Abstract
Serofluid dish (or Jiangshui, in Chinese), a traditional food in the Chinese culture for thousands of years, is made from vegetables by fermentation. In this work, microorganism community of the fermented serofluid dish was investigated by the culture-independent method. The metagenomic data in this article contains the sequences of fungal internal transcribed spacer (ITS) regions of rRNA genes from 12 different serofluid dish samples. The metagenome comprised of 50,865 average raw reads with an average of 8,958,220 bp and G + C content is 45.62%. This is the first report on metagenomic data of fungal ITS from serofluid dish employing Illumina platform to profile the fungal communities of this little known fermented food from Gansu Province, China. The Metagenomic data of fungal internal transcribed spacer can be accessed at NCBI, SRA database accession no. SRP067411.
Keywords: Serofluid dish, Jiangshui, Fungal ITS, Cultivation-independent, Microbial diversity
| Specifications | |
|---|---|
| Organism/cell line/tissue | Fungal internal transcribed spacer from serofluid dish (Jiangshui) |
| Sex | Not applicable |
| Sequencer or array type | Illumina Hiseq 2500 |
| Data format | Raw data: FASTQ file |
| Experimental factors | Environmental sample |
| Experimental features | ITS region of serofluid dish (Jiangshui) was amplified using fungal ITS specific primers from isolated metagenome followed by pyrosequencing using Illumina Hi-Seq technology. |
| Consent | Not applicable |
| Sample source location | A total of 12 serofluid dish samples were collected from 10 handmade areas from Gansu Province, China. |
1. Direct link to deposited data
2. Experimental design, materials and methods
Chinese traditional fermented vegetable foods, such as serofluid dish (also called Jiangshui, in Chinese), have served as low-calorie food that have been one of the major carbohydrate and vitamin sources in Chinese diet for more than two thousand years [1]. In recent years, fermented vegetable foods have attracted considerable interest due to their remarkable nutritional value. They are also well known as functional foods due to their antihypertensive effect, improvement in the intestinal function, and preventing diabetes and anticancer properties [2]. Therefore, they are predicted to become significant for human health [3].
Although diverse eukaryotic microbes were found in serofluid dish by cultivation and microscopy, the abundance and diversity of naturally occurring fungi have not been well characterized [4]. Culture-independent methods based on the rRNA genes of microbes were usually used to study the microbial communities in natural habitats [5]. The internal transcribed spacer (ITS) regions of fungal rRNA genes have been identified as particular targets for molecular biology analysis of fungal communities and their high sequence variability relative to the flanking sequences that makes them valuable for genus- and species-level identification [6]. Therefore, the present research was designed to investigate the fungal communities' composition by using Illumina based metagenomic approach in serofluid dish, which may represent novel species within this genus which was reported for the first time in association with serofluid dish.
A total of 26 serofluid dish samples were collected from 10 handmade areas across Gansu Province, China (35.22°–36.33°N, 103.59°–105.10°E). The fungal internal transcribed spacer was amplified using ITS1 and ITS4 primer combination (5′-GGAAGTAAAAGTCGTAACAAGG-3′ and 5′-GCTGCGTTCTTCATCGATGC-3′, respectively). A 6-bp error-correcting tag was added to the forward primer [7]. A total of 12 serofluid dish samples were sent to Novogene Bioinformatics Technology (Beijing, China) to perform amplicon sequencing according to Hiseq protocols. Amplicon pyrosequencing was performed on the Illumina HiSeq PE250 platforms at Novogene Bioinformatics Technology (Beijing, China). Bioinformatic analysis was performed by QIIME data analysis package [8]. Complete data sets are submitted to the NCBI Short Read Archive under accession numbers SRR3003942, SRR3003943, SRR3003944, SRR3003945, SRR3003947, SRR3003948, SRR3004055, SRR3004201, SRR3004236, SRR3004274, SRR3004275 and SRR3009902, respectively.
