Abstract
Cardiovascular and renal diseases are associated with many risk factors, of which hypertension is one of the most prevalent. Worldwide, blood pressure control is only achieved in ~50% of those treated for hypertension, despite the availability of a considerable number of antihypertensive drugs from different pharmacological classes. Although many reasons exist for poor blood pressure control, a likely contributor is the inability to predict to which antihypertensive drug an individual is most likely to respond. Hypertension pharmacogenomics and other ‘omics’ technologies have the potential to identify genetic signals that are predictive of response or adverse outcome to particular drugs, and guide selection of hypertension treatment for a given individual. Continued research in this field will enhance our understanding of how to maximally deploy the various antihypertensive drug classes to optimize blood pressure response at the individual level. This Review summarizes the available literature on the most convincing genetic signals associated with antihypertensive drug responses and adverse cardiovascular outcomes. Future research in this area will be facilitated by enhancing collaboration between research groups through consortia such as the International Consortium for Antihypertensives Pharmacogenomics Studies, with the goal of translating replicated findings into clinical implementation.
Hypertension affects an estimated 80 million Americans and 1 billion adults globally1. It is the most prevalent modifiable risk factor for global disease burden among men and women, regardless of ancestry, geographic region or income, and contributes substantially to disability and mortality2–5. Available evidence clearly demonstrates that use of antihypertensive drugs to reduce chronically elevated blood pressure is associated with a significant reduction in mortality and other adverse sequelae, including stroke, coronary heart disease and chronic kidney disease6–10.
Hypertension is the most common chronic condition for which medications are prescribed5, and antihypertensive agents are among the most commonly prescribed drugs in the USA. In patients receiving antihypertensive medications, blood pressure is monitored in response to treatment and dosage adjusted on the basis of blood pressure response and adverse effects. End-organ damage that can result from inadequate blood pressure control leads to premature mortality and disability and is associated with marked economic and societal costs11–13. African Americans are disproportionately affected by hypertension and its complications, and are more likely than other US populations to have uncontrolled hypertension14 and to need multiple drugs to achieve blood pressure control15.
Although the 2014 JNC 8 panel recommendations for the management of hypertension16 suggested raising the blood pressure target in older patients with hypertension, most clinicians still use a blood pressure target of <140/90 mmHg in most patients younger than 80 years of age17–19. This target has been recommended for decades on the basis of a multitude of clinical trials and many public health campaigns focused on improving hypertension awareness, treatment and control. Despite these public awareness campaigns and the availability of countless blood-pressure-lowering drugs within multiple drug classes, only about half of patients with treated hypertension achieve appropriate blood pressure control2.
This Review describes some of the reasons for the elusive nature of blood pressure control and discusses the potential for personalized antihypertensive drug therapy through a pharmacogenomics approach. Such a personalized approach could ultimately improve overall blood pressure control and as a result, reduce the prevalence of hypertension-related adverse outcomes.
Key points.
Hypertension is an important, modifiable risk factor for cardiovascular and kidney disease
Rates of blood pressure control are substantially below the desired levels globally, with many factors contributing to poor blood pressure control
Pharmacogenomics and other ‘omics’ approaches could help to identify useful biomarkers for a more personalized or precision approach to antihypertensive treatment strategies
Validating and replicating antihypertensive pharmacogenomics signals will require large sample sizes and will probably not yield a single signal with a large effect size, but rather multiple signals with small effect sizes
As technology continues to evolve and genetic and other ‘omics’ data become available from collaborative studies, identification of biomarkers of blood pressure response might be possible
Personalized treatment approaches
The poor global rates of blood pressure control probably have many causes, including poor adherence to therapy, insufficient effort on the part of the clinician to achieve blood pressure control in their patients, and poor response to the prescribed antihypertensive therapy. An important element underlying poor responses to prescribed treatments is the relative lack of predictive approaches for selecting the most appropriate therapy for individual patients.
Clinical predictors of response
A number of studies have attempted to identify clinical factors that can be used to guide treatment decisions. The influence of age and ancestry on response to first-line antihypertensive therapy has been recognized since the 1980s, particularly stemming from the results of the Veterans Affairs Cooperative Studies20. These differences in response to treatment are often described by categorizing first line antihypertensive drug classes as ABCD, referring to angiotensin-converting enzyme (ACE) inhibitors and/or angiotensin-receptor blockers (ARBs; A), β-blockers (B), calcium-channel blockers (C) and diuretics (D), where responses to classes A and B tend to be similar, as are responses to classes C and D21. With regard to age and ancestry, patients who are young and of European ancestry tend to respond better to classes A and B, whereas older hypertensive patients and those of African ancestry are more likely to respond to classes C and D20,22,23.
