axonemal, dynein heavy chain axonemal, tektin-2 etc. participate in testis development, spermatogenesis, sperm construction and energy metabolism. The female-biased AEGs, on the contrary, including nanos homolog 3, wee1-like protein kinase 2, egg envelop protein, choriogenin l, zona pellucida glycoprotein, zona pellucida sperm-binding protein, vitellogenin receptorand bucky ball etc. are closely related to meiosis, ovary development, oogenesis, egg construction and oocyte polarity formation. We hope that the gonad transcriptome and those gender-enriched transcripts of rock bream can provide some insight into the understanding of genome-wide transcriptome profile of teleost gonad tissue and give useful information in fish gonad development.
The following is the supplementary data related to this article.
List of the differential expressed genes in male (testis) and female (ovary) gonads of the rock bream.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (41106114; 41476127) and the Project of Zhejiang Province of China (2014F10004; 2014F50021; 2015F10001).
Contributor Information
Bao Lou, Email: loubao6577@163.com.
Chung-Der Hsiao, Email: cdhsiao@cycu.edu.tw.
References
- 1.Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 2.Cordero F., Beccuti M., Arigoni M., Donatelli S., Calogero R.A. Optimizing a massive parallel sequencing workflow for quantitative miRNA expression analysis. PLoS One. 2012;7:e31630. doi: 10.1371/journal.pone.0031630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devlin R.H., Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208:191–364. [Google Scholar]
- 4.Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hattori R.S., Murai Y., Oura M., Masuda S., Majhi S.K., Sakamoto T., Fernandino J.I., Somoza G.M., Yokota M., Strussmann C.A. A Y-linked anti-Mullerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2955–2959. doi: 10.1073/pnas.1018392109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamiya T., Kai W., Tasumi S., Oka A., Matsunaga T., Mizuno N., Fujita M., Suetake H., Suzuki S., Hosoya S. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu) PLoS Genet. 2012;8:e1002798. doi: 10.1371/journal.pgen.1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langmead B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics. 2010;Chapter 11(Unit 11):17. doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J., Luan P., Zhang X., Xue S., Peng L., Mahbooband S., Sun X. Gonadal transcriptomic analysis of yellow catfish (Pelteobagrus fulvidraco): identification of sex-related genes and genetic markers. Physiol. Genomics. 2014;46:798–807. doi: 10.1152/physiolgenomics.00088.2014. [DOI] [PubMed] [Google Scholar]
- 10.Mackie M., Lewis P. 2001. Assessment of Gonad Staging Systems and Other Methods Used in the Study of the Reproductive Biology of Narrow-Barred Spanish Mackerel, Scomberomorus commerson, in Western Australia (Citeseer) [Google Scholar]
- 11.Matsuda M., Nagahama Y., Shinomiya A., Sato T., Matsuda C., Kobayashi T., Morrey C.E., Shibata N., Asakawa S., Shimizu N. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- 12.Nanda I., Kondo M., Hornung U., Asakawa S., Winkler C., Shimizu A., Shan Z., Haaf T., Shimizu N., Shima A. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11778–11783. doi: 10.1073/pnas.182314699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandra G.E., Norma M.M. Sexual determination and differentiation in teleost fish. Rev. Fish Biol. Fish. 2010;20:101–121. [Google Scholar]
- 14.Sun L., Wang C., Huang L., Wu M., Zuo Z. Transcriptome analysis of male and female Sebastiscus marmoratus. PLoS One. 2012;7:e50676. doi: 10.1371/journal.pone.0050676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tao W., Yuan J., Zhou L., Sun L., Sun Y., Yang S., Li M., Zeng S., Huang B., Wang D. Characterization of gonadal transcriptomes from Nile tilapia (Oreochromis niloticus) reveals differentially expressed genes. PLoS One. 2013;8:e63604. doi: 10.1371/journal.pone.0063604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu D., Lou B., Bertollo L.A., Cioffi Mde B. Chromosomal mapping of microsatellite repeats in the rock bream fish Oplegnathus fasciatus, with emphasis of their distribution in the neo-Y chromosome. Mol. Cytogenet. 2013;6:12. doi: 10.1186/1755-8166-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu D., Lou B., Bertollo L.A.C., Cioffi M.d.B. Chromosomal mapping of microsatellite repeats in the rock bream fish Oplegnathus fasciatus, with emphasis of their distribution in the neo-Y chromosome. Mol. Cytogenet. 2013;6:1–6. doi: 10.1186/1755-8166-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu D., Lou B., Li S., Sun X., Zhan W., Chen R., Mao G. A novel sex-linked microsatellite marker for molecular sexing in rock bream fish Oplegnathus fasciatus. Biochem. Syst. Ecol. 2015;62:66–68. [Google Scholar]
- 19.Xu D., Lou B., Xu H., Li S., Geng Z. Isolation and characterization of male-specific DNA markers in the rock bream Oplegnathus fasciatus. Mar. Biotechnol. 2013;15:221–229. doi: 10.1007/s10126-012-9480-1. [DOI] [PubMed] [Google Scholar]
- 20.Yano A., Guyomard R., Nicol B., Jouanno E., Quillet E., Klopp C., Cabau C., Bouchez O., Fostier A., Guiguen Y. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 2012;22:1423–1428. doi: 10.1016/j.cub.2012.05.045. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z., Wang Y., Wang S., Liu J., Warren W., Mitreva M., Walter R.B. Transcriptome analysis of female and male Xiphophorus maculatus Jp 163 A. PLoS One. 2011;6:e18379. doi: 10.1371/journal.pone.0018379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of the differential expressed genes in male (testis) and female (ovary) gonads of the rock bream.