Abstract
In recent years, DNA adenine methyltransferase identification (DamID) has emerged as a powerful tool to profile protein-DNA interaction on a genome-wide scale. While DamID has been primarily combined with microarray analyses, which limits the spatial resolution and full potential of this technique, our group was the first to combine DamID with sequencing (DamID-Seq) for characterizing the binding loci and properties of a transcription factor (Tox) (sequencing data available at NCBI's Gene Expression Omnibus under the accession number GSE64240). Our approach was based on the combination and optimization of several bioinformatics tools that are here described in detail. Analysis of Tox proximity to transcriptional start sites, profiling on enhancers and binding motif has allowed us to identify this transcription factor as an important new regulator of neural stem cells differentiation and newborn neurons maturation during mouse cortical development. Here we provide a valuable resource to study the role of Tox as a novel key determinant of mammalian somatic stem cells during development of the nervous and lymphatic system, in which this factor is known to be active, and describe a useful pipeline to perform DamID-Seq analyses for any other transcription factor.
Keywords: DamID-Seq, Neural stem cells, SICER
| Specifications | |
|---|---|
| Organism/cell line/tissue | Human Embryonic Kidney 293 (HEK-293 T) cells and mouse neuroblastoma (Neuro-2a) cells |
| Sex | not applicable |
| Sequencer or array type | Illumina HiSeq 2500 |
| Data format | Raw and analyzed |
| Experimental factors | Fusion Dam-Tox vs. Dam alone |
| Experimental features | DamID-Seq of the HMG-box transcription factor protein Tox |
| Consent | not applicable |
| Sample source location | not applicable |
1. Direct link to deposited data
Data were deposited in Gene Expression Omnibus (GEO) datasets under reference number GSE64240.
2. Experimental design
During embryonic development of the mammalian cortex, neuroepithelial stem cells expand and generate neurogenic progenitors that in turn divide to give rise to neurons [18]. In an attempt to identify genes involved in controlling this process, our group has generated a double reporter mouse line to isolate the three cell populations present in the developing mouse brain and, by transcriptome analyses, determined their molecular signature [3], [4]. The transcription factor Tox was identified among those transcripts that were named “off-switches” since, during corticogenesis, revealed to be highly expressed in neural stem cells, transiently downregulated in neurogenic progenitors, and reinduced in neurons. Many genes essential during neural development are indeed switch genes, showing differential expression between neurogenic progenitors as compared to both neural stem cells and neurons [3]. Interestingly, Tox shared a similar pattern of expression with several such master regulators of corticogenesis. In addition, Tox has been previously associated with differentiation of T-cells in the lymphatic system [1]. Yet, no function was ever reported for this transcription factor during corticogenesis, which leads us to further investigate its role(s) during mammalian brain development.
Acute manipulations of Tox expression in the mouse developing cortex revealed its multiple functions during corticogenesis in different cell types. In fact, Tox negatively regulated neurogenesis by inhibiting differentiation of neural stem cells while, at the same time, controlled neuronal specification and neurites outgrowth of newborn neurons [6]. In order to gain insight into the possible molecular mechanisms by which Tox performs its functions, we then sought to identify its downstream targets by determining binding sites on the genome.
The main experimental approaches to investigate chromatin binding profiles on large scale are chromatin immunoprecipitation (ChIP) and DNA adenine methyltransferase identification (DamID). ChIP has been for many years the gold standard for protein–chromatin interaction profiling [15]. Briefly, ChIP relies on the crosslinking of DNA-protein complexes followed by immunoprecipitation using an antibody recognizing the protein of interest and identification of the co-precipitated DNA sequences. As main drawback, the key determinant for a good ChIP experiment is therefore the availability of an antibody with high affinity and specificity. As alternative antibody-independent method, DamID exploits methylation to label sequences of the genome that are bound by a specific protein. In such a case, the protein of interest is fused to the prokaryotic Dam methylase and expressed in eukaryotic cells at extremely low levels as not to saturate methylation and increase specificity. Dam methylates adenines, modification normally not occurring in eukaryotes, that can therefore be recognized by a restriction enzyme, DpnI, cutting its specific recognition sequence GATC only when the adenine is methylated [16], [20], [22], [23].
