Abstract
Most studies investigating the biology of Hepatitis C virus (HCV) have used the human hepatoma cell line Huh-7 or subclones thereof, as these are the most permissive cell lines for HCV infection and replication. Other cell lines also support replication of HCV, most notably the human hepatoblastoma cell line HuH6. HCV replication in cell culture is generally highly sensitive to interferons (IFNs) and differences in the IFN-mediated inhibition of virus replication may reflect alterations in the IFN-induced antiviral response inherent to different host cells. For example, HCV replication is highly sensitive to IFN-γ treatment in Huh-7, but not in HuH6 cells. In this study, we used microarray-based gene expression profiling to compare the response of Huh-7 and HuH6 cells to stimulation with IFN-α and IFN-γ. Furthermore, we determined whether the resistance of HCV replication in HuH6 cells can be linked to differences in the expression profile of IFN-regulated genes. Although both cells lines responded to IFNs with rapid changes in gene expression, thereby demonstrating functional type I and type II signaling pathways, differences were observed for a number of genes. Raw and normalized expression data have been deposited in GEO under accession number GSE68927.
Keywords: Interferon, IFN-α, IFN-γ, Huh-7, HuH6, Gene expression profile
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens; cell lines HuH6 and Huh-7 |
| Sequencer or array type | Whole Genome Oligo Microarray (Affymetrix GeneChip HG-U133 Plus 2.0) |
| Data format | Raw: CEL files; normalized data: XLS files |
| Experimental factors | HuH6 and Huh-7 cells treated with IFN-α or IFN-γ |
| Experimental features | We performed a microarray-based transcriptome analysis to compare the induction of IFN-stimulated genes in the human hepatoma cell line Huh-7 and the human hepatoblastoma cell line HuH6 in response to IFN-α and IFN-γ. |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. Cell culture experiments and RNA isolation
The hepatoma cell line Huh-7 is the most commonly used cell line to study the biology of the human pathogen Hepatitis C virus (HCV) [1]. In these cells, HCV replication is strongly inhibited by both interferon-α (IFN-α) [2] as well as interferon-γ (IFN-γ) [3]. In the hepatoblastoma cell line HuH6, however, HCV replication is largely resistant to treatment with IFN-γ but not IFN-α [4]. To analyze the IFN-γ and IFN-α response in Huh-7 and HuH6 cells, we decided to perform a microarray-based gene expression analysis. To this end, Huh-7 and HuH6 cells, that were grown in Dulbecco's modified minimal essential medium (Life Technologies, Karlsruhe, Germany) supplemented with 10% fetal calf serum, 2 mM l-glutamine, nonessential amino acids, 100 U of penicillin G/mL, and 100 μg of streptomycin/mL at 37 °C and 5% CO2, were plated in 10-cm cell culture petri dishes at 80% cell confluence and treated with either 1000 IU/mL IFNα-2a (PBL Laboratories, Acris, Herford, Germany), 1000 IU/mL IFN-γ (Roche, Basel, Switzerland), or remained untreated for 24 h. Total RNA was extracted from these samples by a GITC-based protocol [5]. RNA integrity was confirmed by agarose gel electrophoresis and RNA concentration was determined by measurement of OD at 260 nm on a NanoDrop Lite (Thermo Scientific, Braunschweig, Germany).
