Abstract
The adenomatous polyposis coli (APC) tumour suppressor gene is mutated in about 80% of colorectal cancers (CRC) Brannon et al. (2014) [1]. APC is a large multifunctional protein that regulates many biological functions including Wnt signalling (through the regulation of beta-catenin stability) Reya and Clevers (2005) [2], cell migration Kroboth et al. (2007), Sansom et al. (2004) [3], [4], mitosis Kaplan et al. (2001) [5], cell adhesion Faux et al. (2004), Carothers et al. (2001) [6], [7] and differentiation Sansom et al. (2004) [4]. Although the role of APC in CRC is often described as the deregulation of Wnt signalling, its other biological functions suggest that there are other factors at play that contribute to the onset of adenomas and the progression of CRC upon the truncation of APC. To identify genes and pathways that are dysregulated as a consequence of loss of function of APC, we compared the gene expression profiles of the APC mutated human CRC cell line SW480 following reintroduction of wild-type APC (SW480 + APC) or empty control vector (SW480 + vector control) Faux et al. (2004) . Here we describe the RNA-seq data derived for three biological replicates of parental SW480, SW480 + vector control and SW480 + APC cells, and present the bioinformatics pipeline used to test for differential gene expression and pathway enrichment analysis. A total of 1735 genes showed significant differential expression when APC was restored and were enriched for genes associated with cell polarity, Wnt signalling and the epithelial to mesenchymal transition. There was additional enrichment for genes involved in cell–cell adhesion, cell–matrix junctions, angiogenesis, axon morphogenesis and cell movement. The raw and analysed RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE76307. This dataset is useful for further investigations of the impact of APC mutation on the properties of colorectal cancer cells.
Keywords: Adenomatous polyposis coli, Wnt signalling, Colorectal cancer, RNA-seq, Gene expression
| Specification | |
|---|---|
| Organism/cell line/tissue | SW480 Human epithelial cell line |
| Sex | Male |
| Sequencer or array type | Illumina HiSeq RNASEQ |
| Data format | Raw and analysed |
| Experimental factors | Colorectal cancer cell line with/without functional APC |
| Experimental features | Determine gene expression alterations in the presence/absence of APC gene in the colorectal cancer context. |
| Consent | NA |
| Sample source location | Melbourne, Australia |
1. Direct link to deposited data
2. Experimental design, material and methods
2.1. Cell culture and RNA-extraction
The SW480, SW480 + APC (SW480APC.15) and SW480 + control (SW480control.7) cells have been previously described [6]. The cells were thawed from liquid nitrogen and grown in RPMI supplemented with 0.001% thioglycerol, 1 μg/ml hydrocortisone, 0.025 U/ml insulin, 10% foetal calf serum and 1% penicillin/streptomycin (SW480 cells) plus 1.5 mg/ml G418 (SW480 + APC and SW480 + control cells). Cells were passaged three times before being plated onto 100 mm tissue culture plates at a density of 3.35 × 105 for SW480 and SW480 + control cells and 2 × 105 for SW480 + APC cells, in triplicate. Seventy two hours later RNA was extracted from the cells using the RNAspin Mini RNA isolation kit (Illustra 25-0500-70).
