Abstract
Chromatin and the chromatin modulation machinery not only provide a regulatory matrix for enabling cellular functions such as DNA replication and transcription but also regulate the infectious cycles of many DNA viruses. Elucidation of the components and mechanisms involved in this regulation is providing targets for the development of new antiviral therapies. Initiation of infection by herpes simplex virus (HSV) requires the activity of several cellular chromatin modification enzymes including the histone demethylases LSD1 and the family of JMJD2 proteins that promote transcriptional activation of the initial set of viral genes. Depletion of the JMJD2 members or inhibition of their activity with a new drug results in repression of expression of viral immediate early genes and abrogation of infection. This inhibitor also represses the reactivation of HSV from the latent state in sensory neurons. Like HSV, the β-herpesvirus human cytomegalovirus also requires the activity of LSD1 and the JMJD2s to initiate infection, thus demonstrating the potential of this chromatin-based inhibitor to be useful against a variety of different viruses.
Introduction
Chromatin modulation or epigenetic regulation represents a supra-regulatory overlay beyond the specificity determined by DNA binding factors. Hence, chromatin affects all aspects of cellular function from gene transcription and DNA repair to cellular specification and development. Advancements in the elucidation of the mechanistic control of chromatin have identified protein and enzymatic complexes dedicated to histone modification, recognition, and remodeling that result in regulated access to DNA for other effector components. This complex set of modulation components and molecular machines provides targets for developing new therapeutics. Targeting chromatin components has been clearly demonstrated in oncology (1–3), but this approach also has potential for the treatment of a wide range of diseases, including infectious diseases.
Similar to the control of cellular functions, many virus pathogens that invade the host cell nucleus are subject to the regulatory overlay of chromatin that determines the progression of infection (4). Viruses, ranging from DNA viruses (for example, herpesviruses, adenoviruses, and papillomaviruses) to those whose life cycle depends on a DNA transition state (for example, HIV and other lentiviruses), must contend with and use the host cell chromatin for successful infection (5–15). Notably, chromatin modulation is also a determinant of the regulation of viral latency and recurrent reactivation.
For α-herpesviruses such as herpes simplex virus (HSV) and varicella zoster virus (VZV), infection results in the assembly of nucleosomes on the invading viral genome that bear repressive chromatin marks (for example, histone H3–lysine 9 methylation) (16, 17). Successful expression of the viral immediate early (IE) genes requires cellular and viral transcriptional activators functioning synergistically at IE gene enhancer-promoter domains (15). An important element of this complex regulation is the recruitment of an HCF-1 cellular coactivator complex containing both the histone H3K4 methyltransferase Set1/MLL1 (18, 19) and the histone H3K9 demethylase LSD1 (16) that function to reduce repressive and increase activating chromatin marks. The requirement for these chromatin modulation components opens avenues for the development of new antivirals that target the initiation of infection.
The importance of developing epigenetic-based inhibitors that block the initiation of viral infection is illustrated by the limitations of available antiherpetic therapies that target viral DNA replication during late-stage infection. These therapies, although a significant advance in treatment of herpesvirus-associated disease, are not completely effective at (i) preventing herpetic keratitis (remaining the leading cause of blindness in the United States), (ii) preventing congenital infections that can result in blindness and persistent neurological issues, (iii) preventing damage resulting from immune-mediated inflammation, (iv) avoiding escape of resistant viral mutants, and (v) suppressing subclinical reactivation and viral shedding (20, 21).
As proof of principle for the use of epigenetic inhibitors targeting initiation of viral infection, inhibition of the activity of the histone demethylase LSD1 results in accumulation of repressive chromatin on the HSV IE gene regions, suppression of viral lytic infection, and repression of viral reactivation from latency (16). However, LSD1 only removes H3K9 mono- and dimethylation marks (22, 23), and a second set of demethylases is necessary to remove trimethylation (24, 25). Here, we demonstrate that the JMJD2 H3K9 demethylase family members synergistically function to activate HSV IE gene expression. Depletion of these proteins results in increased repressive chromatin on viral IE genes and suppression of viral infection. Furthermore, a new JMJD2 inhibitor potently blocks both the initiation of viral lytic infection and HSV-1 reactivation in the sensory ganglia of latently infected mice. In addition to the α-herpesvirus HSV-1, the β-herpesvirus human cytomegalovirus (hCMV) also requires both LSD1 and JMJD2 proteins for efficient expression of its IE genes and is similarly repressed by inhibitors of these enzymes. These results demonstrate the potential for targeting specific chromatin modulation components to control multiple viral pathogens at an early stage of infection.
Results
Depletion of JMJD2 family members reduces viral IE gene expression
The genomes of many DNA viruses, including the α-herpesviruses, must both contend with and use the host cell's chromatin modulation machinery. Successful progression of infection requires the HCF-1 transcriptional coactivator complex containing the histone H3K4 methyltransferase Set1 (or MLL1) and the histone H3K9 demethylase LSD1. This complex contains some of the required enzymatic activities to limit the accumulation of repressive chromatin marks on viral genes and convert these marks to those that promote transcriptional activation (for example, H3K4-me3).
In addition to LSD1, the removal of the repressive H3K9 trimethylation (H3K9-me3) requires a cooperating set of JMJD2 family demethylases. To investigate the role of these enzymes in promoting HSV-1 infection, HeLa cells were depleted of JMJD2A, JMJD2B, JMJD2C, or JMJD2D and infected with HSV-1 (Fig. 1, A to D). Depletion resulted in reduced viral IE gene expression (Fig. 1B; ICP4 protein and ICP0 and ICP27 mRNAs) without significant impact on expression of control cellular genes [Fig. 1, B and C; tubulin and TATA binding protein (TBP); S15 and TBP mRNAs]. Similar impacts on viral IE gene expression were also observed after JMJD2 depletion in the fetal lung fibroblast line MRC5 (fig. S1, A to C). In these cells, depletion of JMJD2A, JMJD2B, and JMJD2D resulted in clear reductions in viral IE mRNAs, whereas depletion of JMJD2C resulted in a consistent but less marked decrease.
Fig. 1.