2.2. Microarray experiments
For first-strand cDNA synthesis, 13.5 μg total RNA was incubated as published before [6] with polyadenylated control RNAs and T7-oligo (dT)24 primer [5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-(dT24)-3′] at 70 °C for 10 min and put on ice. Next, the first-strand buffer mix (4 μL of 5 × the first-strand buffer, 2 μL 0.1 M dithiothreitol (DTT), and 1 μL 10 mM dNTPs) was preincubated at 42 °C for 2 min. After addition of 2 μL (200 units) Superscript II (Life Technologies, Karlsruhe, Germany), incubation was continued at 42 °C for 1 h. For second-strand synthesis, 30 μL 5 × the second-strand buffer, 91 μL RNase-free water, 3 μL 10 mM dNTPs, 4 μL (40 U) Escherichia coli DNA polymerase I (Life Technologies), 1 μL (12 U) E. coli DNAligase (TaKaRa, Gennevilliers, France), and 1 μL (2 U) RNase H (TaKaRa) were added, and the mix was incubated at 16 °C for 2 h. Then 2.5 μL (10 U) T4 DNA polymerase I (TaKaRa) were added at 16 °C for 5 min. The reaction was stopped by the addition of 10 μL 0.5 M EDTA, double-stranded (ds) cDNA was extracted with phenol/chloroform, and the aqueous phase was recovered by phase-lock gel separation (Eppendorf, Hamburg, Germany). After precipitation, the cDNA was restored in 12 μL RNase-free water. Five microliters ds cDNA were used to synthesize biotinylated cRNA using the BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics, NY). Labeled cRNA was purified using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Fragmentation and hybridization of 10 μg cRNA to GeneChip HG-U133 Plus 2.0 microarrays (Affymetrix, Santa Clara, CA) for 16 h at 45 °C, as well as washing and staining on a Fluidics Station 450 (Affymetrix), and scanning of the arrays in a GeneArray Scanner 2500 (Agilent, Palo Alto, CA) were performed according to the Affymetrix Gene Expression Analysis Technical Manual.
2.3. Signal processing and normalization
Raw Affymetrix data (.CEL-files) were further processed using the Chipster software, version 3.4.0 [7]. Signals were normalized within one cell line (three treatment groups per cell line: mock, IFN-α and IFN-γ) using the RMA method and expressed on a log2-transformed scale. Probe-sets were remapped to 19,674 Entrez Gene IDs using a custom CDF file (version 12, Thompson and Meng microarray lab, University of Michigan) [8] and expression calls (A: absent, M: moderate, or P: present) were calculated using the MAS5 algorithm. Without further processing and filtering, data for Huh-7 and HuH6 cells were merged and exported into an Excel file for user-friendly interactive filtering (Supplementary data file 1).
2.4. Analysis of IFN-induced genes
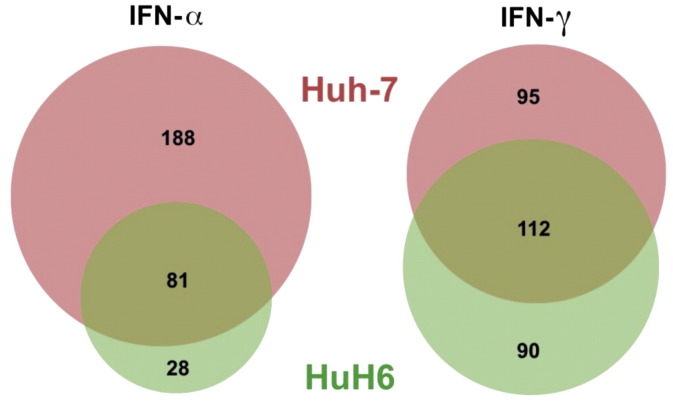
Albeit genes induced by IFN, termed IFN-stimulated genes (ISGs), have been defined and known for a long time (e.g. http://www.interferome.org [10]) [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], the exact set and number of genes that are regulated by IFN varies between different cell types and might substantially affect the cells' capacity to interfere with the replication of specific viruses. We therefore compared the ISG profile of the two hepatocyte-derived cell lines Huh-7 and HuH6, which are widely employed for research on hepatitis viruses, especially Hepatitis C virus (HCV). For this analysis, we defined ISGs as (1) being called “present” in the IFN-treated sample and (2) being upregulated at least 1.5-fold (0.58 on a log2-scale) upon IFN-treatment. For IFN-α, we found 269 ISGs in Huh-7 and 109 ISGs in HuH6, with 81 being common to both (Fig. 1). For IFN-γ, we found 207 ISGs in Huh-7 and 202 in HuH6, with 112 genes in common to both cell lines (Fig. 1). The genes that were most strongly induced upon IFN-α treatment in both cell lines include well-known prototype ISGs, such as MX1, IFIT1 (ISG56), IFIT3, IFI6 and OAS1-3 (Table 1). For IFN-γ, too, well-established prototype ISGs were strongly upregulated, such as GBP1, GBP3 or TAP1 (Table 1).
Fig. 1.
Number of IFN-induced genes in Huh-7 versus HuH6 cells. ISGs were defined as being at least 1.5-fold upregulated and being “present” upon IFN-treatment. Venn diagrams were generated using BioVenn (http://www.cmbi.ru.nl/cdd/biovenn/) [9].