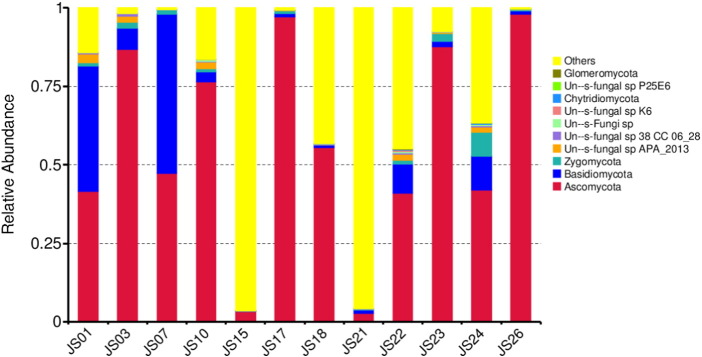
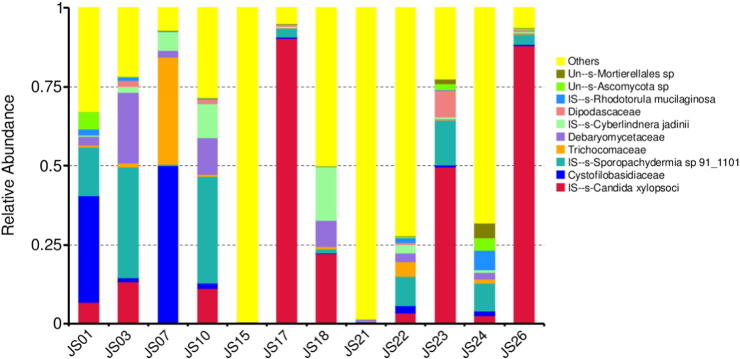
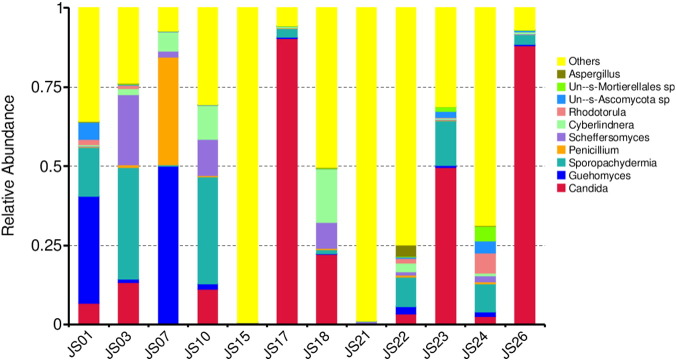
The output file comprised 151 MB data with a total of 610,379 high quality reads having 45.62% GC content. In the serofluid dish sample studied, the classified OTUs represent distinct phyla dominated by Ascomycota (56.62%), Basidiomycota (10.55%), Zygomycota (1.54%) and others (31.29%) (Fig. 1). At the family level, Candida xylopsoci (23.96%) was predominant followed by Sporopachydermia sp. (10.36%), Cystofilobasidiaceae (7.73%), Debaryomycetaceae (4.46%), Trichocomaceae (3.69%), Cyberlindnera (3.40%), Dipodascaceae (1.17%), Ascomycota sp. (1.06%), Rhodotorula mucilaginosa (0.94%), Mortierellales sp. (0.55%) and others (42.68%) (Fig. 2). The leading genera were Candida (23.96%), Sporopachydermia (10.36%), Guehomyces (7.72%), Scheffersomyces (4.0%), Cyberlindnera (3.40%), Penicillium (3.16%), Ascomycota sp. (1.06%) and others (44.44%) (Fig. 3). The remaining sequences were not associated to any known fungi or fungal sequences in the public database.
Fig. 1.
Taxonomy classification of reads at phylum level. Only top 10 enriched class categories are shown in the figure.
Fig. 2.
Taxonomy classification of reads at family level. Only top 10 enriched class categories are shown in the figure.
Fig. 3.
Taxonomy classification of reads at genera level. Only top 10 enriched class categories are shown in the figure.
Our data provide the first scientific report on the composition of the fungal communities, using Illumina sequencing method, from the underdeveloped serofluid dish located in northwest China region. The most dominated phylum in this study was Ascomycota and Basidiomycota which are known to produce beneficial metabolites useful for human health, such as organic acid, antibiotic, polysaccharide, and vitamin. The metagenome analysis of fungal ITS regions from serofluid dish revealed the presence of fungal key functional characteristics. Therefore, these results indicate that both serofluid dish formation complex and distinct fungal communities structure which were not reported before.
Nucleotide sequence accession number
Metagenome sequence data are available on NCBI, SRA database accession no. SRP067411.
Competing interests
The authors declare there are no competing interests.
Acknowledgments
This work was supported by Gansu Province Science Foundation for Distinguished Young Scholars (Grant No. 1308RJDA014), Longyuan Support Project for Young Creative Talents (Grant No. GANZUTONGZI [2014] no.4), Technology Program of Lanzhou City (Grant No. 2015-3-142, Grant No. 2015-3-97), and The Fundamental Research Funds for the Central Universities of China (Grant No. lzujbky-2015-57).
Contributor Information
Peng Chen, Email: chenpeng@lzu.edu.cn.
Hongyu Li, Email: lihy@lzu.edu.cn.
References
- 1.Liu R., Dang S., Yan H., Wang D., Zhao Y., Li Q., Liu X. Association between dietary protein intake and the risk of hypertension: a cross-sectional study from rural western China. Hypertens. Res. 2013;36(11):972–979. doi: 10.1038/hr.2013.71. [DOI] [PubMed] [Google Scholar]
- 2.Li X., Li J., Li M., Meng X. Screening and functional properties of cholesterol-degrading lactic acid bacteria from Jiangshui. Wei Sheng Wu Xue Bao. 2015;55(8):1001–1009. (in Chinese) [PubMed] [Google Scholar]
- 3.Hou Z., Yang J., Jia H., Lu X., Wang L. Research overview and prospect of traditional fermented food — serofluid dish. China Condiment. 2015;40(02):132–136. (in Chinese) [Google Scholar]
- 4.Zhang Y., Wang Y., Chen X., Zhao P. Isolation and initiative identification of microorganism from traditional fermentative food—Jiangshui. Food Sci. 2007;28(01):219–222. (in Chinese) [Google Scholar]
- 5.Dudhagara P., Ghelani A., Bhavsar S., Bhatt S. Metagenomic data of fungal internal transcribed spacer and 18S rRNA gene sequences from Lonar lake sediment, India. Data Brief. 2015;4:266–268. doi: 10.1016/j.dib.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai X., Cao L., Tan H., Fang S., Huang Y., Zhou S. Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J. 2007;1(8):756–762. doi: 10.1038/ismej.2007.51. [DOI] [PubMed] [Google Scholar]
- 7.Hamady M., Walker J.J., Harris J.K., Gold N.J., Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods. 2008;5(3):235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]