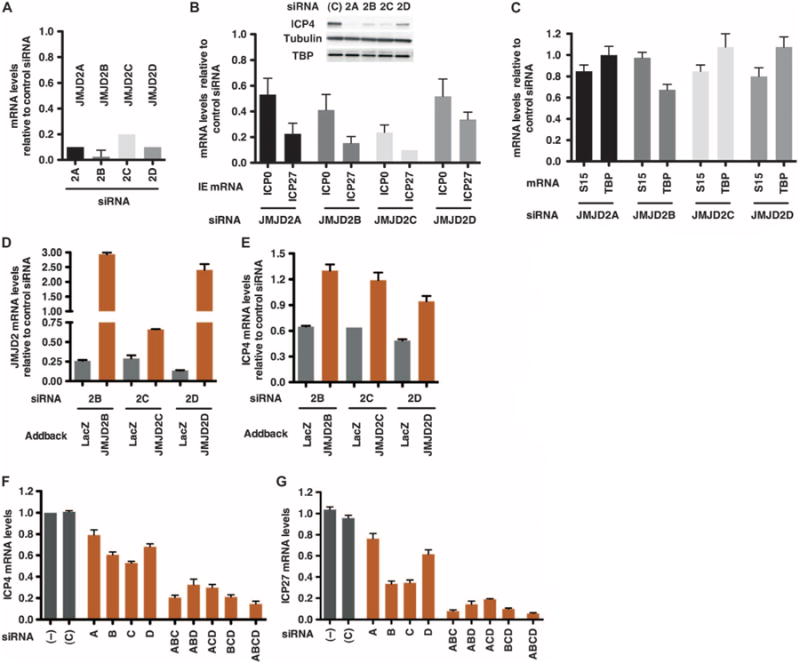
JMJD2 family members synergistically regulate HSV-1 IE gene expression. HeLa cells, depleted of JMJD2A, JMJD2B, JMJD2C, and JMJD2D, were infected with HSV-1 [0.1 plaque-forming unit (PFU) per cell] for 2 hours. mRNA levels are relative to the levels in control siRNA–transfected cells. (A) JMJD2 mRNA levels in the homologous siRNA-transfected cells. (B and C) HSV IE (ICP0 and ICP27) and cellular control (S15 and TBP) mRNA levels in JMJD2-depleted cells. HSV IE ICP4 protein and cellular controls (tubulin, TBP, and Western blot) in cells transfected with control (C) or JMJD2 siRNAs. (D and E) Cells expressing control (LacZ) or siRNA-resistant JMJD2B, JMJD2C, or JMJD2D were transfected with siRNAs to the homologous JMJD2 and infected with HSV-1 (0.1 PFU per cell). mRNA levels of the respective JMJD2 (D) and viral IE ICP4 genes (E) were normalized to cellular S15 and are relative to control siRNA–transfected cells. (F and G) HeLa cells were transfected with 1 nM of control, individual JMJD2, or combinations of JMJD2 siRNAs, followed by infection with HSV-1 (0.1 PFU per cell) for 2 hours. mRNA levels of each JMJD2 (fig. S1, A to D), viral IE (ICP4 and ICP27; F and G), and cellular controls [glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and TBP; fig. S1, F and G] are relative to control-transfected cells. (–), no siRNA; (C), control siRNA. Data are reflective of two independent experiments and represented as means ± SEM.
Recovery of viral IE gene expression by exogenous expression of small interfering RNA–resistant JMJD2s
To verify that small interfering RNA (siRNA) depletion of the JMJD2 members was responsible for the impact on viral IE expression, cell lines expressing control LacZ or siRNA-resistant JMJD2 constructs (JMJD2B, JMJD2C, or JMJD2D) were transfected with the appropriate JMJD2 or control siRNAs, followed by infection with HSV-1. Depletion of endogenous JMJD2B, JMJD2C, or JMJD2D in the control LacZ lines resulted in a decrease in the respective JMJD2 mRNA levels (Fig. 1D) and a reduction in viral IE mRNA levels (ICP4, Fig. 1E). In contrast, exogenous expression of the homologous siRNA-resistant JMJD2 gene resulted in recovery of the respective JMJD2 mRNA levels and restoration of viral IE transcription.
Synergistic impact of combinatorial JMJD2 depletions
Given that the JMJD2 family members are mechanistically redundant (25) and that depletion of each member of the family resulted in decreased HSV IE gene expression, the synergistic impacts on viral IE gene expression were investigated. Depletion of each individual JMJD2 resulted in a modest decrease in JMJD2 mRNA levels (fig. S1D) and a parallel decrease in viral IE gene mRNA (ICP4 and ICP27; Fig. 1, F and G) and protein (ICP4, ICP0, and ICP27; fig. S1E) levels. In turn, depletion of combinations of three JMJD2s further reduced the levels of IE gene mRNAs, whereas depletion of all four JMJD2s resulted in maximal suppression of IE expression. No significant impacts were seen on the levels of the cellular control mRNAs (GAPDH and TBP; fig. S1, F and G). Together, the results demonstrate that the JMJD2 demethylase family is required for efficient viral IE gene expression.
Depletion of JMJD2 family members results in accumulation of repressive chromatin on IE promoters
Upon infection, nucleosomes are assembled on the viral IE promoters bearing repressive histone H3K9-me3, possibly representing an innate cellular defense mechanism to repress genomes of invading viral pathogens. Induction of viral IE genes depends upon the recruitment of the histone demethylase LSD1 to limit the accumulation of this repressive mark. On this basis, the JMJD2 family members would also be required. As shown in Fig. 2, A and B, infection of cells depleted of only JMJD2C resulted in a modest decrease in viral IE gene expression and a correlating increase (∼1.5-fold) in the levels of repressive H3K9-me3 on the viral IE promoters. Depletion of all four JMJD2 proteins (JMJD2A, JMJD2B, JMJD2C, and JMJD2D) resulted in substantial reduction in viral IE proteins and a substantial increase in the levels of repressive marks (two- to fourfold). Additionally, the total levels of histones (represented by histone H3) were elevated, likely as a result of the inability to promote efficient remodeling of the promoter nucleosomes coupled with continued deposition. It is, however, important to note that the fold increase in H3K9-me3 is consistently greater than the increase in total H3 in JMJD2-depleted cells, indicating a higher repressive mark burden on the total nucleosome population. The specificity of the H3K9-me3 chromatin immunoprecipitation (ChIP) is demonstrated by the low level of the repressive mark associated with the active gene (RPL30) and the high (∼30-fold) level associated with the repressed locus (SATa; Fig. 2, C and D).
Fig. 2.
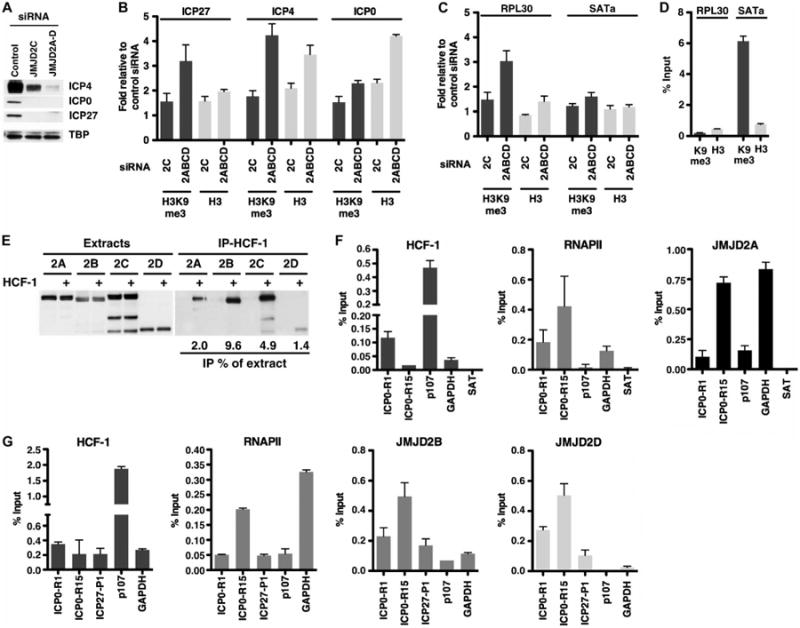
JMJD2 family members interact with HCF-1, are recruited to IE promoters, and are required to prevent the accumulation of repressive chromatin on IE gene promoters. HeLa cells, transfected with siRNAs to JMJD2C or to the four JMJD2s, were infected with HSV-1 (0.1 PFU per cell) for 3 hours. ChIP assays used control immunoglobulin G (IgG), H3K9-me3, or histone H3 antibodies. (A) The levels of viral IE proteins (ICP4, ICP0, and ICP27) and cellular control TBP were monitored by Western blot. (B and C) Levels of H3K9-me3 and histone H3 associated with viral IE promoters (B) and cellular controls (C) relative to cells transfected with control siRNAs. (D) Specificity of the H3K9-me3 ChIP, illustrated by a low level of H3K9-me3 associated with the active RPL30 loci and a high level associated with the repressed SATa region. Data are reflective of two independent experiments. (E) Western blot of extracts and V5 immunoprecipitates (HCF-1) of 293F cells expressing HA-tagged JMJD2 or coexpressing HA-tagged JMJD2 and HCF-1–V5. The amounts of coimmunoprecipitated JMJD2 proteins are represented as percent of extract levels. (F and G) ChIP assays using antibodies to HCF-1, RNAPII, and JMJD2 proteins. Occupancy levels were determined for IE gene promoters (ICP0 and ICP27) and cellular controls (p107, GAPDH, and SATa). Data are means ± SEM.