Plasma renin activity has long been touted as an important biomarker for predicting blood pressure response to antihypertensive therapy. Moreover, the differences in blood pressure response to these drugs noted between young and elderly patients, and between patients of European and African ancestry, could be caused in large part by differences in plasma renin activity24. In a cohort of white and African American patients with hypertension who were treated with either hydrochlorothiazide, atenolol or a combination of the two, plasma renin activity was the only biochemical parameter that consistently significantly contributed to the prediction of systolic and diastolic response to monotherapy or combination therapy25. Beyond age, ancestry and plasma renin activity, limited data exist on clinical factors or biomarkers that are predictive of response to antihypertensive drugs. Although the age and ancestry of a patient are often considered in selecting antihypertensive therapy, plasma renin activity is rarely measured to guide treatment decisions in the clinical setting. Moreover, although plasma renin activity is associated with blood pressure response to treatment, additional factors beyond blood pressure lowering might influence the ability of a drug to prevent cardiovascular events, stroke and kidney failure.
The potential of pharmacogenomics
Since the completion of the Human Genome Project in 2001 there has been substantial interest in identifying genetic biomarkers that help to define an individual’s risk of disease and response to medications. The use of genetic biomarkers to assess response to medications is called pharmacogenomics and work in the pharmacogenomics of hypertension medications is driven largely by the quest to identify genetic or other ‘omic’ biomarkers that could be used to personalize treatments for affected patients.
Technologies for genotyping and genome sequencing have advanced at an unprecedented pace, such that it is now possible to sequence the human genome for about US$1,000, and in hours to days, in contrast to the >10 years and >$2 billion that was spent sequencing the first human genome. These technological advances have led to the projection that in the next decade it will be possible to obtain genome-wide genotype, whole exome or whole genome sequence data for any patient. The considerable challenges associated with these platforms will relate not to the generation of data, but rather to the interpretation and storage of the data, and to defining the genetic markers that are sufficiently predictive to be useful in the clinical setting.
Many arguments exist as to why a pharmacogenomics approach may be useful in patients with hypertension. First, as noted above, rates of blood pressure control are far from optimal, and the ability to identify a priori a therapy or combination of therapies that might be most effective for an individual has potential to be beneficial. More importantly, the risk of future cardiovascular events and kidney failure cannot be defined for an individual and thus the availability of sets of genetic biomarkers that can predict the likelihood of better outcomes with a particular therapy in a particular individual would present certain advantages. The risks associated with a genetic-guided approach would be relatively low as the current method for selection of antihypertensive therapy is largely empirical, and often involves a trial and error approach that disenfranchises patients with hypertension, which is largely an asymptomatic disease.
Pharmacogenetics data are increasingly used to guide treatment decisions, and is now common for many anticancer drugs. The international Clinical Pharmacogenetics Implementation Consortium (CPIC) evaluates the literature to define when and how genetic data should be used to guide drug therapy for individual patients. Their guidelines are freely available online26. Although no guidelines currently exist for the use of pharmacogenomic data to guide antihypertensive drug therapy, CPIC guidelines are available for other cardiovascular drugs, namely clopidogrel27, warfarin28 and simvastatin29. In our institution we have implemented genotype-based guidance for clopidogrel therapy in patients undergoing percutaneous coronary intervention with stent placement30,31.
Hypertension pharmacogenomics
CPIC guidelines have been developed to recommend use of genetic data to guide treatment or dosing decisions for indications in which the specific genetic polymorphisms have a very large effect size. From the available data on hypertension genetics, it seems clear that no genetic polymorphisms with very large effect sizes exist. It is unlikely, therefore, that analysis of only one or two genes will be useful for guiding treatment decisions. Rather, a combination of modest effect size variants will probably need to be defined if we are to reach the goal of genetic-guided antihypertensive therapy. Although no examples of hypertension pharmacogenomics have been sufficiently validated for clinical use, the available findings suggest promise and are highlighted below.