Table 1.
Top 20 induced ISGs upon IFN treatment in Huh-7 and HuH6 cells. ISGs were sorted for the magnitude of their induction (fold-change, FC) within each sample. The 20 genes most strongly induced by IFN-α (left) or IFN-γ (right) are listed for Huh-7 and HuH6, respectively. Fold-changes are given on a log2 scale (i.e. FC 7.35 translates to a 163-fold upregulation).
| IFN-α |
IFN-γ |
||||||
|---|---|---|---|---|---|---|---|
| Gene symbol | FC Huh-7 | Gene symbol | FC HuH6 | Gene symbol | FC Huh-7 | Gene symbol | FC HuH6 |
| MX1 | 7.35 | IFIT1 | 8.4 | GBP1 | 8.46 | GBP1 | 6.55 |
| IFIT1 | 6.93 | CMPK2 | 7.3 | PSMB9 | 7.49 | PSMB9 | 6.04 |
| CMPK2 | 5.93 | IFI27 | 6.91 | TAP1 | 5.85 | UBE2L6 | 6.01 |
| DDX60 | 5.9 | IFI44L | 6.25 | PSMB8 | 5.74 | GBP3 | 5.72 |
| IFI6 | 5.82 | IFIH1 | 5.99 | RARRES3 | 5.67 | TAP1 | 5.37 |
| IFIH1 | 5.81 | DDX60 | 5.62 | UBD | 5.58 | TRIM22 | 5.21 |
| IFIT3 | 5.33 | OAS3 | 5.6 | GBP3 | 5.1 | NMI | 4.97 |
| ISG15 | 5.21 | IFI6 | 5.56 | IFIT3 | 5.04 | RARRES3 | 4.75 |
| OAS1 | 5.17 | IFIT3 | 5.37 | EPSTI1 | 4.97 | ERAP2 | 4.75 |
| OAS3 | 4.92 | OAS1 | 5.11 | CXCL10 | 4.62 | CMPK2 | 4.69 |
| HERC6 | 4.84 | IFI44 | 4.92 | DDX60 | 4.29 | UBD | 4.62 |
| EPSTI1 | 4.79 | IFIT5 | 4.47 | NNMT | 4.28 | SERPING1 | 4.5 |
| IRF9 | 4.2 | UBE2L6 | 4.4 | UBE2L6 | 4.23 | IFI27 | 4.5 |
| OAS2 | 4.13 | OAS2 | 4.37 | TRIM22 | 3.81 | IFIT3 | 4.45 |
| IFI27 | 4.11 | ISG15 | 4.03 | PARP14 | 3.78 | PSMB8 | 4.12 |
| DDX58 | 3.8 | DTX3L | 3.86 | CXCL9 | 3.74 | DTX3L | 3.88 |
| DDX60L | 3.62 | DDX58 | 3.8 | IRF9 | 3.67 | PARP14 | 3.53 |
| IFITM1 | 3.55 | HERC6 | 3.48 | LGALS3BP | 3.59 | PARP9 | 3.47 |
| IFI44L | 3.44 | IRF9 | 3.43 | IFIH1 | 3.47 | BATF2 | 3.4 |
| LGALS3BP | 3.42 | PARP9 | 3.42 | CXCL11 | 3.44 | DDX60 | 3.37 |
2.5. ISGs differentially induced in Huh-7 versus HuH6
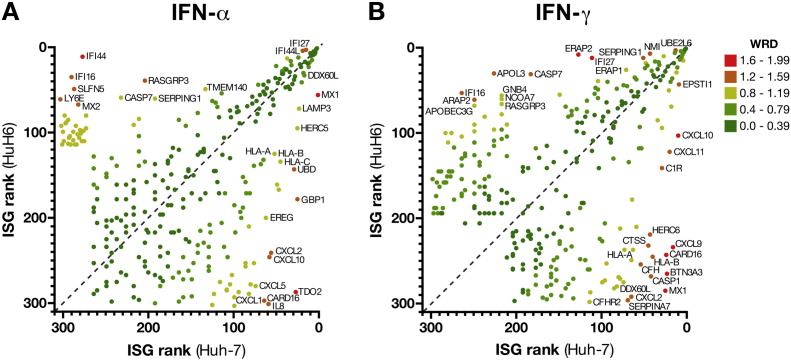
We then analyzed the induced ISG profiles of the two cell lines for differentially regulated genes, i.e. genes whose transcriptional regulation upon IFN-treatment is significantly different between Huh-7 and HuH6. For this analysis, we defined IFN-α or IFN-γ ISGs as being “present” and upregulated by at least 1.5-fold in either Huh-7 or HuH6 upon treatment with the respective IFN, resulting in 301 ISGs for IFN-α and 292 ISGs for IFN-γ (excluding Affymetrix controls, see Supplementary data file 1). To generate ISG profiles for Huh-7 and HuH6, ISGs were then ranked based on their fold-change within the respective cell line, with rank 