Association of JMJD2 proteins with the cellular coactivator HCF-1
IE genes are regulated in a complex combinatorial manner that is dependent upon the cellular coactivator HCF-1 (26) in association with numerous transcription factors, coactivators, and chromatin modulation components, including Set1/MLL1 and LSD1 (16, 18). JMJD2 proteins complement the substrate specificity of LSD1 and co-occupy promoters of some androgen-regulated cellular genes (27, 28). To investigate whether the JMJD2s might also be associated with the HCF-1 coactivator, cells were transfected with constructs expressing HA-tagged JMJD2 proteins to express the JMJD2 proteins at equivalent levels and control for differences in antibody affinity, or were cotransfected with a construct expressing a V5-tagged HCF-1. As shown (Fig. 2E), each JMJD2 protein was coimmunoprecipitated with HCF-1 (V5). No JMJD2 protein was detected in V5 immunoprecipitates from control cells expressing only the JMJD2 proteins.
Recruitment of the JMJD2 family members to viral IE gene promoters
Given the association of the JMJD2 proteins with the HCF-1 coactivator and the phenotypic increase in H3K9-me3 associated with the viral IE promoters upon JMJD2 depletion, ChIP assays were done using available ChIP-competent JMJD2A, JMJD2B, and JMJD2D antibodies. As shown (Fig. 2F), viral IE promoter occupancy of the coactivators HCF-1, RNAPII, and JMJD2A could be readily detected. JMJD2A occupancy was highest within the promoter-proximal domain (ICP0-R15) relative to the enhancer domain (ICP0-R1). Similarly, JMJD2B and JMJD2D (Fig. 2G) were also enriched within the promoter-regulatory domains of the IE genes. HCF-1, JMJD2A, and JMJD2B, but not JMJD2D, were detected at the HCF-1–dependent cellular p107 promoter and the GAPDH promoter, indicating promoter-specific recruitment of JMJD2 proteins. In contrast, the negative control SATa exhibited no significant HCF-1, RNAPII, or JMJD2A occupancy.
Impacts of inhibition of the JMJD2 family catalytic activity parallel those of siRNA depletion
The activities of the JMJD2 family members can be inhibited by DMOG (dimethyloxalylglycine) (29). As a corollary to depletion of JMJD2 proteins, human foreskin fibroblast (HFF) cells were treated with DMOG and infected with HSV-1. DMOG treatment resulted in (i) decreased IE gene expression in a dose-dependent manner [median inhibitory concentration (IC50) ∼0.75 mM] (Fig. 3A), (ii) reduced viral yields (3.5 logs at 24 hours after infection, Fig. 3B), and (iii) an increase in the levels of the repressive H3K9-me3 marks (3- to 4-fold) and total histone H3 (1.5- to 2-fold) on IE gene promoters early in infection (Fig. 3C). The impacts on the cellular controls (RPL30 and SATa) mimicked those observed in JMJD2-depleted cells.
Fig. 3.
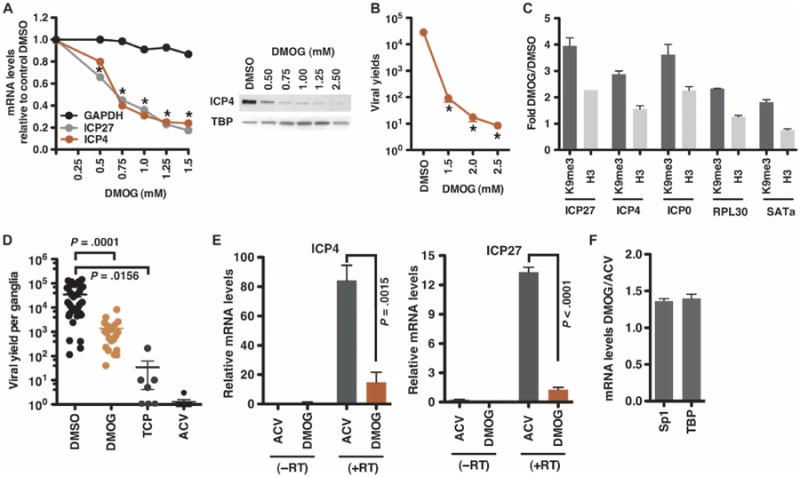
Inhibition of the JMJD2 family catalytic activities results in reduced lytic HSV IE expression and reduced reactivation from latency. (A) HFF cells, treated with the indicated concentrations of the JMJD2 inhibitor DMOG for 4 hours, were infected with HSV-1 (0.1 PFU per cell). Levels of viral and cellular control mRNAs are expressed relative to cells treated with dimethyl sulfoxide (DMSO). *P < 0.0001, two-way analysis of variance (ANOVA) with Dunnett's post hoc test. Viral IE (ICP4) and cellular (TBP) proteins were monitored by Western blot. (B) Viral yields from HFF cells infected with HSV-1 (0.1 PFU per cell) for 24 hours in the presence of DMSO or DMOG. *P < 0.0001, one-way ANOVA with Dunnett's post hoc test. (C) HeLa cells treated with DMSO or 2.5 mM DMOG for 4 hours were infected with HSV-1 (0.1 PFU per cell) for 3 hours. ChIP assays were performed with IgG, H3K9-me3, or histone H3 antibodies. Levels of H3K9-me3 and histone H3 associated with IE promoters and cellular controls are expressed relative to cells treated with DMSO. The data are representative of two independent experiments. (D) HSV-1 latently infected trigeminal ganglia were explanted in the presence of DMSO, 2 mM DMOG, 2 mM TCP, or 100 mM acyclovir (ACV) for 2 days. Viral yields are the titer per ganglia (DMSO versus DMOG: P = 0.0001, Wilcoxon matched-pairs signed rank test; n = 20; DMSO versus TCP: P = 0.0156; n = 7). (E and F) HSV-1 latently infected trigeminal ganglia were explanted in the presence of 100 μM ACV or 2 mM DMOG for 6 hours. The levels of (E) viral IE (ICP4 and ICP27) and (F) cellular (Sp1 and TBP) mRNAs were determined by nested reverse transcription–polymerase chain reaction (RT-PCR) or quantitative RT-PCR, respectively, and are relative to levels in 3T3 cells infected with HSV-1 [6.5 × 10−5 multiplicity of infection (MOIs), 4 hours] (paired two-tailed t test, P values as indicated, n = 4). (−RT), control cDNA synthesis reactions without reverse transcriptase. Data are means ± SEM.
