Abstract
Myeloid-derived suppressor cells (MDSCs) are potently immunosuppressive innate immune cells that accumulate in advanced cancer patients and actively inhibit anti-tumor T lymphocyte responses [1]. Increased numbers of circulating MDSCs directly correlate with melanoma patient morbidity and reduced anti-tumor immune responses [2], [3]. Previous studies have revealed that monocyte-derived macrophage migration inhibitory factor (MIF) is necessary for the immune suppressive function of MDSCs in mouse models of melanoma [4], [5]. To investigate whether MIF participates in human melanoma-induced MDSC differentiation and/or suppressive function, we have established an in vitro MDSC induction model using primary, normal human monocytes co-cultured with human melanoma cell lines in the presence or absence of the MIF antagonist—4-IPP [4], [6], [7], [8], [9]. To identify potential mechanistic effectors, we have performed transcriptome analyses on cultured monocytes and on melanoma-induced MDSCs obtained from either untreated or 4-IPP-treated A375:monocyte co-cultures. Here, we present a detailed protocol, which can facilitate easy reproduction of the microarray results (NCBI GEO accession number GSE73333) published by Yaddanapudi et al. (2015) in Cancer Immunology Research [10].
Keywords: Melanoma, MDSC, MIF, Immunesuppression, Trancriptome analysis
| Specifications | |
|---|---|
| Organism/cell line/tissue | Homo sapiens/A375 melanoma cell line |
| Sex | NA |
| Sequencer or array type | Affymetrix Primeview® Human Gene Expression Arrays |
| Data format | Raw and Normalized Data |
| Experimental factors | Monocytes vs. untreated tumor-monocyte co-cultures (A375-MDSC) vs. MIF inhibitor-treated tumor-monocyte co-cultures (A375-MDSC + 4-IPP) |
| Experimental features | Expression profiling of healthy donor CD14+ monocytes cultured for 64 h in the absence of (cultured monocytes), or in the presence of A375 melanoma cells (A375-MDSC) or treated with MIF inhibitor, 4-IPP (MDSC + 4-IPP; 100 μM, day 0 and 50 μM, day 2). |
| Consent | All samples were obtained after informed consent |
| Sample source location | Louisville, KY |
1. Direct link to deposited data
Deposited data can be found here: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73333.
2. Experimental design, materials, and methods
2.1. Monocyte isolation and cell lines
Peripheral blood was collected from healthy donors. Peripheral blood mononuclear cells (PBMCs) were isolated from the peripheral blood by density gradient centrifugation (Ficoll-Hypaque, GE, Piscataway, NJ). Monocytic CD14+ cells were isolated from PBMCs using anti-CD14 magnetic microbeads and the autoMACS Pro Separator (Miltenyi Biotec, Auburn, CA), per manufacturer's instructions. Purity of isolated cell populations was > 90%, as evaluated by flow cytometry. Melanoma cell line [A375] (ATCC® CRL-1619™) was purchased from ATCC (Manassas, VA) and maintained in DMEM containing 10% (v/v) FBS. We do not culture this cell line for longer than 6–8 weeks and all cell-stocks came from thawed vials that were frozen at passage two after receiving from ATCC. A375 cell line was authenticated by ATCC cell bank using the Short Tandem Repeat (STR) profiling.
