Abstract
Prostate cancer (PCa) is the second most common cancer in men. The Androgen Receptor (AR) is the major driver of PCa and the main target of therapy in the advanced setting. AR is a nuclear receptor that binds the chromatin and regulates transcription of genes involved in cancer cell proliferation and survival. In a study by Stelloo et al. (1) we explored prostate cancer on the level of transcriptional regulation by means of Formaldehyde-Assisted Isolation of Regulatory Elements and Chromatin Immunoprecipitation coupled with massive parallel sequencing (FAIRE-seq and ChIP-seq, respectively). We employed these data for the assessment of differences in transcriptional regulation at distinct stages of PCa progression and to construct a prognostic gene expression classifier. Genomics data includes FAIRE-seq data from normal prostate tissue as well as primary, hormone therapy resistant and metastatic PCa. Furthermore, ChIP-seq data from primary and resistant PCa were generated, along with multiple input controls. The data are publicly available through NCBI GEO database with accession number GSE65478. Here we describe the genomics and clinical data in detail and provide comparative analysis of FAIRE-seq and ChIP-seq data.
Keywords: Prostate cancer, androgen receptor, ChIP-seq, FAIRE-seq
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo Sapiens |
| Sex | Male |
| Sequencer or array type | Illumina Hiseq 2000 genome analyzer |
| Data format | Raw: SRA study; processed: BED |
| Experimental factors | Normal, primary and therapy resistant tumors, lymph node metastases |
| Experimental features | FAIRE-seq and Androgen Receptor ChIP-seq |
| Consent | Leftover anonymized tissue (not traceable back to the patient and not interfering with care and/or prognosis) used for research purposes. |
| Sample source location | Samples were from prostate cancer patients, treated at the Erasmus University Medical Center (EMC; Rotterdam, The Netherlands), The Netherlands Cancer Institute/Antoni van Leeuwenhoek hospital (Amsterdam, The Netherlands) |
Direct link to deposited data https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65478
1. Experimental design, materials and methods
1.1. Clinical samples and experimental design
Fresh frozen tissue samples were obtained through postoperative needle biopsies targeting both tumor and normal areas of prostatectomy specimens at The Netherlands Cancer Institute (Amsterdam, The Netherlands). Tissue samples from androgen deprivation resistant tumors (from transurethral resection of the prostate (TURP)) and lymph node metastases were obtained from the Erasmus University Medical Center (Rotterdam, The Netherlands). Slides stained with hematoxylin and eosin (H&E) of the cases were reviewed by our pathologists. Clinical and pathological parameters of the selected patients are provided in Table 1. Leftover anonymized tissue, which cannot be traced back to the patient and does not interfere with care and/or prognosis, and would have been discarded otherwise, has been used in accordance with the Code of Conduct of the Federation of Medical Scientific Societies in The Netherlands. NKI and Erasmus MC institutional medical ethics committees have approved the study.
Table 1.
Patient and tumor characteristics of the selected samples.