DMOG blocks viral IE gene expression and reactivation from latency
A characteristic of all herpesviruses is the ability to establish lifelong latency after an initial primary infection. For HSV-1, latency is established in the neurons of sensory ganglia and is punctuated by episodes of lytic reactivation (20). Previous studies have demonstrated that the state of the viral chromatin correlates with the state of the genome in the lytic-latency cycle; that is, repressive chromatin marks (H3K9-me2/3 and H3K27-me3) are associated with lytic gene promoters during latency and transition to activating marks (H3K4-me3 and H3K9-acetyl) during lytic reactivation (5, 6, 30–34). In addition to the chromatin state of the viral genome, the subcellular localization of the coactivator HCF-1 also correlates with viral latency reactivation (35, 36). In unstimulated sensory neurons, HCF-1 is sequestered in the cytoplasm but is rapidly transported to the nucleus and recruited to the viral IE gene promoters (37) upon stimulation that results in viral reactivation. Accordingly, it was proposed that HCF-1 and its associated chromatin modulation components are critical to the initiation of viral reactivation from latency.
Therefore, the potential for inhibition of JMJD2 protein activities to block viral reactivation from latency was assessed in paired ganglia explant analyses. Trigeminal ganglia from HSV-1 latently infected mice were bisected, and halves were explanted in the presence of the JMJD2 inhibitor DMOG and control vehicle (DMSO) for 48 hours. As previously demonstrated (16, 38), inhibition of LSD1 activity via tranylcypromine (TCP) or viral DNA replication via the widely used pharmaceutical ACV resulted in a significant decrease in the resulting viral titers (Fig. 3D). In a similar manner, ganglia explanted in the presence of DMOG exhibited a ∼1.5-log (P = 0.0001) decrease in viral yields, indicating that inhibition of the JMJD2 family reduces viral reactivation.
IE gene expression was also analyzed by explanting ganglia for 6 hours in the presence of either DMOG or ACV, which blocks viral DNA replication but does not affect the earlier stages of infection or reactivation [that is, IE or E gene expression (20, 39–41)]. As shown in Fig. 3E, IE mRNA levels were substantially reduced in the presence of DMOG relative to ACV-treated ganglia. In contrast, the levels of control cellular mRNAs (Sp1 and TBP) were not significantly affected (Fig. 3F). Thus, the inhibition of viral reactivation by DMOG is due to inhibition of viral IE gene expression.
New JMJD2 inhibitors block HSV IE gene expression and viral reactivation from latency
Although DMOG reduces IE gene expression both during lytic infection and in ganglia explant reactivation studies, its efficacy is low (IC50 ∼0.75 mM). Therefore, a series of new JMJD2 inhibitors were tested for their ability to inhibit viral IE gene expression. Of these, N-(3-(dimethylamino)propyl)-4-(8-hydroxyquinolin-6-yl)benzamide (designated ML324) was highly effective in reducing IE gene expression with an IC50 ∼10 μM (Fig. 4A). ML324 was ∼75-fold more efficient than DMOG (compare Figs. 3A and 4A). Additionally, ML324 (i) reduced viral yields in a dose-dependent manner (∼5 logs at 50 μM, Fig. 4B); (ii) blocked viral IE gene transcription and reduced viral yields when cells were infected at high MOI (Fig. 4, C and D); and (iii) suppressed IE gene transcription, even when added at times after infection (>50% reduction at 2 hours after infection, Fig. 4E). As further evidence for the efficacy of ML324, cells were infected with HSV to allow for a productive cycle of infection, followed by treatment with DMSO, ACV, or ML324. ML324 markedly suppressed the formation of HSV plaques (Fig. 4F and fig. S2, A and B) and the spread of infection to adjacent cells as visualized by immunofluorescence staining for the HSV DNA replication protein UL29 (Fig. 4G).
Fig. 4.
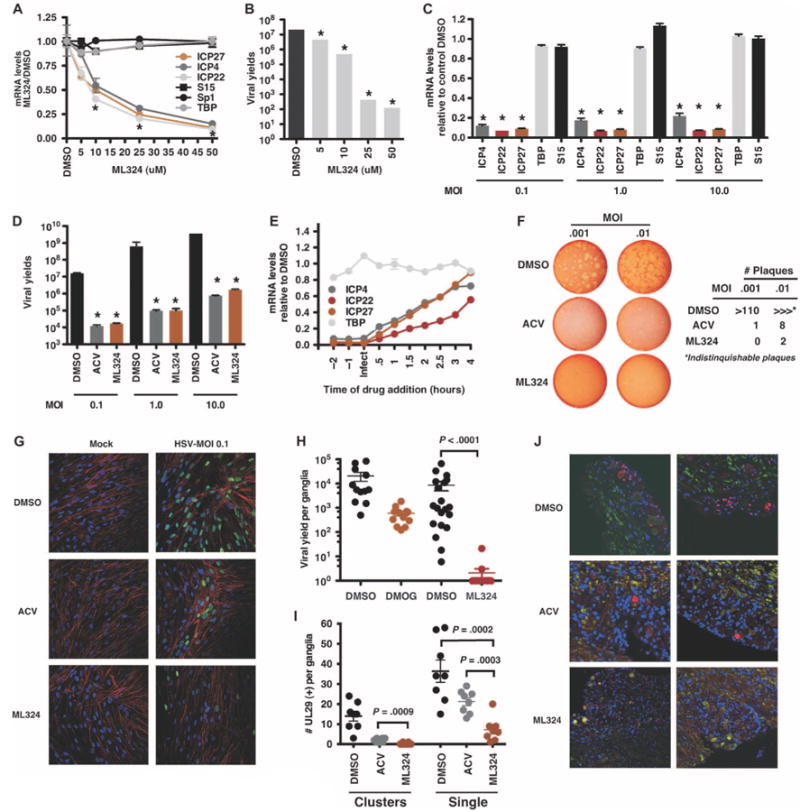
A new JMJD2 inhibitor blocks HSV IE gene expression and reactivation from latency with increased potency. (A) Viral IE (ICP4, ICP27, and ICP22) and control (S15, Sp1, and TBP) mRNA levels in HFF cells treated with DMSO or the indicated concentrations of ML324 for 3 hours and infected with HSV-1 (0.1 PFU per cell) for 3 hours. *P < 0.0001, two-way ANOVA with Tukey's post hoc test. (B) Viral yields from HFF cells treated with DMSO or ML324 for 3 hours and infected with HSV-1 (0.1 PFU per cell) for 24 hours in the presence of the drugs. *P < 0.004, one-way ANOVA with Dunnett's post hoc test. (C and D) MRC-5 cells treated with DMSO or 50 μM ML324 were infected with HSV-1 at the indicated MOI. Viral IE and cellular mRNA levels were quantitated at 2 hours after infection; viral yields were determined at 24 hours after infection. *P < 0.0001, two-way ANOVA with Tukey's post hoc test. (E) MRC-5 cells were treated with DMSO or ML324 at the indicated time relative to infection with HSV-1 (1.0 MOI). Viral and cellular mRNA levels were determined at 4 hours after infection. (F) Plaque assay of Vero cells infected with HSV-1 at the indicated MOI for 12 hours, followed by the addition of DMSO, ACV (100 μM), or ML324 (50 μM) for 48 hours. (G) MRC-5 cells were infected with HSV-1 (0.1 MOI) for 8 hours, followed by the addition of DMSO, ACV (100 μM), or ML324 (50 μM) for 12 hours. Cells were stained with anti-UL29 (green), 4′,6-diamidino-2-phenylindole (DAPI) (blue), and phalloidin-647 (F-actin, red). (H) Viral yields from HSV-1 latently infected trigeminal ganglia explanted in the presence of DMSO, or 2 mM DMOG or 50 μM ML324 for 48 hours (DMSO versus DMOG: P = 0.0015, Wilcoxon matched-pairs signed rank test; n = 12; DMSO versus ML324: P < 0.0001; n = 20). (I and J) Latently infected trigeminal ganglia were explanted in the presence of DMSO, 100 μM ACV, or 50 μM ML324 for 48 hours. Sections were costained with anti-UL29 (red), neurofilament 200 (green), and DAPI (blue) and scored for UL29+ cell clusters (Clusters) and individual neurons (Single) (ACV versus ML324; two-tailed t test; n = 48 sections representing eight ganglia). The total number of UL29+ per ganglia is graphed (I) and representative sections are illustrated (J). Data are means ± SEM.