2.2. Sequencing, mapping and normalisation
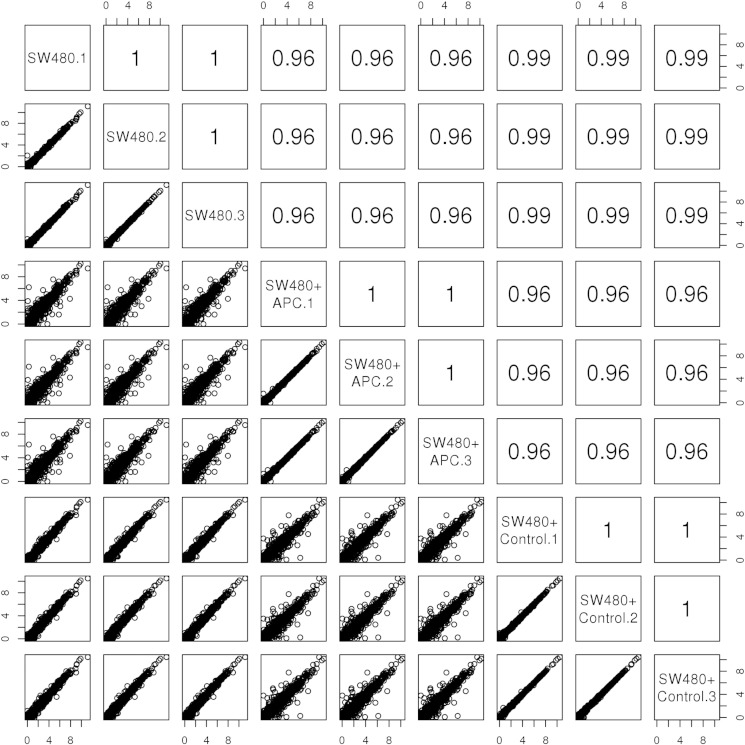
RNA samples were prepared for sequencing using the Illumina TruSeq RNA Library Preparation Kit v2. Libraries were pooled (9 samples per pool) and clustered using the Illumina cBot system with TruSeq SR Cluster Kit v3 reagents, followed by sequencing on the Illumina HiSeq 2000 system with TruSeq SBS Kit v3 reagents (101 cycles) at the Australian Genome Research Facility. Each sample was sequenced to a depth of approximately 20 million reads (see Table 1). Sequencing reads were quality assessed and trimmed for any remaining sequencing adaptor using Trimmomatic (v0.22) [8]; reads smaller than 50 bp were removed. Reads were subsequently aligned to human genome build Hg19 using Tophat (v2.0.6.Linux_x86_64) [9] with parameters -g 1 and the corresponding transcript gtf file. Reads aligning to transcripts were counted and quantified by RPKM using the RNASEQC software [10]. The expression level of the canonical (longest) transcript was taken as representative of gene level expression. Sample expression levels between replicates showed a high correlation (Pearson correlation coefficient > 0.9) (Fig. 1).
Table 1.
Summary sequencing statistics for RNA-seq data.
| Sample | Replicate | Raw reads | Trimmed reads | Aligned reads |
|---|---|---|---|---|
| SW480 + APC.1 | 1 | 22,518,756 | 21,966,108 | 21,050,021 |
| SW480 + APC.2 | 2 | 21,381,045 | 20,806,066 | 19,682,638 |
| SW480 + APC.3 | 3 | 20,825,899 | 20,295,588 | 19,438,311 |
| SW480 + Control.1 | 1 | 22,596,671 | 22,056,529 | 21,039,650 |
| SW480 + Control.2 | 2 | 19,832,005 | 19,354,879 | 18,551,827 |
| SW480 + Control.3 | 3 | 23,352,479 | 22,793,011 | 21,855,257 |
| SW480.1 | 1 | 22,075,530 | 21,528,124 | 20,699,835 |
| SW480.2 | 2 | 22,389,667 | 21,835,786 | 20,798,952 |
| SW480.3 | 3 | 22,563,469 | 22,007,875 | 21,000,302 |
Fig. 1.
Scatter plots of log2 expression values (RPKM) for 1000 randomly selected genes between cell line samples. Pearson correlation coefficients are indicated in the top half of quadrant.
2.3. Differential gene expression
Differential gene expression analysis was conducted using read counts with the Bioconductor edgeR package [11]. Genes represented with a frequency of > 1 read per million in at least one sample were considered as expressed, limiting the analysis to 13,965 genes. The edgeR GLM approach was subsequently applied to determine the differential expression between groups using TMM normalisation [12]. Three comparisons were performed:
-
1.
SW480 + APC v SW480 (restored APC against defective APC).
-
2.
SW480 + control v SW480 (control vector against defective APC).
-
3.
SW480 + APC v SW480 + control (restored APC against control vector).
FDR adjustment was performed to account for multiple testing. Genes with an adjusted two-sided P-value of less than 0.05 and showing a greater than 2-fold change in expression were considered differentially expressed.