| Characteristic | Number of patients |
|||
|---|---|---|---|---|
| Normal |
Primary |
Resistant |
Metastasis |
|
| 4 | 4 | 4 | 3 | |
| Treatment type | ||||
| Untreated | 3 | 4 | 0 | 2 |
| Bicalutamide/cyproteron acetate | 1 | 0 | 0 | 0 |
| Bicalutamide/LHRH analogue | 0 | 0 | 1 | 0 |
| Cyproteron acetate + LHRH analogue | 0 | 0 | 1 | 0 |
| LHRH analogue | 0 | 0 | 2 | 0 |
| LHRH analogue/Cyproteron Acetate | 0 | 0 | 0 | 1 |
| Gleason score | ||||
| 6 | 1 | 0 | 0 | 0 |
| 7 | 2 | 2 | 0 | 0 |
| 8 | 0 | 0 | 1 | 1 |
| 9 | 1 | 2 | 0 | 0 |
| 10 | 0 | 0 | 3 | 2 |
| Initial PSA (ng/ml) | ||||
| Mean | 8.7 | 19.6 | 149.5 | 135.5 |
| Range | 5.3–13.0 | 8.5–38.0 | 6.5–511.0 | 17.0–254.0 |
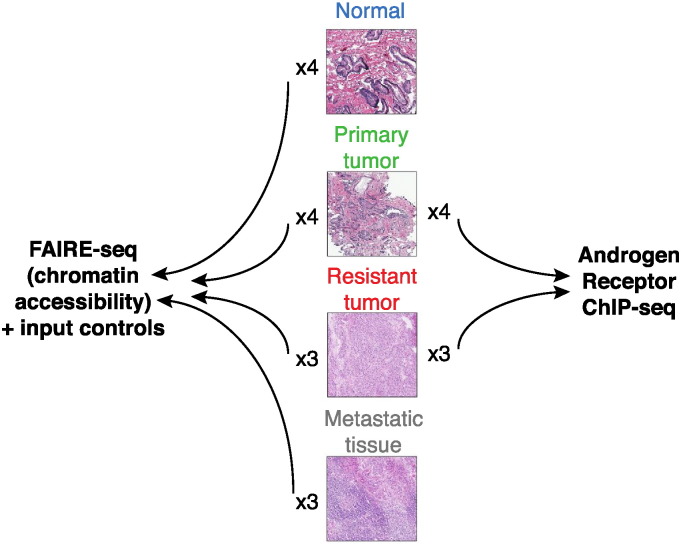
FAIRE-seq was performed on four normal samples, four primary, three therapy resistant tumors and three lymph node metastases (Fig. 1). Androgen Receptor ChIP-seq was carried out on four primary and three resistant tumors (Fig. 1).
Fig. 1.
FAIRE-seq and ChIP-seq analyses were performed on normal prostate tissue and prostate cancer samples from different stages of the disease.
1.2. Formaldehyde-assisted isolation of regulatory elements (FAIRE)
FAIRE was performed as previously described [2]. Briefly, fresh frozen tissues were cross-linked with 1% formaldehyde for 20 min. After washing, nuclei were isolated as described before [3]. Afterwards chromatin was sonicated, cleared by centrifugation and subjected to three consecutive phenol–chloroform–isoamyl alcohol (25:24:1) extractions. Reverse cross-linking was performed at 65 °C overnight. Subsequently, samples were treated with RNase A and proteinase K and purified by using a PCR purification kit (Roche).
1.3. Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation was carried out as described before [3], [4]. 10 μg of AR-N20 (sc-618; Santa Cruz) antibody was used for immunoprecipitation, with 100 μl of Protein A magnetic beads (Invitrogen).
1.4. DNA sequencing
Libraries were prepared according to Illumina DNA Sample Kit instructions. Sequencing was performed on the Illumina HiSeq 2000 Genome Analyzer using 51-bp reads. Reads were aligned to the Human Reference Genome (assembly hg19, February 2009) using bwa 0.5.9.
1.5. Data analysis
Reads that map uniquely to the genome, with MAPQ quality score above 20, were used for the analysis. FAIRE-seq and ChIP-seq peaks were called with two algorithms, MACS 1.4 [5] and DFilter 1.0 [6], against mixed input controls corresponding to each group. MACS was run with default parameters, except for p = 10− 7 for ChIP-seq data. DFilter was run with bs = 100, ks = 50 for FAIRE-seq data and bs = 50, ks = 30, refine, nonzero for ChIP-seq data. Peaks detected by both algorithms were used for further analysis. Sequencing read depths and number of called peaks can be found in Table 2.
Table 2.
Sequencing and peak calling details.