Commercially available Tox antibodies were not validated for ChIP and we could not achieve immunoprecipitation with any of the antibodies tested. Therefore, we decided to employ DamID to profile Tox genome binding sites.
Detailed reviews on the advantages and disadvantages of DamID versus ChIP have been reported elsewhere (e.g. [8]). It is however important to point out that DamID typically provided a lower spatial resolution than ChIP-Seq in defining binding domains [14]. Inspired by a report on nuclear envelop proteins combining DamID with high-throughput sequencing [24], we explored the use of DamID-Seq to detect narrow regions of chromatin binding by a transcription factor [6]. Our approach allowed the fine characterization of the binding sites, including binding motif prediction, whose pipeline is provided below.
Experiments were performed according to the standard DamID protocol [23] and using HIV lentiviruses that trigger the expression of Tox-Dam or Dam alone at very low concentrations. Ensuring a low expression level of the ectopic genes is particularly critical to obtain a high signal/background ratio by avoiding saturation of methylation by Dam. In order to achieve this, expression of Tox-Dam fusion gene, and Dam as negative control, was put under the regulation of two inducible promoters neither of which was induced, resulting only in minimal “double-leakyness” of transgene expression. As such, ectopic proteins were undetectable by both immunofluorescence and Western blot (data not shown).
DamID was followed by Illumina next-generation sequencing and bionformatic comparison of Tox-Dam versus Dam alone to identify Tox binding targets. This was performed on both HEK-293T (human embryonic kidney cell line) and Neuro-2a (mouse neuroblastoma cell line). The rationale behind using both cell lines was that in the former case we wanted to use a brain-unbiased system to assess all possible Tox targets while the latter provided a cell line recapitulating the physiology of neural stem cells in which Tox may have additional properties.
Biological duplicates performed on HEK-293T cells revealed to be successful as we could observe substantial differences between Dam-Tox and Dam control samples. This allowed us to identify ca. 13,000 chromatin regions bound by Tox. Conversely, by using the Neuro-2a cell line we did not observe any substantial enrichment of Tox binding as compared to the Dam background. Although we did not perform experiments to explain this discrepancy between cell lines, we find it reasonable that in Neuro-2a the ectopic Tox-Dam fusion protein would compete with the endogenous Tox for its binding to chromatin and since the former is expressed at minimal levels it would be outcompeted by the latter resulting in no differential methylation pattern relative to Dam control [6]. In support to this hypothesis, in HEK-293T cells endogenous Tox expression was undetectable by transcriptome analysis [17] and, hence, ectopic Tox-Dam would be free to bind its targets. These may be important considerations to keep in mind while choosing the cell system to perform DamID, as it could influence the final outcome of the experiments. Tox binding loci, including proximity to transcriptional start sites, profiling on enhancers and binding motif were subsequently analyzed by several bioinformatic tools with some of these predictions being later validated in vivo [6].
3. Materials and methods
3.1. Lentiviral transfection of DamID construct
Total RNA was extracted from E13.5 mouse lateral cortices and used as template for RT-PCR amplification of Tox cDNA that was cloned in the pLgwV5EcoDam (Dam construct) [23] to generate the Tox-Dam construct. To ensure that the fusion protein (Tox-Dam) would not display aberrant expression pattern or localization as compared to the native Tox, plasmid coding for the Tox-Dam or the Tox wt protein were transfected in HEK-293T cells and their subcellular localization determined by Western blot and immunocytochemistry [6].
3.2. DamID-Seq
pLgwV5Eco-ToxDam and pLgxV5Dam were used to produce Tox-Dam and Dam control lentiviruses, respectively, as described in [5], [7]. HEK-293T and Neuro-2a cells were infected with Tox-Dam or Dam viral supernatant diluted 1:2 or 1:10 and DamID performed as described [23]. Briefly, 48 h after infection genomic DNA was extracted, digested with DpnI, ligated to adaptors and PCR amplified. Experiments were performed in duplicates for each cell line and condition. Sequencing libraries were prepared according to a standard Illumina protocol and subjected to 75 bp single read sequencing on a HiSeq 2000 machine, resulting in ca. 20 million reads per sample (DNA libraries of replicates were sequenced separately).