ML324 was next assessed in the mouse sensory ganglia explant model where it was highly effective at reducing the level of viral reactivation at a significantly lower concentration (4.5-log reduction, 50 μM, P < 0.0001) than DMOG (∼1.5-log reduction, 2 mM, P = 0.0015) (Fig. 4H). Furthermore, ML324 suppressed the primary viral reactivation events as determined by immunofluorescence staining of explanted ganglia sections (Fig. 4, I and J). Ganglia treated with DMSO exhibited viral lytic gene (UL29) expression in isolated neurons and in clusters of neurons and support cells (112 clusters in 48 ganglia sections), illustrating the spread of the reactivated viral infection in the ganglia. ACV-treated ganglia exhibited a reduced number of UL29+ cell clusters (15 clusters in 48 ganglia sections) and isolated neurons (primary reactivating events) relative to DMSO. ML324-treated ganglia exhibited the most significant reduction in UL29+ cell clusters (2 clusters in 48 ganglia sections) and isolated UL29+ neurons relative to both DMSO (P = 0.0002) and ACV (P = 0.0003) (Fig. 4, I and J). Reduction in reactivation by ML324 was not due to the inability of the ganglia to support viral replication as demonstrated by robust viral replication after ML324 withdrawal (fig. S2B). Together, the data indicate that new potent JMJD2 inhibitors may provide a new means for pharmacological control of HSV infection and recurrent disease.
Depletion of HCF-1, LSD1, or the JMJD2 family members represses hCMV IE gene expression
The β-herpesvirus hCMV is an important human pathogen that can cause severe disease and mortality in immunocompromised patients (individuals with HIV-AIDS and organ transplant recipients) and is a major viral cause of birth defects (42, 43). Although members of the herpesvirus groups have unique regulatory mechanisms and cellular tropisms, a common element of these viruses is that they undergo chromatin assembly and modulation, which contributes to their lytic replication and latency reactivation cycles. For hCMV, histone modifications correlate with the ability of the virus to replicate during lytic infection as well as reactivation from latency (7, 44–48).
Given the similarities in the chromatin modulation of hCMV to that of the α-herpesviruses (HSV-1 and VZV), the potential requirements for the coactivator HCF-1 and its associated demethylases (LSD1 and JMJD2 family members) were investigated in a culture model of hCMV infection. Cells depleted of HCF-1 and infected with HSV-1 or hCMV exhibited a decrease in both HSV (∼75% reduction; ICP4 and ICP27) and hCMV IE (∼55% reduction; IE72 and US3) mRNA levels (Fig. 5A). Similarly, depletion of the demethylase LSD1 reduced hCMV IE mRNA levels (Fig. 5, B and C; ∼45% reduction). To investigate the role of the JMJD2 family members, cells were depleted for each JMJD2 family member (Fig. 5, B to D). In contrast to the pattern exhibited by HSV-1 (Fig. 1B), depletion of JMJD2A or JMJD2B had minimal but consistent impact on hCMV IE gene expression (∼20 to 25% reduction, Fig. 5C). Depletion of JMJD2C modestly reduced viral IE expression (∼40 to 45% reduction), whereas depletion of JMJD2D had the most impact (60 to 70% reduction). The data indicate that HSV and hCMV herpesviruses use similar components such as HCF-1, LSD1, and JMJD2 members to regulate the expression of their IE genes.
Fig. 5.
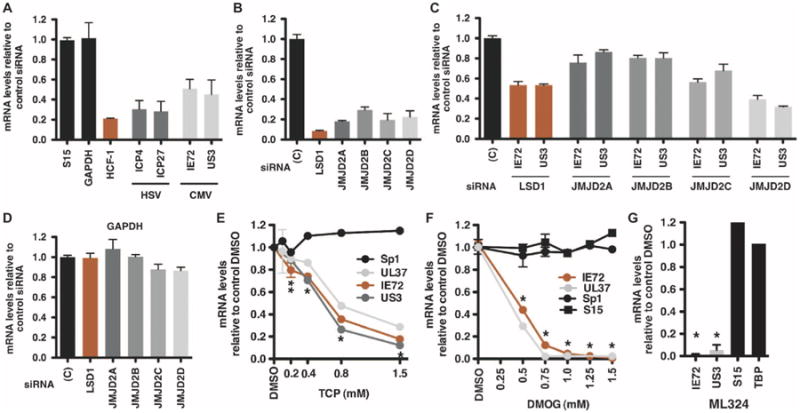
Depletion of the coactivator HCF-1, LSD1, or the members of the JMJD2 family reduces hCMV IE gene expression. (A to D) MRC-5 cells, transfected with control siRNA or siRNAs to HCF-1 (A), LSD1, or the JMJD2 members (B to D), were infected with HSV-1 or hCMV (0.1 PFU per cell) for 4 hours. JMJD2 and LSD1 (B), viral IE (HSV-1 ICP4, ICP27) (A), (hCMV IE72, US3) (C), and control cellular mRNA levels (A and D) were normalized to cellular control S15 mRNA levels and expressed as the ratios to cells transfected with control siRNA. (E and F) MRC-5 cells were treated with DMSO or the indicated concentrations of LSD1 inhibitor TCP or DMOG for 3 hours, followed by infection with hCMV (0.1 PFU per cell) for 5 hours. The levels of viral IE (IE72, UL37, and US3) and control cellular (Sp1 and S15) mRNAs are expressed relative to cells treated with DMSO. *P < 0.0001, **P < 0.002, two-way ANOVA with Dunnett's post hoc test. (G) MRC-5 cells were treated with DMSO or 50 μM ML324 for 3 hours, followed by infection with hCMV (0.1 PFU per cell) for 5 hours. mRNA levels of viral and cellular controls are expressed relative to cells treated with DMSO. Data are means ± SEM. *P < 0.0001, one-way ANOVA with Tukey's post hoc test.