| GEO accession | Experiment | Tissue | Total number of reads | Mapped reads | % mapped reads | No. peaks |
|---|---|---|---|---|---|---|
| GSM1598204 | FAIRE-seq | Normal | 19,147,127 | 17,986,187 | 93.94 | 50 |
| GSM1598205 | FAIRE-seq | Normal | 21,599,945 | 19,883,501 | 92.05 | 472 |
| GSM1598206 | FAIRE-seq | Normal | 26,080,719 | 25,043,481 | 96.02 | 61 |
| GSM1598207 | FAIRE-seq | Normal | 23,167,347 | 22,177,458 | 95.73 | 2837 |
| GSM1598208 | FAIRE-seq | Primary | 36,827,373 | 34,441,896 | 93.52 | 6450 |
| GSM1598209 | FAIRE-seq | Primary | 18,306,926 | 17,002,416 | 92.87 | 1579 |
| GSM1598210 | FAIRE-seq | Primary | 32,197,589 | 30,568,523 | 94.94 | 13,348 |
| GSM1598211 | FAIRE-seq | Primary | 28,992,853 | 27,590,961 | 95.16 | 2243 |
| GSM1598212 | FAIRE-seq | Resistant | 37,452,682 | 35,655,681 | 95.2 | 80 |
| GSM1598213 | FAIRE-seq | Resistant | 28,372,546 | 26,836,918 | 94.59 | 3497 |
| GSM1598214 | FAIRE-seq | Resistant | 27,545,618 | 26,061,843 | 94.61 | 5754 |
| GSM1598215 | FAIRE-seq | Metastasis | 39,562,972 | 37,594,752 | 95.03 | 2043 |
| GSM1598216 | FAIRE-seq | Metastasis | 29,130,845 | 27,291,106 | 93.68 | 281 |
| GSM1598217 | FAIRE-seq | Metastasis | 27,253,810 | 25,789,354 | 94.63 | 1313 |
| GSM1598218 | AR ChIP-seq | Primary | 13,782,549 | 12,232,556 | 88.75 | 754 |
| GSM1598219 | AR ChIP-seq | Primary | 18,146,927 | 16,009,388 | 88.22 | 402 |
| GSM1598220 | AR ChIP-seq | Primary | 13,040,014 | 11,254,994 | 86.31 | 17,511 |
| GSM1598221 | AR ChIP-seq | Primary | 9,928,626 | 7,080,840 | 71.32 | 3278 |
| GSM1598222 | AR ChIP-seq | Primary | 12,243,485 | 11,160,623 | 91.16 | 7932 |
| GSM1598223 | AR ChIP-seq | Resistant | 16,518,987 | 14,727,645 | 89.16 | 739 |
| GSM1598224 | AR ChIP-seq | Resistant | 16,382,421 | 14,441,817 | 88.15 | 238 |
| GSM1598225 | AR ChIP-seq | Resistant | 15,621,538 | 13,967,477 | 89.41 | 1779 |
| GSM1598226 | Input | Resistant | 28,171,838 | 26,825,849 | 95.22 | |
| GSM1598227 | Input | Metastasis | 24,117,145 | 22,902,755 | 94.96 | |
| GSM1598228 | Input | Primary | 23,982,305 | 22,739,491 | 94.82 | |
| GSM1598229 | Input | Primary | 27,642,177 | 26,387,234 | 95.46 |
FAIRE-seq, ChIP-seq data and clinical annotation of the samples that are deposited in NCBI GEO under accession number GSE65478.
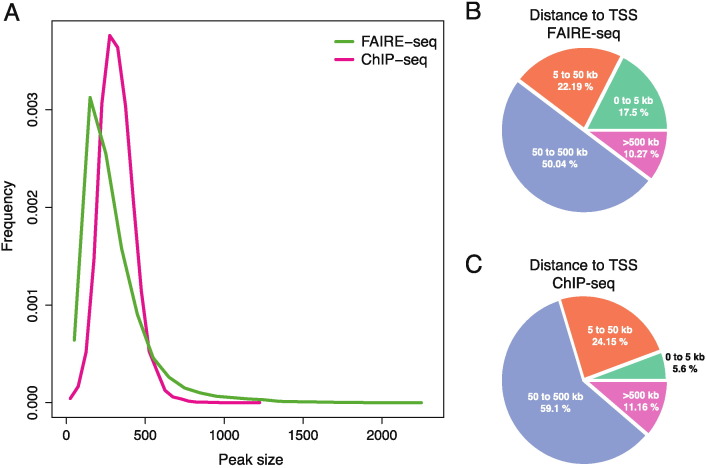
For further analysis, a merged list of peaks present in all samples from each technique was generated. The number of peaks detected by FAIRE-seq was 25,797, while 20,703 peaks were detected by ChIP-seq. The AR binding sites had a median width of 350 bp and peak size did not vary strongly with the largest peak size of 1202 bp (Fig. 2A). In contrast, FAIRE-seq peaks had a larger spread in size with a median size of 255 bp. The largest peak size of FAIRE-seq data was 2300 bp and a higher proportion of both small and large peaks was present (Fig. 2A). The distance to the nearest transcription start site (TSS) was determined by the GREAT tool (http://great.stanford.edu/) [7]. The number of peaks within 5 kb from the nearest TSS was significantly higher in FAIRE-seq data as compared to ChIP-seq data and the number of peaks further than 50 kb from a TSS was higher in ChIP-seq data than in FAIRE-seq (p < 10− 15 Fisher's exact; Fig. 2B-C). This is in accordance with AR binding mainly distant enhancer elements [8], while accessible regions detected by FAIRE-seq include not only enhancers, but also promoters [9].