3.3. Primary processing of sequencing data
Raw read quality was evaluated using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Read alignment to the genome (Homo sapiens ensembl67 or Mus musculus ensembl61) was performed with bowtie v0.12.7 [10] using the default parameters with the options “--best” and “-m 1” to retain only uniquely mapped reads. Replicate reproducibility was tested using bamCorrelate from the NGS analysis suite deepTools [13], with the custom options bamCorrelate bins --fragmentLength 200 --corMethod pearson. Pair of replicates displayed Pearson correlation coefficients > 0.80 and therefore the alignments corresponding to 2 replicates of the same condition were merged before peak calling. Since experiments performed in Neuro-2a cells did not show any significant difference between conditions (Tox-Dam vs. Dam), our further analyses were based only on data obtained from HEK-293T cells.
3.4. Identification of Tox binding sites
To identify genomic regions of Tox-Dam enrichment we used the peak caller SICER [27] version 1.1, which has been previously used to detect enrichment in DamID experiments [24]. SICER has been initially developed to detect enrichment (ChIP over input) of diffuse histone modifications. Differently from transcription factors, which usually bind at very localized genomic loci and therefore lead to strong and localized signals, histone modification signals are more diffused and lack well defined peaks. SICER is therefore an algorithm designed to deal with more diffused enrichment spreads over extended genomic regions, rather than strong local enrichment [27]. Since DamID-methylation could spread for some distance from the actual binding site (ca. 2 kb) [20], we thought that SICER would be more appropriate than other methods for the detection of Tox enriched regions.
Briefly, SICER first models the distribution of random reads in a genomic background to determine read spatial clusters that are unlikely to appear by chance. Based on this “random model”, clusters of enrichment in a given experimental condition can be identified and scored, and comparison with the control used to determine the statistical significance of such enrichments. To do this, SICER first divides the genome into non-overlapping, contiguous windows of a defined size (w). Then, it identifies “eligible” windows, as those showing reads enrichment, determining “islands” as clusters of eligible windows separated by a gap of a certain size (g) of ineligible windows. SICER finally retains candidate islands, which represent clusters of eligible windows unlikely to appear by chance, and further filters them against the control sample. The result of the analysis is then highly influenced by the two key setting parameters: the window size w and the gap size g. Since SICER has not been originally developed for analysis of DamID-Seq data, these two settings, window and gap size, had to be determined empirically and further optimized for our data.
In general, by selecting large window sizes, regions of lower enrichment are also included in an island (peak) with consequent reduction in resolution. On the other hand, by choosing too narrow window size, the same island risks to be fragmented into many intervals of windows and gaps. Bearing this in mind, we tested two window sizes, 50 and 200 bp. These sizes are often suitable for transcription factors and histone modifications, respectively, as suggested by the authors of SICER [25].
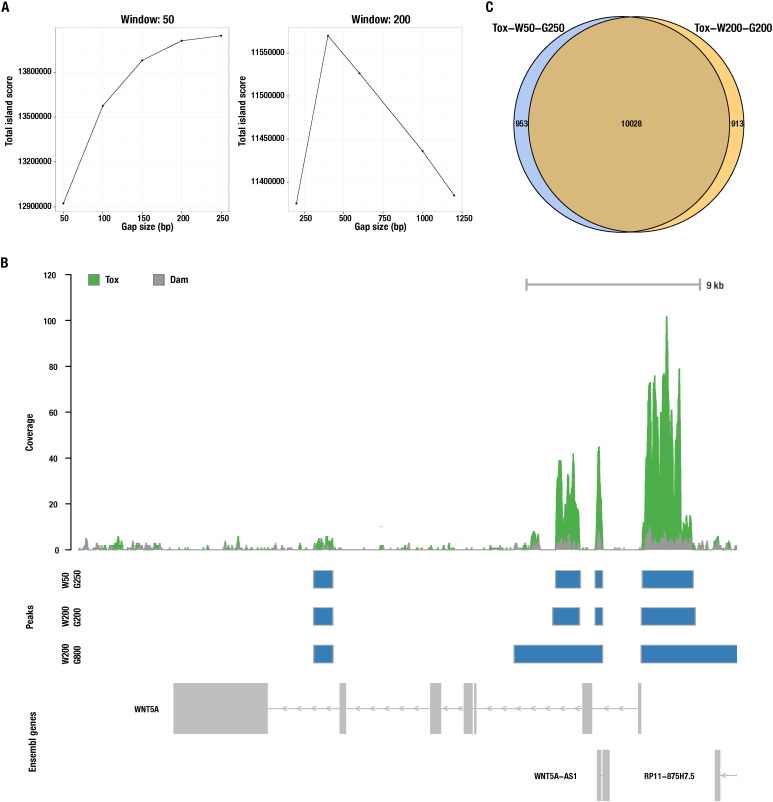