1 being awarded to the gene with highest induction. Plotting the ranked profiles against each other allows for easy visual identification of the ISGs most strongly differing in their regulation between Huh-7 and HuH6 (Fig. 2). Surprisingly, for IFN-α, MX1, being one of the most strongly induced and best characterized ISGs, only ranks 56th in HuH6 (rank 1 in Huh-7), whereas IFI44 is a top-ISG in HuH6 (rank 11) but hardly induced in Huh-7 (rank 277). Further, CXCL-type chemokines CXCL1 (Gro-α), CXCL2 (Gro-β) and CXCL10 (IP-10) are upregulated in Huh-7, whereas they are not induced in HuH6 (Fig. 2). For IFN-γ, the CXCL chemokines, in particular CXCL10 and 11, are again strongly upregulated in Huh-7 (ranking 10th and 20th, respectively) and only to a much lesser extend in HuH6 (ranking above 100). Further, the MHC genes HLA-A, B and C are strongly induced in Huh-7 but substantially less so in HuH6. Conversely, IFI27 (rank 12) and ERAP2 (rank 8) are prominent ISGs in HuH6 but not in Huh-7 (ranking above 100).
Fig. 2.
Correlation of ISGs induced in Huh-7 and HuH6. ISGs were ranked by their respective induction upon IFN-treatment (fold-change); rank 1 corresponds to the highest induction. Ranks in Huh-7 were plotted against ranks in HuH6. Genes were color-coded according to their weighted rank difference (WRD) and genes with high WRD values are labeled.
As a quantitative measure to score differentially induced ISGs, we calculated a weighted rank difference (WRD) for each gene. The WRD was defined as the difference in a gene's rank between Huh-7 and HuH6, normalized to the gene's mean rank: (with rn being the rank in the respective cell line), and therefore 0 ≤ WRD < 2. Normalization to the mean rank accounts for the increasing possible variation for lower ranks; simply put, a rank difference of 50 is less significant in case of a gene ranking 250th and 300th (WRD = 0.18) as compared to a gene ranking 1st and 51st (WRD = 1.92). Table 2 lists the most significant (WRD > 1) differentially regulated ISGs between Huh-7 and HuH6 for IFN-α and IFN-γ in Huh-7 (full list of ISGs with WRDs in Supplementary data file 1).
Table 2.
ISGs differentially induced in Huh-7 versus HuH6. ISGs were defined as “present” and upregulated at least 1.5-fold (0.58 on a log2 scale) in either Huh-7 or HuH6 and sorted for their weighted rank difference (WRD). Highest WRD corresponds to the strongest difference in induction between Huh-7 and HuH6 (see text). FC: fold-change (log2 scale).