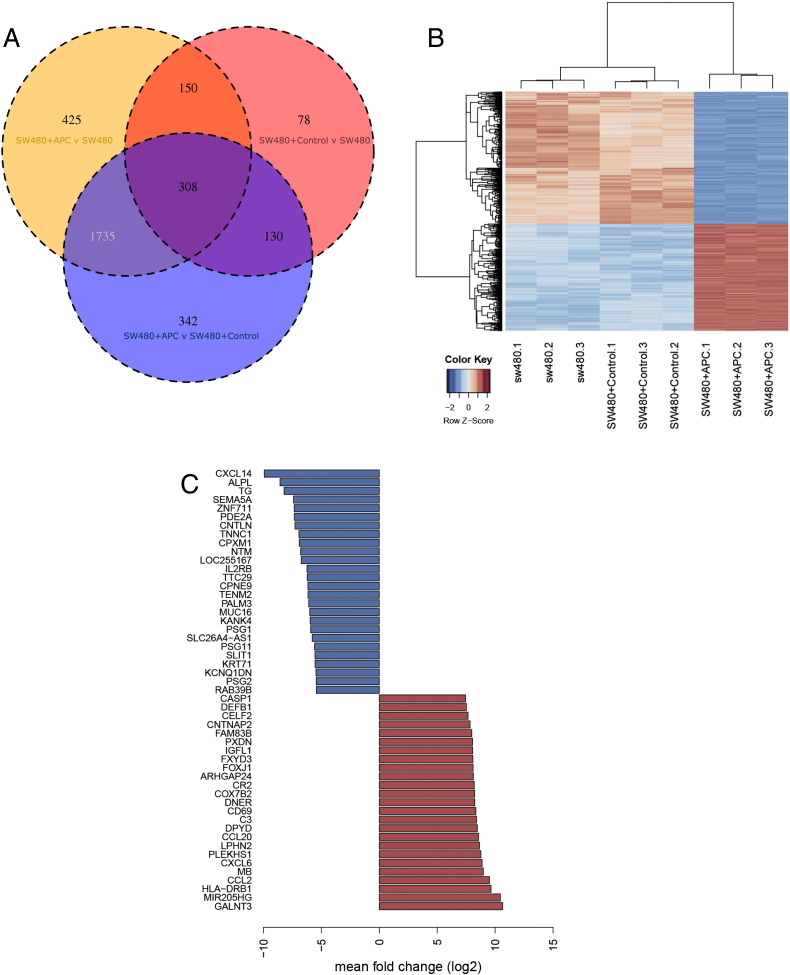
Gene expression changes associated with loss of the APC gene and unrelated to introduction of empty vector were identified by overlapping the differential gene sets from the three comparisons as shown in the Venn diagram in Fig. 2a. We identified a total of 1735 genes specific to APC loss, all of which showed concordant up- or down-regulation in SW480 + APC v SW480 and SW480 + APC v SW480 + control cells, represented as a heatmap in Fig. 2b. The top 25 upregulated and downregulated genes comparing SW480 + APC cells to the average expression score of SW480 and SW480 + control cells are shown as a barplot in Fig. 2c. All differentially expressed genes with their associated log2 fold change values cross-referenced to the Venn diagram are summarised in Supplementary Table 1. Upregulated genes in the SW480 + APC cells include the Rho GTPase-activating protein 24, ARHGAP24, a protein involved in cell polarity, cell morphology and cytoskeletal organisation [13] and the mir-205 host gene, MIR205HG, an established tumour suppressor [14]. Downregulated genes in the SW480 + APC cells include semaphorin 5A, SEMA5A, an axonal regulator molecule associated with tumour growth, invasion and metastasis [15].
Fig. 2.
(a) Venn diagram indicating differentially expressed genes overlapping between the samples. (b) A heatmap displaying the differentially expressed genes in SW480 + APC compared to SW480 and SW480 + control (from the 1735 subset shown in the Venn diagram). The heatmap was drawn using log2 (+ 1 offset) expression values, mean centred and scaled by gene. Gene and sample dendrograms were generated using divisive hierarchical clustering (DIANA). (c) Barplot of top 25 upregulated (red) and downregulated (blue) genes in SW480 + APC compared to SW480 and SW480 control cells. Values plotted are mean (log2) fold change in gene expression of SW480 + APC vs SW480 and SW480 + APC vs SW480 + control.
2.4. Pathway enrichment analysis
Functional category enrichment analysis was performed using DAVID [16] to test gene ontology (GO) categories. Enriched GO categories describing the same function were combined to within a single cluster to reduce redundancy in the results. The enrichment score was calculated as per the DAVID cluster enrichment score; by calculating the mean -log10 GO category P-value within a cluster. A cluster enrichment score threshold of 1.3 was applied, corresponding to a significant cluster enrichment cut-off of P < 0.05. The top 15 highest scoring clusters are shown in Table 2 and include functions important in cell–cell adhesion, cell–matrix junctions, angiogenesis, axon morphogenesis and cell movement. Gene details pertaining to all significant GO clusters are available in Supplementary Table 2.