Gap size must be equal or higher than the windows size. In order to determine the optimal gap size, it is possible to plot the aggregate score of all the significant islands (with island score representing the negative logarithm of the probability of finding reads in the island if the reads should be randomly distributed in the genome with equal probability) as a function of the gap size (Fig. 1A). The gap size for which the aggregate score is higher, or closer to saturation, should obviously be selected [25]. We tested increasing gap sizes (Fig. 1A) with maximal scores being obtained at 250 bp and 200 bp, for window size 50 bp and 200 bp, respectively. Gap size influenced the peak size (w200-g200 versus w200-g600) (Fig. 1B) with regions of enrichment being merged together and extending to poorly enriched locations.
Fig. 1.
Effect of SICER gap and window parameters in peak calling. (A) Effect of gap size on total island score. The gap size with the highest total score, for a given window size, was chosen as the optimal gap size. (B) Choosing an optimal combination of gap/window size ensured that detected regions were well defined and narrow. The upper panel is a genomic view of Tox and Dam coverage (read density). Below are represented peaks identified with different settings (blue boxes). (C) Overlap of genes identified as Tox targets by different SICER parameters.
Using the software suite BEDTools [12] we have also determined how many genes were associated with Tox in the two setting combinations chosen (i.e. w50, g250 and w200, g200) by assigning each peak to a certain transcript if the peak was located within 15 kb upstream of the transcription start site, or inside the transcript body (Fig. 1B). The intersection was done with transcripts from the UCSC ensGene table (hg19), with the command bedtools intersect -a islands-summary-FDR0.01 -b ensGenes -wo, and the gene locations extended with slopBed -s -l 15000 -r 0 before intersection. The number of identified genes in both settings was nearly identical (10,981 and 10,941 for w50, g250 and w200, g200, respectively) with 10,028 in common being identified by both settings (Fig. 1C). Since peak width was generally narrower for w50 and this setting is recommended for transcription factors [25], we decided to use peaks identified with this window size and g250 for further analysis. The peak width is of particular importance for motif analysis, where the region analyzed should be as narrow as possible, ideally the size of the Tox binding footprint, improving the accuracy of results. To remove poorly enriched peaks from our dataset, only peaks supported by more than 50 reads and with at least a two-fold enrichment of Tox reads over Dam control, were retained for further analysis.
3.5. Peak characterization
To identify which genes are associated with Tox, the gene table ensGene was retrieved from UCSC (hg19), each transcript extended 15 kb upstream with slopBed -i -s -l 15000 -r 0 (BEDTools) and intersected with the significant peaks identified by SICER (intersectBed). Thus, genes/transcripts that have Tox peak(s) within 15 kb upstream of the transcription start site, or inside the transcript body, were matched.
The Bioconductor package ChIPseeker [26] was used to annotate the genomic features (e.g. introns, exons, 5′UTR, and 3′UTR) of each peak. Specifically, the function annotatePeak matches peaks with genomic features extracted from hg19 (ensGene) and calculates the proportion of peaks matching each feature. Gene ontology enrichment in genes associated with Tox, was determined with the function getEnrichedGO of the Bioconductor package ChIPpeakAnno [28].