| IFN-α |
IFN-γ |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene symbol | FC Huh-7 | FC HuH6 | rank Huh-7 | rank HuH6 | WRD | Gene symbol | FC Huh-7 | FC HuH6 | rank Huh-7 | rank HuH6 | WRD |
| MX1 | 7.35 | 1.49 | 1 | 56 | 1.93 | ERAP2 | 1.08 | 4.75 | 126 | 8 | 1.76 |
| IFI44 | 0.47 | 4.92 | 277 | 11 | 1.85 | CXCL9 | 3.74 | 0.31 | 16 | 234 | 1.74 |
| TDO2 | 3.07 | − 0.19 | 27 | 287 | 1.66 | BTN3A3 | 3.28 | 0.04 | 23 | 265 | 1.68 |
| IFI16 | 0.06 | 2.56 | 290 | 35 | 1.57 | MX1 | 3.16 | − 0.1 | 26 | 285 | 1.67 |
| GBP1 | 3.11 | 0.19 | 25 | 178 | 1.51 | CXCL10 | 4.62 | 1.06 | 10 | 103 | 1.65 |
| SLFN5 | 0.08 | 1.66 | 287 | 49 | 1.42 | CARD16 | 3.27 | 0.24 | 24 | 243 | 1.64 |
| RASGRP3 | 0.68 | 2.13 | 204 | 39 | 1.36 | IFI27 | 1.21 | 4.5 | 111 | 12 | 1.61 |
| IL8 | 1.7 | − 0.36 | 59 | 301 | 1.34 | APOL3 | 0.37 | 2.84 | 228 | 30 | 1.53 |
| IFI27 | 4.11 | 6.91 | 15 | 3 | 1.33 | CASP1 | 2.69 | 0.02 | 42 | 268 | 1.46 |
| LY6E | − 0.29 | 1.4 | 303 | 61 | 1.33 | NMI | 2.61 | 4.97 | 43 | 7 | 1.44 |
| UBD | 3.04 | 0.33 | 29 | 143 | 1.33 | HLA-B | 2.75 | 0.21 | 40 | 245 | 1.44 |
| IFI44L | 3.44 | 6.25 | 19 | 4 | 1.30 | CXCL11 | 3.44 | 0.88 | 20 | 122 | 1.44 |
| CARD16 | 1.59 | − 0.28 | 64 | 297 | 1.29 | CASP7 | 0.69 | 2.8 | 184 | 31 | 1.42 |
| CXCL2 | 1.89 | − 0.01 | 56 | 241 | 1.25 | CTSS | 2.57 | 0.34 | 45 | 232 | 1.35 |
| CXCL10 | 1.75 | − 0.03 | 58 | 246 | 1.24 | IFI16 | 0 | 1.98 | 268 | 53 | 1.34 |
| MX2 | 0.23 | 1.23 | 282 | 67 | 1.23 | HERC6 | 2.59 | 0.43 | 44 | 219 | 1.33 |
| CASP7 | 0.64 | 1.44 | 232 | 59 | 1.19 | C1R | 3.06 | 0.75 | 29 | 141 | 1.32 |
| HERC5 | 3.11 | 0.77 | 25 | 95 | 1.17 | EPSTI1 | 4.97 | 2.42 | 9 | 43 | 1.31 |
| CXCL5 | 1.38 | − 0.15 | 74 | 280 | 1.16 | CFH | 2.35 | 0.13 | 54 | 254 | 1.30 |
| LOC100506113 | − 0.08 | 0.92 | 298 | 80 | 1.15 | CXCL2 | 1.99 | − 0.26 | 65 | 292 | 1.27 |
| RARRES3 | 2.12 | 0.22 | 47 | 167 | 1.12 | UBE2L6 | 4.23 | 6.01 | 13 | 3 | 1.25 |
| MYH4 | 1.29 | − 0.2 | 82 | 289 | 1.12 | SERPINA7 | 1.95 | − 0.34 | 69 | 296 | 1.24 |
| SLC26A3 | 1.34 | − 0.14 | 80 | 278 | 1.11 | SERPING1 | 2.45 | 4.5 | 51 | 12 | 1.24 |
| ERAP2 | − 0.04 | 0.86 | 294 | 85 | 1.10 | ARAP2 | 0.21 | 1.76 | 250 | 61 | 1.22 |
| TAP2 | 0.58 | 0.92 | 273 | 80 | 1.09 | GNB4 | 0.5 | 1.89 | 217 | 56 | 1.18 |
| PROCR | − 0.06 | 0.82 | 296 | 88 | 1.08 | LGALS3BP | 3.59 | 1.53 | 18 | 69 | 1.17 |
| SH3BGRL | − 0.07 | 0.81 | 297 | 89 | 1.08 | HLA-A | 2.03 | 0.29 | 63 | 237 | 1.16 |
| B3GALT2 | 1.37 | − 0.05 | 76 | 253 | 1.08 | PSMB8 | 5.74 | 4.12 | 4 | 15 | 1.16 |
| BTN3A2 | 2.08 | 0.24 | 49 | 161 | 1.07 | NCOA7 | 0.42 | 1.79 | 224 | 60 | 1.15 |
| HSH2D | − 0.23 | 0.78 | 302 | 92 | 1.07 | IL7 | 1.97 | 0.17 | 67 | 249 | 1.15 |
| EREG | 1.63 | 0.14 | 62 | 200 | 1.05 | NLRC5 | 1.87 | − 0.03 | 74 | 275 | 1.15 |
| SERPING1 | 0.71 | 1.41 | 192 | 60 | 1.05 | APOBEC3G | 0.19 | 1.56 | 251 | 68 | 1.15 |
| LAMP3 | 3.23 | 1.09 | 23 | 72 | 1.03 | CD274 | 2.15 | 0.58 | 60 | 210 | 1.11 |