2.2. In vitro generation of human MDSC and MIF-inhibitor treatment
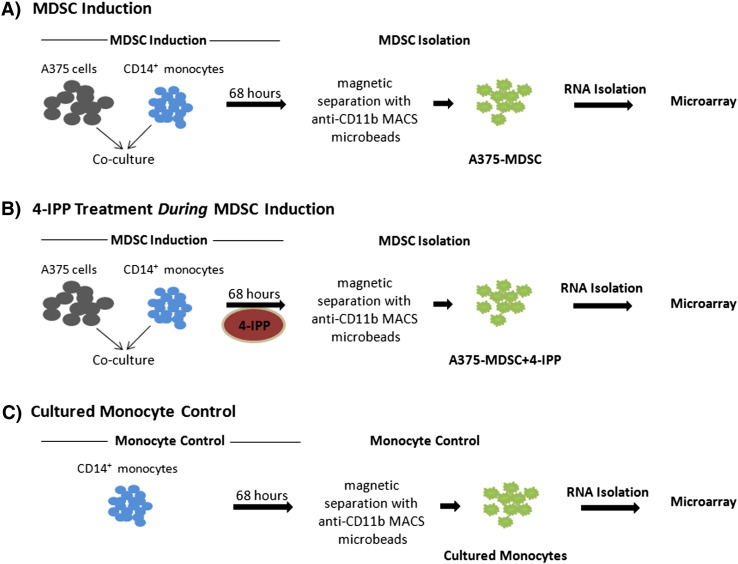
We established an in vitro model of melanoma cell-line-induced MDSCs that faithfully recapitulates patient-derived CD14+ HLADR−/low monocytic MDSC phenotype and function [9], [10]. CD14+ cells (1 × 106 cells/well), isolated from PBMCs obtained from healthy donors, were co-cultured with A375 tumor cells (5 × 105 cells/well) in complete IMDM (supplemented with 10% human AB serum [Sigma-Aldrich, St. Louis, MO], L-glutamine [2 mM], and penicillin/streptomycin) per well in a 6-well plate (BD Falcon). Tumor/monocyte co-cultures were treated twice with the small molecule inhibitor of MIF, 4-IPP (100 μM on day 0 and 50 μM on day 2), or 0.1% DMSO (vehicle control). A375 co-cultured monocytes (both vehicle-treated and 4-IPP-treated) and control monocytes cultured without tumor cells (cultured monocytes) were gently harvested after 64–68 h of culture and CD11b+ cells (A375-MDSCs) were purified with anti-CD11b+ microbeads and autoMACS separator (Miltenyi Biotec) (Fig. 1). Purity of isolated cell populations was > 90% as evaluated by flow cytometry. We refer to the CD11b+ cells isolated from the A375 monocyte co-cultures as A375-MDSCs although we are cognizant of the fact that these cells represent a heterogenous population of cells [2]. The percentage of CD14+ CD11b+ CD33+ HLADR−/low cells are substantially increased in A375-monocyte co-cultures in comparison to that of monocytes cultured in the absence of melanoma cells—an expression signature that closely corresponds to monocytic MDSCs isolated from late stage melanoma patients [10], [11], [12]. Furthermore, in contrast to tumor cell-free cultured monocytes, A375-MDSCs exhibit potent suppressive activity against autologous T cell proliferation and IFN-γ production [9], [10].
Fig. 1.
Schematic representation of the experimental design for the microarray analysis. (A) Normal donor CD14+ monocytes were co-cultured with human A375 melanoma cells for 68 h (MDSC induction). CD11b+ A375-MDSCs were isolated from the co-cultures by anti-CD11b+ microbead labeling and magnetic column separation (MDSC isolation). RNA for the microarray analysis was subsequently isolated from the purified A375-MDSCs; (B) 4-IPP (50 μM) was added during the A375:monocyte MDSC induction phase (A375-MDSC + 4-IPP), which was followed by the isolation of RNA from the purified 4-IPP-treated MDSCs; and (C) RNA was isolated from monocytes that were cultured for 68 h without tumor cells (cultured monocytes control).
2.3. Microarray Analysis
Gene expression profiles of vehicle-treated A375-MDSCs or 4-IPP-treated A375-MDSCs (A375-MDSC + 4-IPP) were compared to that obtained using monocytes cultured without tumor cells (Fig. 1). Total RNA from cultured monocytes, vehicle-treated, and 4-IPP-treated A375-MDSCs (n = 3) were isolated post-68 h in vitro culture and analyzed over a microarray. For RNA extraction, cells were lysed in buffer RLT (Qiagen, Valencia, CA), homogenized, and purified using RNeasy mini kit (Qiagen) following manufacturer's instructions. Integrity of RNA was checked using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Biotinylated cRNA was prepared from 100 ng of total RNA according to the standard protocol for Affymetrix 3′ IVT Express Plus Reagent kit. Following fragmentation, cRNA was hybridized for 16 h at 45 °C to Affymetrix Primeview Human arrays according to the GeneChip 3′ Array Hybridization User Manual from Affymetrix. The arrays were processed following the manufacturer recommended wash and stain protocol on an Affymetrix FS-450 fluidics station and scanned on an Affymetrix GeneChip scanner. The resulting .cel files were imported into Partek Genomics Suite 6.6 and transcripts were normalized and summarized using RMA as normalization and background correction method. Contrasts in a 1-way ANOVA were set up to compare the treatments of interest. Step-up False Discovery Rate was chosen as multiple test correction for the resulting p-values. The microarray datasets discussed in current study have been deposited in NCBI's Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession number GSE73333.