The majority of studies in this field have either evaluated the association between genotype and blood pressure lowering or sought to identify treatment-related outcomes that differ by genotype. Most of the studies that have tested genetic associations with blood pressure response were designed specifically to test pharmacogenetic hypotheses. In contrast, data on the pharmacogenetics of treatment-related outcomes have arisen from large randomized controlled trials that had a primary clinical question, and for which genetic samples were collected to enable conduct of genetic studies. An overview of the approaches used in genetic and pharmacogenetic studies can be found in BOX 1. The following discussion focuses on the most promising findings from these studies. Specifically, we focus on examples where significant associations have been identified in more than one cohort, in the absence of a number of studies showing no association (TABLE 1). Studies have also investigated genetic associations with adverse metabolic effects, including new-onset diabetes; however, uncertainty exists as to whether these genetic signals are promising. Finally, a relatively new focus on the genetic predictors of resistant hypertension seems to hold promise, and could help to identify patients in whom an aggressive approach to treatment will be needed32.
Box 1. Basic approaches in pharmacogenomics research.
Candidate gene studies
Focuses on a gene or single nucleotide polymorphisms (SNPs) within a gene that are selected on the basis of existing knowledge
Hypothesis-driven
In pharmacogenomics, the focus is usually a protein target of a drug, a signal transduction cascade or proteins involved in pharmacokinetics, particularly drug metabolizing enzymes
This approach has been more successful in pharmacogenetics than in disease genetics
Pharmacological and metabolism pathways for drugs tend to be well defined and limited to a handful of genes or proteins
Effect sizes can be relatively large, perhaps due to an absence of selection pressure on drug responses and drug metabolism
Genome-wide association studies
Test SNPs across the human genome
Agnostic approach; enables discovery of novel genes and/or pathways
Early genome-wide association study (GWAS) chips had 100,000 SNPs; GWAS chips with >5 million SNPs are now available; imputation to 1,000 Genomes data can result in >10 million SNPs being tested
Multiple comparisons are a major statistical issue
A P value <5 × 10−8 is required to achieve ‘genome-wide significance’
Replication in an independent sample is essential
To meet statistical challenges, large sample sizes are needed
Most disease genetics GWAS have required thousands to tens of thousands of samples
Larger effect sizes in pharmacogenomics have enabled replicable discoveries with smaller sample sizes
Targeted sequencing, whole exome sequencing and whole genome sequencing
Sequence-based approaches that test targeted genes, the whole exome or the whole genome
Increasingly applied in disease genetics and the typical approach for highly penetrant genetic diseases
Typically an extreme phenotype sample set is used to discover rare, large effect variants
Few examples of this approach in pharmacogenomics for primary discovery exist to date; to our knowledge no examples exist in hypertension pharmacogenomics
Table 1.
Top pharmacogenomic signals associated with blood pressure response and/or cardiovascular outcomes
| Association | Drug class | Locus (SNP) | Associated allele | Magnitude of effect |
|---|---|---|---|---|
| BP response and CV outcomes | Thiazide diuretic | NEDD4L (rs4149601) | G allele | BP lowering: G allele associated with greater BP response (−19.5 versus −15.0 mmHg SBP and −15.4 versus −14.0 mmHg DBP)38 |
| Adverse CV outcomes: G allele associated with reduced risk of CV outcomes in NORDIL β-blocker/diuretic arm (OR 0.52, P <0.0001)38 | ||||
| G allele (one or two copies) associated with increased risk of CV outcomes when a thiazide diuretic was not included in treatment (OR 8.94–10.7, P = 0.051–0.022)39 | ||||
| β-blocker | ADRB1 (rs1801253 [Arg389Gly]) | C allele | C allele (arginine) homozygotes had greater BP response to metoprolol: −6.5 mmHg in 24 h DBP more than G (glycine) allele carriers (P = 0.0018)45 | |
| ADRB1 (rs1801252 [Ser49Gly]) | A allele | A allele (serine) homozygotes had similar trend for greater DBP response to metoprolol (P = 0.08)45 | ||
| ADRB1 Ser49/Arg389 haplotype | Haplotype | Ser49Arg389/Ser49Arg389 haplotype associated with greater BP response to metoprolol (−14.7 mmHg versus −0.5 mmHg in patients with the Gly49Arg389/Ser49Gly389 haplotype; P = 0.006)45 | ||