| CCR6 | 0.12 | 0.78 | 285 | 92 | 1.02 | TRIM31 | 1.8 | 0 | 79 | 271 | 1.10 |
| MT1M | 1.15 | − 0.46 | 99 | 303 | 1.01 | TLR3 | 1.75 | − 0.06 | 82 | 280 | 1.09 |
| KLF10 | 1.19 | − 0.19 | 94 | 287 | 1.01 | RASGRP3 | 0.38 | 1.64 | 225 | 66 | 1.09 |
| LOC731779 | − 0.2 | 0.71 | 300 | 99 | 1.01 | LAMP3 | 1.78 | 0.01 | 80 | 269 | 1.08 |
| DDX60L | 1.68 | − 0.07 | 85 | 282 | 1.07 | ||||||
| ACY3 | 1.91 | 0.29 | 73 | 237 | 1.06 | ||||||
| TAPSAR1 | 3.01 | 1.12 | 30 | 94 | 1.03 | ||||||
3. Discussion
In this study, we describe the experimental details used to compare the IFN-induced changes in gene expression in the two human liver derived cell lines Huh-7 and HuH6. A large number of genes have been classified as ISGs in the past [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21]. The ISG database http://www.interferome.org [10] currently lists 3185 type I IFN regulated human genes in total, however with substantial differences between different cell or tissue types. For non-immune cells lines, typically 200 to 400 genes are upregulated robustly by IFN [14], [15], [16], [17], [20], depending on the chosen cut-off. This is in good agreement with our findings for Huh-7 (IFN-α: 269; IFN-γ: 207 genes > 1.5-fold) and HuH6 (IFN-α: 109; IFN-γ: 202 genes > 1.5-fold). Moreover, the genes that were most strongly upregulated in both cell lines (see Table 1) comprise prominent ISGs found in various earlier studies [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21]; of the 20 highest ranking ISGs induced by IFN-α in our study, 16 (Huh-7) or 17 (HuH6) can be found in the manually curated list of ISGs described by Schoggins and colleagues [20]. Remarkably, IFN-α induced more ISGs in Huh-7 cells than in HuH6 cells. The reason for this apparent lack of IFN-α-stimulated ISGs in HuH6 cells likely reflects the arbitrary cutoff of 1.5-fold upregulation used to identify ISGs. However, comparing the 269 ISGs from Huh-7 to the 269 most upregulated genes in HuH6 (disregarding the fold-change cutoff) it only marginally increases the overlap between the two cell lines from 88 genes (see Fig. 1) to 102 genes, with the remaining 167 being unique to Huh-7 or HuH6, respectively. This argues against the hypothesis that HuH6 has a similar ISG profile, but lower overall induction values. In fact, the most strongly induced ISGs exhibit comparable fold-inductions in both cell lines (see Table 1). Furthermore, the ability of HuH6 cells to mount a functional antiviral response is not compromised in general, as these cells are capable to inhibit the replication of several viruses upon treatment with IFN-α [4]. In contrast to type I IFN, the number of genes induced upon IFN-γ treatment (by at least 1.5-fold) was similar in Huh-7 and HuH6 cells, and the overlap of induced genes was more substantial (see Fig. 1). Nonetheless, even for IFN-γ, roughly half of the ISGs were unique to the respective cell line, again highlighting the cell-type dependence of the induced ISG profile.