3. Conclusions
A microarray analysis of normal monocytes, vehicle-treated-A375-MDSCs, and 4-IPP-treated A375-MDSCs was performed to identify downstream MIF effectors that could be responsible for MIF-dependent phenotypic and functional contributions to human MDSCs. Melanoma cell line-induced MDSC transcriptome reveals a unique gene expression profile that is largely reversed to normal monocyte levels by MIF inhibition. Inflammatory cytokines, chemokines/chemokine receptors, matrix metalloproteases, angiogenic growth factors, and arachidonic acid/prostaglandin-generating enzymes are all differentially expressed in A375-MDSCs and restored to “normal” levels by MIF inhibition. These results are discussed in detail in the article published by Yaddanapudi et al. (2015) in Cancer Immunology Research [10].
Funding
This work was supported in part by NIH CA186661 and NIH CA102285. Part of this work was performed with assistance of the Genomics Facility (University of Louisville), which is supported by NIH P20GM103436 (KY IDeA Networks of Biomedical Research Excellence), NIH P30GM106396 (J. G. Brown Cancer Center, University of Louisville, Phase III CoBRE), and the James Graham Brown Foundation, and user fees.
Contributor Information
Robert A. Mitchell, Email: robert.mitchell@louisville.edu.
Kavitha Yaddanapudi, Email: kavitha.yaddanapudi@louisville.edu.
References
- 1.Gabrilovich D.I., Ostrand-Rosenberg S., Bronte V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012;12(4):253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gros A. Myeloid cells obtained from the blood but not from the tumor can suppress T-cell proliferation in patients with melanoma. Clin. Cancer Res. 2012;18(19):5212–5223. doi: 10.1158/1078-0432.CCR-12-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weide B. Myeloid-derived suppressor cells predict survival of patients with advanced melanoma: comparison with regulatory T cells and NY-ESO-1- or melan-A-specific T cells. Clin. Cancer Res. 2014;20(6):1601–1609. doi: 10.1158/1078-0432.CCR-13-2508. [DOI] [PubMed] [Google Scholar]
- 4.Yaddanapudi K. Control of tumor-associated macrophage alternative activation by macrophage migration inhibitory factor. J. Immunol. Mar 15 2013;190(6):2984–2993. doi: 10.4049/jimmunol.1201650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson K.D., Templeton D.J., Cross J.V. Macrophage migration inhibitory factor promotes tumor growth and metastasis by inducing myeloid-derived suppressor cells in the tumor microenvironment. J. Immunol. 2012;189(12):5533–5540. doi: 10.4049/jimmunol.1201161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winner M. A novel, macrophage migration inhibitory factor suicide substrate inhibits motility and growth of lung cancer cells. Cancer Res. 2008;68(18):7253–7257. doi: 10.1158/0008-5472.CAN-07-6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadjeva M. Inhibition of macrophage migration inhibitory factor ameliorates ocular Pseudomonas aeruginosa-induced keratitis. PLoS Pathog. 2010;6(3):e1000826. doi: 10.1371/journal.ppat.1000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kindt N. Pharmacological inhibition of macrophage migration inhibitory factor interferes with the proliferation and invasiveness of squamous carcinoma cells. Int. J. Oncol. 2013;43(1):185–193. doi: 10.3892/ijo.2013.1944. [DOI] [PubMed] [Google Scholar]
- 9.Mao Y. Melanoma-educated CD14 + cells acquire a myeloid-derived suppressor cell phenotype through COX-2-dependent mechanisms. Cancer Res. 2013;73(13):3877–3887. doi: 10.1158/0008-5472.CAN-12-4115. [DOI] [PubMed] [Google Scholar]
- 10.Yaddanapudi K. MIF is necessary for late-stage melanoma patient MDSC immune suppression and differentiation. Cancer Immunol. Res. 2015 doi: 10.1158/2326-6066.CIR-15-0070-T. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poschke I. Immature immunosuppressive CD14+ HLA-DR −/low cells in melanoma patients are Stat3hi and overexpress CD80, CD83, and DC-sign. Cancer Res. 2010;70(11):4335–4345. doi: 10.1158/0008-5472.CAN-09-3767. [DOI] [PubMed] [Google Scholar]
- 12.Lechner M.G., Liebertz D.J., Epstein A.L. Characterization of cytokine-induced myeloid-derived suppressor cells from normal human peripheral blood mononuclear cells. J. Immunol. 2010;185(4):2273–2284. doi: 10.4049/jimmunol.1000901. [DOI] [PMC free article] [PubMed] [Google Scholar]