3.6. Binding motif
Tox putative binding motif was predicted with gimmeMotifs v0.65 [19] that summarizes the results from motif prediction tools such as GADEM, MEME, Improbizer and others, producing a metrics report to evaluate the predicted motifs. Two common assumptions valid for ChIP-Seq data may not apply to DamID-Seq data: (i) the binding site may not be at the center of the SICER island and (ii) many enriched regions can be relatively large (more than 1 Kb). For (i) most programs run by gimmeMotifs do not take the peak location into consideration and (ii) was taken into account by using only islands with a 2-fold enrichment (Tox-Dam/Dam reads), width of less than 1 kb and changing the program setting to include the option --width=1000 to prevent truncation of input peaks to 200 bp (the default). The final command was gimme_motifs.py --analysis = medium --genome=hg19 --fraction=0.3 --width=1000 --localization_width=1000. Genomic sequences corresponding to input peaks were retrieved with bedtools getfasta.
3.7. Tox binding profile
The genomic location of telencephalic enhancers [21] were downloaded from GEO (GSM1052710) and converted from hg18 to hg19 using liftOver [9]. Locations of other cell specific enhancers [2] were obtained from http://enhancer.binf.ku.dk/Pre-defined_tracks.html. A set of random genomic locations to use as background, matched to telencephalic enhancers, was created with bedtools shuffleBed with the settings -i telencephalon_enhancers -seed 927442958 -chrom -noOverlapping. The average size of enhancers was also previously calculated with a custom AWK script. Using the tools from DeepTools [13] we first normalized Tox-Dam read coverage to Dam by binning the genome, counting the number of reads in each bin for either condition and calculating the log2 of the ratio. This was accomplished with bamCompare -bs -f 200 –missingDataAsZero no –ratio log2 –normalizeTo1x 2451960000 which generates a bigwig of normalized read density. The normalized Tox read density over the enhancers was summarized with computeMatrix scale-regions -a 3000 -b 3000 -m 3000 -bs 10, which scales each enhancer to the average size (3 kb). Finally the profile of Tox binding in enhancers was generated using profiler (DeepTools).
The Tox binding profile at transcription start sites was also generated using the normalized Tox read density calculated with bamCompare. Non-redundant transcription start sites were obtained from the ensGene table of UCSC (hg19) using a combination of AWK and bedtools merge. The normalized Tox read density over such transcription start sites was calculated with computeMatrix reference-point --referencePoint TSS -a 50000 -b 50000 --sortRegions descend --binSize 50 and plotted with profiler.
3.8. Peak conservation
For conservation analysis of Tox peaks, conservation scores of 99 vertebrate genomes, including human, were obtained from the UCSC genome browser (table phastCons100way http://hgdownload.cse.ucsc.edu/goldenpath/hg19/phyloP100way/). Since exons are more conserved than introns [11] and Tox peaks are more likely to be present in exons [6], we looked at the conservation of Tox peaks in a way that was not biased by exonic conservation. To achieve this, exonic regions were masked from the phastCons100way scores in a series of steps. The bigwig was converted to BedGraph with bigWigToBedGraph [9], intersected with the genomic locations of human exons (ensGene table hg19) with intersectBed, conservation scores of overlapping regions converted to zero and finally saved as bigwig with bedGraphToBigWig [9]. As background, matched peak random genomic locations were created with shuffleBed -seed 927442958 -chrom -noOverlapping. The scores were summarized with computeMatrix scale-regions -a 500 -b 500 -m 1500 -bs 10, scaling each peak to the average size (~ 1.5 kb). The plot was generated with profiler --plotType se, to include the standard deviation of conservation scores at each position. It should be noted that masking the conservation scores of exons did not affect the overall result (data not shown).
Acknowledgments
We are grateful to all people providing us with reagents, advise and sharing protocols, particularly Drs. Bas van Steensel, Pompeo Macioce, Joerg Mansfeld, Gabor Bakos, Satoshi Matsuda, Shigeo Koyasu, Sebastian Jaulian and Tony Southall. This work was supported by the CRTD, the TU Dresden, the DFG (CA 893/9-1) and Collaborative Research Center SFB655 (subproject A20). BA is currently supported by a FEBS long-term fellowship.