| Mortality risk was significantly increased among patients with one or two copies of the Ser49/Arg389 haplotype who were randomly assigned to verapamil SR (HR 8.58, 95% CI 2.06–35.8, P = 0.003) but not in patients assigned to atenolol (HR 2.31, 95% CI 0.82–6.55, P = 0.11)51 | ||||
| BP response | Thiazide diuretic | Chromosome 12, LYZ, YEATS4, FRS2 (rs7297610, rs317689, rs315135) | Primarily driven by C allele of rs7297610 | African Americans who were homozygous for CC alleles had a greater mean SBP response (−13 versus −9.6 mmHg) and DBP response (−8 versus −5.2 mmHg) than homozygous carriers of TT alleles57 |
| PRKCA (rs16960228) | A allele | A allele carriers had a 4/4 mmHg greater BP response than GG homozygotes58 | ||
| GNAS–EDN3 rs2273359 | G allele | G allele carriers had a 7/5 mmHg greater BP response than CC homozygotes58 | ||
| β-blocker | GRK4 (rs2960306 [Arg65Leu], rs1024323 [Ala142Val], rs1081058 [Ala486Val]) | T allele for all three SNPs | Increasing copies of the 65Leu–142Val haplotype were associated with significantly reduced atenolol-induced DBP lowering (−9.1 ± 6.8 versus −6.8 ± 7.1 versus −5.3 ± 6.4 mmHg in participants with zero, one, and two copies of 65Leu–142Val, respectively; P = 0.0088)74 | |
| CV outcomes | CCB | SIGLEC12, A1BG, F5 (rs16982743, rs893184, rs4525) | Risk score with one point given for each genotype that conferred a higher risk in the CCB arm than the β-blocker arm | In patients with a genetic risk score of 0 or 1, CCB treatment was associated with lower risk of treatment-related adverse CV outcomes (heart attack, stroke or death; OR 0.60, 95% CI 0.42–0.86); in patients with a genetic risk score of 2–3, CCB treatment was associated with higher risk (OR 1.31, 95% CI 1.08–1.59; meta-analysis interaction P = 2.39 × 10−5)84 |
ARB, angiotensin-receptor blocker; BP, blood pressure; CCB, calcium channel blocker; CV, cardiovascular; DBP, diastolic blood pressure; SBP, systolic blood pressure; SNP, single nucleotide polymorphism, SR, slow release.
Blood pressure and cardiovascular outcomes
Two genes exist for which genetic association studies have documented associations between blood pressure lowering and adverse cardiovascular outcomes with specific classes of drug: NEDD4L with thiazide diuretics and ADRB1 with β-blockers. In both cases the associated genes were identified through candidate gene studies, as they reside within the known pharmacological pathways of the drugs and because the functional effects of polymorphisms in these genes have been well defined. These two genes can therefore be considered to have the strongest levels of evidence to date for effects on antihypertensive agents.
NEDD4L and thiazide diuretics
NEDD4L encodes the NEDD4-2 protein, which has long been purported to have an essential role in controlling the cell surface expression of the epithelial sodium channel, ENaC, thereby influencing epithelial sodium transport33. Studies from the past couple of years, however, suggest that NEDD4-2 ubiquitylates and controls the cell surface expression of several different renal sodium transporters, including the sodium–hydrogen exchanger (NHE3), the sodium–potassium–chloride cotransporter (NKCC2) and the sodium–chloride cotransporter (NCC)33,34.
The genetic polymorphism rs4149601G>A creates a cryptic splice site in NEDD4L, with the G allele resulting in less ENaC downregulation, thereby leading to increased sodium retention. The G allele of this polymorphism is associated with hypertension35–37, an increased risk of cardiovascular disease and cardiovascular mortality37, and greater salt-sensitive hypertension with lower plasma renin activity38. The latter phenotype would be predicted to be associated with a greater blood pressure lowering response to a thiazide diuretic.
The hypothesis that patients with the rs4149601G>A polymorphism will exhibit a greater response to thiazides has in fact been confirmed by several studies. The Nordic Diltiazem (NORDIL) study first showed that the blood pressure lowering effects of combination thiazide/β-blocker treatment were greater in carriers of the G allele compared to carriers of the AA genotype (reduction in systolic blood pressure [SBP] of 19.5 ± 16.8 mmHg versus 15.0 ± 19.3 mmHg; P <0.001 and reduction in diastolic blood pressure [DBP] of 15.4 ± 8.3 versus 14.1 ± 8.4 mmHg; P = 0.02 for carriers of the G allele and AA alleles, respectively). By contrast, no difference in response to the calcium channel blocker diltiazem was observed by NEDD4L genotype39. The Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study then showed greater blood pressure lowering in patients with the G allele in response to hydrochlorothiazide but no difference by genotype in response to the β-blocker atenolol40. This finding is fully consistent with the hypothesis that the rs4149601 G allele leads to greater salt sensitivity and lower plasma renin activity.