In these studies, it was noted that although HSV infection does not differentially affect the levels of JMJD2 mRNAs, hCMV selectively stimulates the accumulation of JMJD2D mRNA (fig. S3). The varied impacts of individual JMJD2 depletion on hCMV viral IE gene expression and the selective induction of JMJD2D mRNA during infection suggest that this virus may manipulate the levels of the specific JMJD2 family member most critical to expression of its IE genes. Thus, although HSV and hCMV use common components, the viruses may selectively use distinct members of the JMJD2 family.
Inhibition of LSD1 or JMJD2 activities blocks hCMV IE gene expression
As further confirmation of the requirement for LSD1 and the JMJD2(s) for hCMV IE gene expression, cells were treated with TCP (LSD1 inhibitor) or the JMJD2 inhibitor DMOG or ML324. Treatment of MRC5 cells with TCP or DMOG resulted in dose-dependent decreases in the expression of the hCMV IE genes (UL37, UL72, and US3) without impact on the expression of the cellular control Sp1 (Fig. 5, E to G). Notably, the new JMJD2 inhibitor ML324 potently inhibited hCMV IE gene expression (Fig. 5G).
Discussion
Research on cellular gene transcription, DNA replication repair, and cell specification has produced a growing body of data to support the significant role of chromatin and chromatin modulation machinery as a regulatory matrix that controls the ability of effector proteins to access the genome. Recently, the importance of this matrix has been extended to the regulation of transcription and replication of many DNA viruses that are also subject to the assembly, modification, and remodeling of nucleosomes. This regulation controls not only the viral lytic replication cycle but also viral latency and reactivation, thus opening up avenues for the development of new antivirals.
The data presented here argue that members of the JMJD2 family of chromatin modification enzymes are required for both HSV and hCMV IE gene expression. For HSV, inhibition of the JMJD2 activities (i) suppresses the initiation of infection, even at high MOI; (ii) blocks the spread of infection to adjacent cells, even when added after primary infection; and (iii) results in suppression of reactivation in latently infected sensory neurons via inhibition of the initiation stage (IE gene expression). Thus, the JMJD2 histone H3K9 demethylases are factors required to modulate chromatin and activate IE transcription during the HSV lytic as well as latency reactivation stages of infection. Because inhibition of the JMJD2 family activities also suppresses the β-herpesvirus hCMV IE gene expression, these enzymes represent epigenetic targets for suppression of both viruses.
In addition to oral and genital lesions, HSV is responsible for congenital infections that result in persistent neurological issues and herpetic keratitis, a leading cause of blindness in the United States (20, 21). hCMV represents an important clinical challenge in immunocompromised individuals during solid organ transplant and remains the leading cause of birth defects (42, 43). Although DNA replication inhibitors (for example, ACV and derivatives) are widely used to treat herpesvirus infections, these pharmaceuticals (i) target late-stage infection (replication), allowing for escape mutants and the rise of resistant strains (20); (ii) do not block the expression of early viral gene products that alter the cellular state and, for some viruses, are oncogenic; and (iii) do not suppress infection before the production of viral proteins that elicit immune-mediated inflammation and tissue damage (20, 21, 42, 43), which is an important component of diseases such as HSV keratitis and hCMV complications in solid organ transplants. Notably, these compounds do not efficiently suppress subclinical reactivation and viral shedding, an important mode of transmission (20).
Thus, there remains a clear need to develop new antivirals. The requirement for common components, such as JMJD2 demethylases, provides opportunity for the development of broad-spectrum epigenetic antivirals that target initiation of infection. Such inhibitors have potential to reduce spontaneous reactivation via inhibition of the activities that modulate the level of viral genome repression and reduce the inflammatory responses resulting from reactivation that mediate additional tissue damage. As proof of principle, MAOI (monoamine oxidase inhibitor)–based LSD1 inhibitors suppress HSV primary infection and function synergistically with ACV in a mouse model. However, these inhibitors (MAOIs for LSD1 and DMOG for JMJD2s) have low specificity and potency. The development of potent cell-permeable inhibitors, such as the JMJD2 inhibitor ML324 described here, represent a new set of antivirals.
Most importantly, HSV and other viruses that are subject to regulation by assembled chromatin use some common mechanisms and components. Inhibition of the activity of either LSD1 or the JMJD2 demethylases suppresses initial stages in both the α-herpesvirus HSV and the β-herpesvirus hCMV infection. Thus, although the regulatory pathways of HSV and hCMV have unique aspects, both viruses can be pharmacologically controlled via inhibition of these common components. It is worth noting that, in addition to LSD1 and the JMJD2(s), other epigenetic activities must be required for efficient viral lytic infection and reactivation. Studies demonstrating the presence of significant levels of both H3K9 and H3K27 repressive methylation on the HSV genome during latency (5, 6, 31, 32) would suggest that inhibition of the enzymatic components that reverse H3K27-me3 would also suppress reactivation. Continued elucidation of the mechanisms and components involved in epigenetic regulation of viral pathogens will lead to additional targets for antiviral development.
Further studies will be required to address tolerance for long-term therapies and the potential to maintain suppression of latent virus. Latent viruses are already subject to epigenetic repression, and the potential impacts of long-term epigenetic therapy for suppression of viral reactivation will need to be assessed in vivo relative to the levels required to maintain that suppression. Additionally, the identification of specific components and the development of target-specific inhibitors (for example, JMJD2D for hCMV) may be important for reducing off-target impacts. Given these considerations, epigenetic approaches have advantages in limiting viral infection-reactivation in clinical situations where the present inhibitors have limitations.
There is intensive focus on the development of inhibitors of epigenetic components for the treatment of cancers and other diseases. The study presented here demonstrates that epigenetic inhibitors can also function as antiviral therapeutic agents. As a different example of epigenetic antiviral approaches, latent pools of the HIV are maintained by specific histone deacetylase (HDAC) complexes and are thus refractory to highly active antiretroviral therapy. Disruption of viral latency using HDAC inhibitors is a new approach to eradicate latent pools of HIV (49). Thus, the continued investigations of the mechanisms involved in chromatin regulation of multiple classes of viruses have the potential to identify specific epigenetic targets as well as broaden the potential use of compounds currently in development.
Materials and Methods
Cells and viruses
HeLa, human embryonic kidney (HEK) 293, MRC-5, Vero, and TERT-immortalized HFF cell lines and viral strains HSV-1-17, HSV-1-F, and hCMV Towne RC256 were propagated according to standard procedures.
Antibodies, primers, and siRNAs
Antibodies, siRNAs, and primers sequences are listed in table S1.
Quantitative RT-PCR
Complementary DNA (cDNA) was produced from total RNA as described (16) and quantitated using an ABI 7900HT (ABI SDS 2.3 software). The levels of LSD1 and JMJD2 mRNAs in selected tissues and cell types are shown in fig. S4.