References
- 1.Aliahmad P., Seksenyan A., Kaye J. The many roles of TOX in the immune system. Curr. Opin. Immunol. 2012;24:173–177. doi: 10.1016/j.coi.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aprea J., Prenninger S., Dori M., Ghosh T., Monasor L.S., Wessendorf E., Zocher S., Massalini S., Alexopoulou D., Lesche M. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013;32:3145–3160. doi: 10.1038/emboj.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aprea J., Lesche M., Massalini S., Prenninger S., Alexopoulou D., Dahl A., Hiller M., Calegari F. Identification and expression patterns of novel long non-coding RNAs in neural progenitors of the developing mammalian cortex. Neurogenesis. 2015:e995524. doi: 10.1080/23262133.2014.995524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Artegiani B., Calegari F. Lentiviruses allow widespread and conditional manipulation of gene expression in the developing mouse brain. Development. 2013;140:2818–2822. doi: 10.1242/dev.093823. [DOI] [PubMed] [Google Scholar]
- 6.Artegiani B., de Jesus Domingues A.M., Bragado Alonso S., Brandl E., Massalini S., Dahl A., Calegari F. Tox: a multifunctional transcription factor and novel regulator of mammalian corticogenesis. EMBO J. 2015;34:896–910. doi: 10.15252/embj.201490061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Artegiani B., Lange C., Calegari F. Expansion of embryonic and adult neural stem cells by in utero electroporation or viral stereotaxic injection. J. Vis. Exp. 2012:e4093. doi: 10.3791/4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aughey G.N., Southall T.D. Dam it's good! DamID profiling of protein-DNA interactions. Wiley Interdiscip. Rev. Dev. Biol. 2016;5:25–37. doi: 10.1002/wdev.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn R.M., Haussler D., Kent W.J. The UCSC genome browser and associated tools. Brief. Bioinform. 2013;14:144–161. doi: 10.1093/bib/bbs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mouse Genome Sequencing C., Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 12.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T. DeepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:187–191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimbo T., Du Y., Grimm S.A., Dhasarathy A., Mav D., Shah R.R., Shi H., Wade P.A. MBD3 localizes at promoters, gene bodies and enhancers of active genes. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1004028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solomon M.J., Larsen P.L., Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 16.Southall T.D., Brand A.H. Chromatin profiling in model organisms. Brief. Funct. Genomic. Proteomic. 2007;6:133–140. doi: 10.1093/bfgp/elm013. [DOI] [PubMed] [Google Scholar]
- 17.Sultan M., Schulz M.H., Richard H., Magen A., Klingenhoff A., Scherf M., Seifert M., Borodina T., Soldatov A., Parkhomchuk D. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 18.Taverna E., Gotz M., Huttner W.B. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu. Rev. Cell Dev. Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
- 19.van Heeringen S.J., Veenstra G.J. GimmeMotifs: a de novo motif prediction pipeline for ChIP-sequencing experiments. Bioinformatics. 2011;27:270–271. doi: 10.1093/bioinformatics/btq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Steensel B., Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 21.Visel A., Taher L., Girgis H., May D., Golonzhka O., Hoch R.V., McKinsey G.L., Pattabiraman K., Silberberg S.N., Blow M.J. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogel M.J., Guelen L., de Wit E., Peric-Hupkes D., Loden M., Talhout W., Feenstra M., Abbas B., Classen A.K., van Steensel B. Human heterochromatin proteins form large domains containing KRAB-ZNF genes. Genome Res. 2006;16:1493–1504. doi: 10.1101/gr.5391806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogel M.J., Peric-Hupkes D., van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 24.Wu F., Yao J. Spatial compartmentalization at the nuclear periphery characterized by genome-wide mapping. BMC Genomics. 2013;14:591. doi: 10.1186/1471-2164-14-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu S., Grullon S., Ge K., Peng W. Spatial clustering for identification of ChIP-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol. Biol. 2014;1150:97–111. doi: 10.1007/978-1-4939-0512-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu G., Wang L.G., He Q.Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 27.Zang C., Schones D.E., Zeng C., Cui K., Zhao K., Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu L.J., Gazin C., Lawson N.D., Pages H., Lin S.M., Lapointe D.S., Green M.R. ChIPpeakAnno: a bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinf. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]