The potential importance of NEDD4L as a genetic marker of the efficacy of antihypertensive agents has also been evaluated in two clinical trials that addressed the influence of the rs4149601 polymorphism on treatment-related outcomes. In the NORDIL trial, as mentioned above, carriers of the G allele had significantly better outcomes in response to thiazide diuretic/β-blocker combination treatment than did patients with the AA genotype (OR 0.52, 95% CI 0.36–0.74, P <0.0001)39. Similarly, in the International Verapamil SR Trandolapril Study (INVEST), patients with a G allele who were not treated with a thiazide diuretic had a significantly higher risk of cardiovascular events than those with the AA genotype, although this increased risk was mitigated by treatment with a thiazide diuretic40.
Collectively, substantial evidence points to a role for NEDD4-2 in regulating sodium reabsorption in the kidney and influencing blood pressure, particularly salt-sensitive elevations in blood pressure. As discussed, genetic polymorphisms in NEDD4L, particularly rs4149601, influence the blood pressure response to thiazide diuretics, and the risk of cardiovascular events is lowered in carriers of the G (risk) allele who are treated with a thiazide diuretic. Although future research is needed to more fully understand whether this polymorphism could be used to guide treatment decisions in patients with hypertension, the available data are certainly compelling.
ADRB1 and β-blockers
ADRB1 encodes the β1-adrenergic receptor, which mediates the physiological response to noradrenaline, and to a lesser extent, adrenaline. The β1-adrenergic receptor is the primary protein target of all β-adrenergic-receptor blockers and the majority of their beneficial effects arise by inhibiting agonist-mediated effects. ADRB1 contains two common and well-studied genetic polymorphisms that lead to changes in the encoded amino acids: rs1801252, which has a minor allele frequency ranging from 0.13 to 0.20 and results in the Ser49Gly variant, and rs1801253, which has a minor allele frequency ranging from 0.2 to 0.5 and results in the Arg389Gly polymorphism41. A substantial amount of evidence suggests that these polymorphisms, particularly rs1801253, are functional. The Arg389 allele is associated with greater coupling of the β1-adrenergic receptor to the second messenger adenylyl cyclase, resulting in greater downstream signalling in response to agonist binding41. Studies have also suggested that Ser49 undergoes less receptor internalization than the glycine variant, also resulting in greater downstream signalling. Data from the last 10–15 years and confirmed again in the past 2 years document that both the Ser49Gly and Arg389Gly variants are functional, although the effect of the Arg389Gly allele is greater and might obscure the functional effect of the codon 49 genotype42. The ancestral alleles (Ser49 and Arg389) are both associated with greater agonist-mediated effects than the variant alleles. The majority of the available literature focuses on the Arg389Gly polymorphism or on the haplotypes resulting from the codon 49 and 389 polymorphisms.
Similar to NEDD4L, ADRB1 has been associated with blood pressure and hypertension in numerous studies. For example, a study of nearly 90,000 individuals showed that the Arg389 allele is associated with hypertension43, and genome-wide association studies (GWAS) involving >60,000 individuals have identified associations between ADRB1 and hypertension44,45. Of note, the polymorphisms that result in the Ser49Gly and Arg389Gly variants are in regions that are very GC rich, which makes them difficult to genotype. Analyses of these regions tend not to yield high quality genotype data from large genotyping arrays, which limits the ability of these specific single nucleotide polymorphisms (SNPs) to be detected in arrays such as GWAS because they typically do not survive the genotyping quality control procedures. In fact because of difficulties with the genotyping assay on arrays, the Ser49Gly and Arg389Gly variants are no longer included in the more recent genome-wide SNP arrays.