Inhibitors
Cells were treated with DMSO, LSD1 inhibitor (TCP, Sigma P8511), or JMJD2 inhibitors [DMOG (Cayman 71210) and ML324] and infected with HSV-1 or hCMV as described in the figure legends. The structure of ML324, developed based on previously reported studies (50), is shown in fig. S5.
siRNA depletion-addbacks
Primers used to produce siRNA-resistant JMJD2 constructs are listed in table S1. HeLa cells were transfected with control pcDNA3-LacZ or pcDNA3-siRNA–resistant JMJD2 expression constructs and subsequently depleted for the endogenous JMJD2s as detailed in Supplementary Materials and Methods.
Western blots
Western blots were done with the antibodies in table S1. Blots were quantitated with a Kodak 4000MM Image Station.
ChIP assays
Histone ChIP assays were done essentially as previously described (16, 26). JMJD2, HCF-1, and RNAPII ChIP assays were done under previously described conditions (51). Details are in Supplementary Materials and Methods.
Coimmunoprecipitations
V5 immunoprecipitations from 293F cell nuclear extracts were done as previously described (52) and as detailed in Supplementary Materials and Methods.
Immunofluorescence
The expression of UL29 in HSV-1–infected MRC-5 and Vero cells was detected with standard immunofluorescence methods. For reactivation, ganglia were explanted in the presence or absence of the indicated compound for 48 hours, fixed with 4% paraformaldehyde, and paraffin-embedded. Sections were subjected to antigen retrieval and stained with the indicated antibodies as detailed in Supplementary Materials and Methods.
Latently infected trigeminal ganglia
BALB/c mice were infected with 5 × 105 PFU HSV-1 (strain F) per eye. Trigeminal ganglia of latently infected mice were harvested 30 days after clearance of the primary infection. Viral yields from paired ganglia analyses and from drug reversals were done as described (16). For mRNA levels, cDNA was prepared from paired latently infected ganglia explanted in the presence of ACV or DMOG for 6 hours and quantitated with the primer sets listed in table S1. Animal care and handling was done in accordance with the National Institutes of Health (NIH) Animal Care and Use Guidelines and the National Institute of Allergy and Infectious Diseases (NIAID) Animal Care and Use Committee.
Statistical analyses
Statistical comparisons were made with paired two-tailed t test (ganglia mRNA), unpaired two-tailed t test (ganglia reactivation-immunofluorescence assay), and Wilcoxon signed rank tests (ganglia viral yields) with a statistical significance of <0.05 (Prism V5.0a). Multiple sample comparisons were done with one- or two-way ANOVA followed by Dunnett's or Tukey's post hoc tests as indicated in the appropriate figure legend. Details of statistical analyses are given in Supplementary Materials and Methods.
Supplementary Material
Fig. S1. Depletion of JMJD2 members resulted in decreased HSV-1 IE mRNAs.
Fig. S2. ML324-mediated inhibition of HSV lytic infection and reactivation from latency.
Fig. S3. Selective induction of JMJD2D transcription upon infection with hCMV.
Fig. S4. Levels of JMJD2 mRNAs in selected tissues and cell types.
Fig. S5. Structure and characteristics of ML324.
Table S1. Antibodies and primers.
Acknowledgments
We thank T. Pierson and members of the Molecular Genetics Section for discussions; J. Skinner (Laboratory of Immunogenetics, NIAID) for expert statistical analyses; NIAID Bld33 Animal Facility staff; W. Ruyechan for UL29 antiseras; C. J. Schofield, U. Oppermann, O. N. F. King, A. Kawamura, A. Tumber, and N. R. Rose (University of Oxford) for assistance with initial ML324 biochemical characterization.
Funding: Supported by the Division of Intramural Research, NIAID, NIH (T.M.K.) and the Molecular Libraries Initiative of the NIH Roadmap for Medical Research, NIH U54MH084681 (G.R., A.J., A.S., and D.J.M.).
Footnotes
Author contributions: Y.L. performed ChIP, qPCR, and animal reactivation studies; J.L.V. performed ChIP, coimmunoprecipitations, and qPCR analyses; J.H.A. performed immunofluorescence assays; G.R., A.J., A.S., and D.J.M. provided the JMJD2 inhibitors and controls; T.M.K. designed the study, performed animal reactivation studies, and wrote the paper. All authors discussed and commented on the manuscript.
Competing interests: The NIH has the following patent applications: (i) Methods of preventing or treating viral infection or reactivation from latency in a host using inhibitors of the LSD1 protein (T.M.K. and Y.L.; U.S. patent application no. 61/083,304; International patent application no. PCT/US2009/051557); (ii) Method of preventing or treating viral infection via inhibition of the JMJD2 proteins (T.M.K., Y.L., and J.L.V.; U.S. patent application no. 61/366,563).
Data and materials availability: Requests for materials should be addressed to T.M.K.. All material requests require a Material Transfer Agreement or Letter of Agreement with the NIAID and/or the National Center for Advancing Translational Sciences (ML324).
References and Notes
- 1.Best JD, Carey N. Epigenetic opportunities and challenges in cancer. Drug Discov Today. 2010;15:65–70. doi: 10.1016/j.drudis.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Copeland RA, Olhava EJ, Scott MP. Targeting epigenetic enzymes for drug discovery. Curr Opin Chem Biol. 2010;14:505–510. doi: 10.1016/j.cbpa.2010.06.174. [DOI] [PubMed] [Google Scholar]
- 3.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lieberman PM. Chromatin regulation of virus infection. Trends Microbiol. 2006;14:132–140. doi: 10.1016/j.tim.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Bloom DC, Giordani NV, Kwiatkowski DL. Epigenetic regulation of latent HSV-1 gene expression. Biochim Biophys Acta. 2010;1799:246–256. doi: 10.1016/j.bbagrm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol. 2008;6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- 7.Sinclair J. Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection. Biochim Biophys Acta. 2010;1799:286–295. doi: 10.1016/j.bbagrm.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Easley R, Van Duyne R, Coley W, Guendel I, Dadgar S, Kehn-Hall K, Kashanchi F. Chromatin dynamics associated with HIV-1 Tat-activated transcription. Biochim Biophys Acta. 2010;1799:275–285. doi: 10.1016/j.bbagrm.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giberson AN, Davidson AR, Parks RJ. Chromatin structure of adenovirus DNA throughout infection. Nucleic Acids Res. 2012;40:2369–2376. doi: 10.1093/nar/gkr1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Günther T, Grundhoff A. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 2010;6:e1000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mok HP, Lever AM. Chromatin, gene silencing and HIV latency. Genome Biol. 2007;8:228. doi: 10.1186/gb-2007-8-11-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takacs M, Banati F, Koroknai A, Segesdi J, Salamon D, Wolf H, Niller HH, Minarovits J. Epigenetic regulation of latent Epstein–Barr virus promoters. Biochim Biophys Acta. 2010;1799:228–235. doi: 10.1016/j.bbagrm.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Tempera I, Lieberman PM. Chromatin organization of gammaherpesvirus latent genomes. Biochim Biophys Acta. 2010;1799:236–245. doi: 10.1016/j.bbagrm.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.You J. Papillomavirus interaction with cellular chromatin. Biochim Biophys Acta. 2010;1799:192–199. doi: 10.1016/j.bbagrm.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kristie TM, Liang Y, Vogel JL. Control of α-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities. Biochim Biophys Acta. 2010;1799:257–265. doi: 10.1016/j.bbagrm.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Inhibition of the histone demethylase LSD1 blocks α-herpesvirus lytic replication and reactivation from latency. Nat Med. 2009;15:1312–1317. doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva L, Cliffe A, Chang L, Knipe DM. Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog. 2008;4:e1000071. doi: 10.1371/journal.ppat.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narayanan A, Ruyechan WT, Kristie TM. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc Natl Acad Sci U S A. 2007;104:10835–10840. doi: 10.1073/pnas.0704351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roizman B, Knipe DM, Whitley RJ. In: Fields Virology. 5. Knipe DM, Howley PM, editors. Vol. 2. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2501–2601. [Google Scholar]