In order to specifically analyze differences between the ISG profiles of Huh-7 and HuH6, we used a ranked list approach, screening for those genes, whose rank differed most between the two profiles. These differentially regulated genes represent highly interesting candidates for future studies on functional differences between the IFN response in Huh-7 and HuH6 cells. For example, it has been shown before, that the IFN-γ response in HuH6 – in contrast to Huh-7 – cells is not capable to efficiently inhibit the replication of HCV [4]. In fact, we could show that one of the genes identified as being differentially regulated by IFN-γ in the two cell lines, DDX60L (WRD = 1.07, see Table 2), functionally contributes to this resistance phenotype of HCV in IFN-γ treated HuH6 cells [22].
The underlying reasons for the observed profound differences in the ISG profiles between Huh-7 and HuH6 cells and previously observed differences between other cell types remain largely elusive [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21]. Subtle differences in the expression of certain “master regulators”, e.g. transcription factors or secreted signaling molecules, such as cytokines, may drive the up- (or down-) regulation of whole sets of effector genes. In line with this hypothesis, in their study across six different virus species, Schoggins and colleagues identified as some of the most broadly active ISGs master regulators, such as IRF1 (impacting all six viral species), RIG-I, MDA5 and IRF7 [20], [23]. In our data, one striking difference between the cell lines was the production of several CXCL-type chemokines in response to IFN-α and IFN-γ, which was observed in Huh-7 but not in HuH6 cells. Furthermore, also the cytokines IL-8 (WRD 1.34 in IFN-α) and IL-7 (WRD 1.15 in IFN-γ), as well as the negative regulatory factor IRF2 (WRD 0.98 in IFN-α, WRD 0.95 in IFN-γ) were differentially regulated between the two cell lines. It is unclear if and to what extent these signaling modulators contribute to the observed differences in ISG expression profiles, but the findings represent interesting starting points for further investigations and may direct future experimentation.
Acknowledgments
This project was funded by grants from the Deutsche Forschungsgemeinschaft (MB: BI 1693/1-1; VL: FOR1202, TP3; RB: FOR1202, TP1). The authors would like to especially thank Rahel Klein and Ulrike Herian for excellent technical assistance.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.gdata.2015.12.017.
Appendix A. Supplementary data
Normalized expression data (Microsoft Excel file).
References
- 1.Lohmann V., Bartenschlager R. On the history of hepatitis C virus cell culture systems. J. Med. Chem. 2014;57:1627–1642. doi: 10.1021/jm401401n. [DOI] [PubMed] [Google Scholar]
- 2.Frese M., Pietschmann T., Moradpour D., Haller O., Bartenschlager R. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 2001;82:723–733. doi: 10.1099/0022-1317-82-4-723. [DOI] [PubMed] [Google Scholar]
- 3.Frese M., Schwarzle V., Barth K., Krieger N., Lohmann V., Mihm S., Haller O., Bartenschlager R. Interferon-gamma inhibits replication of subgenomic and genomic hepatitis C virus RNAs. Hepatology. 2002;35:694–703. doi: 10.1053/jhep.2002.31770. [DOI] [PubMed] [Google Scholar]
- 4.Windisch M.P., Frese M., Kaul A., Trippler M., Lohmann V., Bartenschlager R. Dissecting the interferon-induced inhibition of hepatitis C virus replication by using a novel host cell line. J. Virol. 2005;79:13778–13793. doi: 10.1128/JVI.79.21.13778-13793.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162 doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Durig J., Nuckel H., Huttmann A., Kruse E., Holter T., Halfmeyer K., Fuhrer A., Rudolph R., Kalhori N., Nusch A., Deaglio S., Malavasi F., Moroy T., Klein-Hitpass L., Duhrsen U. Expression of ribosomal and translation-associated genes is correlated with a favorable clinical course in chronic lymphocytic leukemia. Blood. 2003;101:2748–2755. doi: 10.1182/blood-2002-09-2683. [DOI] [PubMed] [Google Scholar]