Fig. 2.
Comparative analysis of FAIRE-seq and Androgen Receptor ChIP-seq data. (A) Size distribution of peaks detected by FAIRE-seq and ChIP-seq in prostate cancer specimens. Pie charts showing the percentage of peaks in categories based on the distance to the nearest transcription start site (TSS) in FAIRE-seq (B) and ChIP-seq (C) data.
2. Conclusions
In conclusion, we provide a unique dataset of genome-wide epigenetic profiling of prostate cancer tissue from different stages of the disease. The dataset consists of two parts: accessible chromatin profiling by FAIRE-seq and genome-wide androgen receptor binding to DNA by ChIP-seq. We previously used this dataset to identify changes in transcriptional regulation in prostate cancer upon acquisition of resistance to hormonal therapy, as well as to derive a prognostic gene expression signature for prostate cancer [1].
Acknowledgment
We thank Geert JLH van Leenders and Guido Jenster from Erasmus MC for providing tissue samples, clinical support and valuable discussions. We thank the NKI Genomics Core Facility for sequencing analyses and the Core Facility Molecular Pathology and Biobanking for help with tissue access.
This work was financially supported by Movember, grant number NKI01, and KWF/Alpe d'HuZes (NKI 2014-6711).
Contributor Information
Andries M. Bergman, Email: a.bergman@nki.nl.
Wilbert Zwart, Email: w.zwart@nki.nl.
References
- 1.Stelloo S., Nevedomskaya E., van der Poel H.G., de Jong J., van Leenders G.J., Jenster G. Androgen receptor profiling predicts prostate cancer outcome. EMBO Mol. Med. 2015;7(11):1450–1464. doi: 10.15252/emmm.201505424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giresi P.G., Lieb J.D. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. Jul 2009;48(3):233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zwart W., Koornstra R., Wesseling J., Rutgers E., Linn S., Carroll J.S. A carrier-assisted ChIP-seq method for estrogen receptor–chromatin interactions from breast cancer core needle biopsy samples. BMC Genomics. 2013;14(1):232. doi: 10.1186/1471-2164-14-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jansen M.P.H.M., Knijnenburg T., Reijm E.A., Simon I., Kerkhoven R., Droog M. Hallmarks of aromatase inhibitor drug resistance revealed by epigenetic profiling in breast cancer. Cancer Res. Nov 15 2013;73(22):6632–6641. doi: 10.1158/0008-5472.CAN-13-0704. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar V., Muratani M., Rayan N.A., Kraus P., Lufkin T., Ng H.H. Uniform, optimal signal processing of mapped deep-sequencing data. Nat. Biotechnol. Jul 2013;31(7):615–622. doi: 10.1038/nbt.2596. [DOI] [PubMed] [Google Scholar]
- 7.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. May 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu J., Yu J., Mani R.-S., Cao Q., Brenner C.J., Cao X. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. May 18 2010;17(5):443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davie K., Jacobs J., Atkins M., Potier D., Christiaens V., Halder G. In: Discovery of Transcription Factors and Regulatory Regions Driving In Vivo Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling. McKinnon P., editor. 11(2) Public Library of Science; Feb 2015. p. e1004994. (PLoS Genet.). [DOI] [PMC free article] [PubMed] [Google Scholar]