- 21.Whitley R, Kimberlin DW, Prober CG. In: Human Herpesviruses Biology, Therapy, and Immunoprophylaxis. Arvin A, Whitley R, editors. Cambridge University Press; Cambridge: 2007. pp. 589–601. [PubMed] [Google Scholar]
- 22.Culhane JC, Cole PA. LSD1 and the chemistry of histone demethylation. Curr Opin Chem Biol. 2007;11:561–568. doi: 10.1016/j.cbpa.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 24.Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 25.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 26.Narayanan A, Nogueira ML, Ruyechan WT, Kristie TM. Combinatorial transcription of herpes simplex virus and varicella zoster virus immediate early genes is strictly determined by the cellular coactivator HCF-1. J Biol Chem. 2005;280:1369–1375. doi: 10.1074/jbc.M410178200. [DOI] [PubMed] [Google Scholar]
- 27.Shin S, Janknecht R. Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochem Biophys Res Commun. 2007;359:742–746. doi: 10.1016/j.bbrc.2007.05.179. [DOI] [PubMed] [Google Scholar]
- 28.Wissmann M, Yin N, Müller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Günther T, Buettner R, Metzger E, Schüle R. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 29.Hamada S, Kim TD, Suzuki T, Itoh Y, Tsumoto H, Nakagawa H, Janknecht R, Miyata N. Synthesis and activity of N-oxalylglycine and its derivatives as Jumonji C-domain-containing histone lysine demethylase inhibitors. Bioorg Med Chem Lett. 2009;19:2852–2855. doi: 10.1016/j.bmcl.2009.03.098. [DOI] [PubMed] [Google Scholar]
- 30.Amelio AL, Giordani NV, Kubat NJ, O'Neil JE, Bloom DC. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times post-explant. J Virol. 2006;80:2063–2068. doi: 10.1128/JVI.80.4.2063-2068.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cliffe AR, Garber DA, Knipe DM. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol. 2009;83:8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwiatkowski DL, Thompson HW, Bloom DC. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J Virol. 2009;83:8173–8181. doi: 10.1128/JVI.00686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neumann DM, Bhattacharjee PS, Giordani NV, Bloom DC, Hill JM. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J Virol. 2007;81:13248–13253. doi: 10.1128/JVI.01569-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A. 2005;102:16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolb G, Kristie TM. Association of the cellular coactivator HCF-1 with the Golgi apparatus in sensory neurons. J Virol. 2008;82:9555–9563. doi: 10.1128/JVI.01174-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kristie TM, Vogel JL, Sears AE. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc Natl Acad Sci U S A. 1999;96:1229–1233. doi: 10.1073/pnas.96.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitlow Z, Kristie TM. Recruitment of the transcriptional coactivator HCF-1 to viral immediate-early promoters during initiation of reactivation from latency of herpes simplex virus type 1. J Virol. 2009;83:9591–9595. doi: 10.1128/JVI.01115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt DM, McCafferty DG. trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry. 2007;46:4408–4416. doi: 10.1021/bi0618621. [DOI] [PubMed] [Google Scholar]
- 39.Coen DM, Richman DD. In: Fields Virology. 5. Knipe DM, Howley PM, editors. Vol. 1. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 447–485. [Google Scholar]
- 40.Nichol PF, Chang JY, Johnson EM, Jr, Olivo PD. Herpes simplex virus gene expression in neurons: Viral DNA synthesis is a critical regulatory event in the branch point between the lytic and latent pathways. J Virol. 1996;70:5476–5486. doi: 10.1128/jvi.70.8.5476-5486.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pesola JM, Zhu J, Knipe DM, Coen DM. Herpes simplex virus 1 immediate-early and early gene expression during reactivation from latency under conditions that prevent infectious virus production. J Virol. 2005;79:14516–14525. doi: 10.1128/JVI.79.23.14516-14525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Britt W. In: Human Herpesviruses Biology, Therapy, and Immunoprophylaxis. Arvin A, Whitley R, editors. Cambridge University Press; Cambridge: 2007. pp. 737–764. [PubMed] [Google Scholar]
- 43.Mocarski ES, Shenk T, Pass RF. In: Fields Virology. Knipe DM, Howley PM, editors. Vol. 5. Vol. 2. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2701–2772. [Google Scholar]
- 44.Cuevas-Bennett C, Shenk T. Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. J Virol. 2008;82:9525–9536. doi: 10.1128/JVI.00946-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ioudinkova E, Arcangeletti MC, Rynditch A, De Conto F, Motta F, Covan S, Pinardi F, Razin SV, Chezzi C. Control of human cytomegalovirus gene expression by differential histone modifications during lytic and latent infection of a monocytic cell line. Gene. 2006;384:120–128. doi: 10.1016/j.gene.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 46.Nitzsche A, Paulus C, Nevels M. Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells. J Virol. 2008;82:11167–11180. doi: 10.1128/JVI.01218-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reeves MB. Chromatin-mediated regulation of cytomegalovirus gene expression. Virus Res. 2011;157:134–143. doi: 10.1016/j.virusres.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeves MB, Lehner PJ, Sissons JG, Sinclair JH. An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J Gen Virol. 2005;86:2949–2954. doi: 10.1099/vir.0.81161-0. [DOI] [PubMed] [Google Scholar]
- 49.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.King ON, Li XS, Sakurai M, Kawamura A, Rose NR, Ng SS, Quinn AM, Rai G, Mott BT, Beswick P, Klose RJ, Oppermann U, Jadhav A, Heightman TD, Maloney DJ, Schofield CJ, Simeonov A. Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors. PLoS One. 2010;5:e15535. doi: 10.1371/journal.pone.0015535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogel JL, Kristie TM. Site-specific proteolysis of the transcriptional coactivator HCF-1 can regulate its interaction with protein cofactors. Proc Natl Acad Sci U S A. 2006;103:6817–6822. doi: 10.1073/pnas.0602109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Depletion of JMJD2 members resulted in decreased HSV-1 IE mRNAs.
Fig. S2. ML324-mediated inhibition of HSV lytic infection and reactivation from latency.
Fig. S3. Selective induction of JMJD2D transcription upon infection with hCMV.
Fig. S4. Levels of JMJD2 mRNAs in selected tissues and cell types.
Fig. S5. Structure and characteristics of ML324.
Table S1. Antibodies and primers.