- 7.Kallio M.A., Tuimala J.T., Hupponen T., Klemela P., Gentile M., Scheinin I., Koski M., Kaki J., Korpelainen E.I. Chipster: user-friendly analysis software for microarray and other high-throughput data. BMC Genomics. 2011;12:507. doi: 10.1186/1471-2164-12-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai M., Wang P., Boyd A.D., Kostov G., Athey B., Jones E.G., Bunney W.E., Myers R.M., Speed T.P., Akil H., Watson S.J., Meng F. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hulsen T., De V.J., Alkema W. BioVenn — a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC.Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H., Chapman R., Hertzog P.J. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brodsky L.I., Wahed A.S., Li J., Tavis J.E., Tsukahara T., Taylor M.W. A novel unsupervised method to identify genes important in the anti-viral response: application to interferon/ribavirin in hepatitis C patients. PLoS.One. 2007;2 doi: 10.1371/journal.pone.0000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Veer M.J., Holko M., Frevel M., Walker E., Der S., Paranjape J.M., Silverman R.H., Williams B.R. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- 13.Hilkens C.M., Schlaak J.F., Kerr I.M. Differential responses to IFN-alpha subtypes in human T cells and dendritic cells. J. Immunol. 2003;171:5255–5263. doi: 10.4049/jimmunol.171.10.5255. [DOI] [PubMed] [Google Scholar]
- 14.Hultcrantz M., Huhn M.H., Wolf M., Olsson A., Jacobson S., Williams B.R., Korsgren O., Flodstrom-Tullberg M. Interferons induce an antiviral state in human pancreatic islet cells. Virology. 2007;367:92–101. doi: 10.1016/j.virol.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 15.Indraccolo S., Pfeffer U., Minuzzo S., Esposito G., Roni V., Mandruzzato S., Ferrari N., Anfosso L., Dell'Eva R., Noonan D.M., Chieco-Bianchi L., Albini A., Amadori A. Identification of genes selectively regulated by IFNs in endothelial cells. J. Immunol. 2007;178:1122–1135. doi: 10.4049/jimmunol.178.2.1122. [DOI] [PubMed] [Google Scholar]
- 16.Lanford R.E., Guerra B., Lee H., Chavez D., Brasky K.M., Bigger C.B. Genomic response to interferon-alpha in chimpanzees: implications of rapid downregulation for hepatitis C kinetics. Hepatology. 2006;43:961–972. doi: 10.1002/hep.21167. [DOI] [PubMed] [Google Scholar]
- 17.Leaman D.W., Chawla-Sarkar M., Jacobs B., Vyas K., Sun Y., Ozdemir A., Yi T., Williams B.R., Borden E.C. Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: greater potency of IFN-beta compared with IFN-alpha2. J. Interf. Cytokine Res. 2003;23:745–756. doi: 10.1089/107999003772084860. [DOI] [PubMed] [Google Scholar]
- 18.Rani M.R., Shrock J., Appachi S., Rudick R.A., Williams B.R., Ransohoff R.M. Novel interferon-beta-induced gene expression in peripheral blood cells. J. Leukoc. Biol. 2007;82:1353–1360. doi: 10.1189/jlb.0507273. [DOI] [PubMed] [Google Scholar]
- 19.Sarasin-Filipowicz M., Oakeley E.J., Duong F.H., Christen V., Terracciano L., Filipowicz W., Heim M.H. Interferon signaling and treatment outcome in chronic hepatitis C. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He X.S., Ji X., Hale M.B., Cheung R., Ahmed A., Guo Y., Nolan G.P., Pfeffer L.M., Wright T.L., Risch N., Tibshirani R., Greenberg H.B. Global transcriptional response to interferon is a determinant of HCV treatment outcome and is modified by race. Hepatology. 2006;44:352–359. doi: 10.1002/hep.21267. [DOI] [PubMed] [Google Scholar]
- 22.Grunvogel O., Esser-Nobis K., Reustle A., Schult P., Muller B., Metz P., Trippler M., Windisch M.P., Frese M., Binder M., Fackler O., Bartenschlager R., Ruggieri A., Lohmann V. DDX60L is an interferon-stimulated gene product restricting hepatitis C virus replication in cell culture. J. Virol. 2015;89:10548–10568. doi: 10.1128/JVI.01297-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoggins J.W., MacDuff D.A., Imanaka N., Gainey M.D., Shrestha B., Eitson J.L., Mar K.B., Richardson R.B., Ratushny A.V., Litvak V., Dabelic R., Manicassamy B., Aitchison J.D., Aderem A., Elliott R.M., Garcia-Sastre A., Racaniello V., Snijder E.J., Yokoyama W.M., Diamond M.S., Virgin H.W., Rice C.M. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Normalized expression data (Microsoft Excel file).