Table 2.
Significantly enriched pathways from DAVID analysis of the SW480/SW480 + control and SW480 + APC cell lines
| Cluster function | Enrichment score |
|---|---|
| Membrane proteins | 7.621 |
| Cell–matrix junctions | 4.610 |
| Cell–cell junctions | 4.348 |
| Angiogenesis | 5.516 |
| Axon/neuron morphogenesis | 3.684 |
| Cell movement | 2.983 |
| Organogenesis/development | 2.566 |
| Immune system development | 2.404 |
| Wnt signalling | 2.388 |
| Alkaloid responses | 2.327 |
| Carbohydrate binding | 2.269 |
| Muscle development | 2.052 |
| Epithelial cell development | 2.136 |
| Cytoskeleton | 1.956 |
| Epidermal cell development | 1.939 |
The following are the supplementary data related to this article.
List of differential genes comparing SW480 + APC cells against SW480 and SW480 + control cells cross-referenced to the Venn diagram in Fig. 2a.
Gene details for significantly enriched GO annotation clusters identified using DAVID analysis.
Acknowledgements
This work was supported by the Ludwig Institute for Cancer Research and the National Health and Medical Research Council (NH&MRC) Australia Program grant #487922. OMS is also supported by (NHMRC) R.D. Wright Biomedical Career Development Fellowship (APP1062226). The authors would also like to acknowledge that the research was supported by the VLSCI's Life Sciences Computation Centre, an initiative of the Victorian Government, Australia hosted at the University of Melbourne.
References
- 1.Brannon A.R., Vakiani E., Sylvester B.E., Scott S.N., McDermott G., Shah R.H., Kania K., Viale A., Oschwald D.M., Vacic V. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 2014;15:454. doi: 10.1186/s13059-014-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reya T., Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 3.Kroboth K., Newton I.P., Kita K., Dikovskaya D., Zumbrunn J., Waterman-Storer C.M., Nathke I.S. Lack of adenomatous polyposis coli protein correlates with a decrease in cell migration and overall changes in microtubule stability. Mol. Biol. Cell. 2007;18:910–918. doi: 10.1091/mbc.E06-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sansom O.J., Reed K.R., Hayes A.J., Ireland H., Brinkmann H., Newton I.P., Batlle E., Simon-Assmann P., Clevers H., Nathke I.S. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplan K.B., Burds A.A., Swedlow J.R., Bekir S.S., Sorger P.K., Nathke I.S. A role for the adenomatous polyposis coli protein in chromosome segregation. Nat. Cell Biol. 2001;3:429–432. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- 6.Faux M.C., Ross J.L., Meeker C., Johns T., Ji H., Simpson R.J., Layton M.J., Burgess A.W. Restoration of full-length adenomatous polyposis coli (APC) protein in a colon cancer cell line enhances cell adhesion. J. Cell Sci. 2004;117:427–439. doi: 10.1242/jcs.00862. [DOI] [PubMed] [Google Scholar]
- 7.Carothers A.M., Melstrom K.A., Jr., Mueller J.D., Weyant M.J., Bertagnolli M.M. Progressive changes in adherens junction structure during intestinal adenoma formation in Apc mutant mice. J. Biol. Chem. 2001;276:39094–39102. doi: 10.1074/jbc.M103450200. [DOI] [PubMed] [Google Scholar]
- 8.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLuca D.S., Levin J.Z., Sivachenko A., Fennell T., Nazaire M.D., Williams C., Reich M., Winckler W., Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta Y., Hartwig J.H., Stossel T.P. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat. Cell Biol. 2006;8:803–814. doi: 10.1038/ncb1437. [DOI] [PubMed] [Google Scholar]
- 14.Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., Goodall G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 15.Sadanandam A., Varney M.L., Singh S., Ashour A.E., Moniaux N., Deb S., Lele S.M., Batra S.K., Singh R.K. High gene expression of semaphorin 5A in pancreatic cancer is associated with tumor growth, invasion and metastasis. Int. J. Cancer. 2010;127:1373–1383. doi: 10.1002/ijc.25166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of differential genes comparing SW480 + APC cells against SW480 and SW480 + control cells cross-referenced to the Venn diagram in Fig. 2a.
Gene details for significantly enriched GO annotation clusters identified using DAVID analysis.