The Arg389 allele and the Ser49Arg389 haplotype are thought to be associated with greater β-blocker efficacy, as these alleles are associated with greater agonist-mediated signalling, and are therefore more likely than variant forms to be inhibited by the β-blocker. Indeed a number of studies46–49, although not all50,51, have shown that the Arg389 allele, particularly the Arg389Arg genotype or the Ser49/Arg389 haplotype, is associated with greater blood pressure lowering in response to β-blockers than are other variants. In one study, individuals who were homozygous for the Arg389 allele had a greater blood pressure response to the β-blocker metoprolol (with a difference in 24 h DBP of −6.5 mmHg compared to carriers of the Gly389 allele, P = 0.0018)46. Similarly, carriers of the Ser49Arg389/Ser49Arg389 haplotype had a greater blood pressure response to metoprolol than did carriers of the Gly49Arg389/Ser49Gly389 haplotype (−14.7 mmHg versus −0.5 mmHg, P = 0.006).
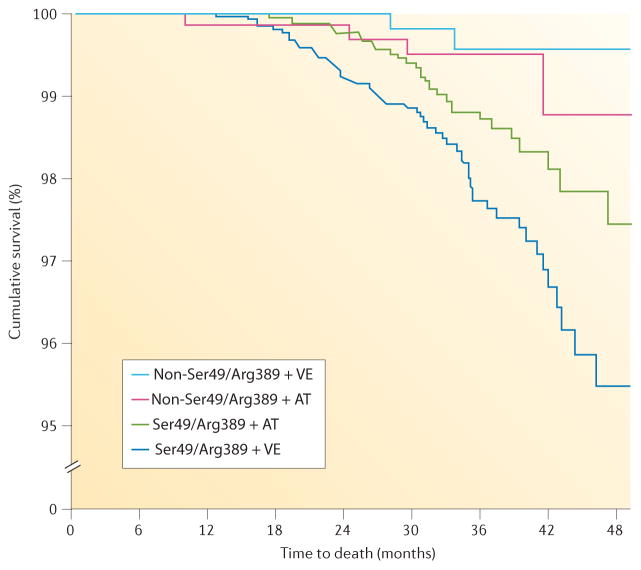
A body of literature suggests that ADRB1 genotype can influence clinical outcomes associated with β-blocker therapy. In the context of hypertension, data from INVEST suggest that compared to other haplotypes, the Ser49/Arg389 haplotype is associated with a significantly increased risk of death in patients treated with the verapamil SR-based treatment strategy (HR 8.58, 95% CI 2.06–35.8). This increased risk was offset by treatment with an atenolol-based treatment strategy (HR 2.31, 95% CI 0.82–6.55)52 (FIG. 1). Similarly, a population cohort study found that individuals with SNPs in the promoter region of ADRB1 had a higher risk of adverse cardiovascular events associated with β-blocker use than did homozygous carriers of the common allele53. The Arg389 allele or the Ser49Arg389 haplotype has also been associated with better clinical outcomes with β-blocker treatment in patients with atrial fibrillation54, ventricular arrhythmias55 and heart failure41,56.
Figure 1. The effect of ADRB1 haplotype on treatment response in patients with hypertension.
The Ser49/Arg389 haplotype is associated with a significantly increased risk of death in patients treated with a verapamil-based strategy. This increased risk is offset by treatment with an atenolol-based treatment strategy. AT, atenolol; VE, verapamil SR. Permission obtained from Wiley Ltd © Pacanowski, M. A. et al. Clin. Pharmacol. Ther. 84, 715–721 (2008).
Collectively these data provide clear evidence that functional variation exists in ADRB1 that is likely to influence blood pressure response and outcomes in patients with hypertension and other cardiovascular diseases. Additional studies are needed to determine whether these genetic associations are sufficiently robust to use this information clinically, including validation in other cohorts, particularly in hypertension studies with cardiovascular outcomes as the response studied.
Other promising gene–drug interactions
A number of studies have focused on genetic associations with response to antihypertensive therapies, particularly thiazide diuretics, with some promising data also for β-blockers. A number of candidate gene studies have investigated genetic associations with response to ACE inhibitors and ARBs, but the data from these studies are highly conflicting such that no candidate gene exists for which the majority of data from genetic associations studies suggest a positive association with response to ACE inhibitors and ARBs. Very few studies have focused on the genetic association of calcium channel blockers with blood pressure lowering.
Pharmacogenomic signals for thiazide diuretics
Chromosome 12 haplotype
The first GWAS on the pharmacogenomics of hypertension treatment was the Genetic Epidemiology of Responses to Antihypertensives (GERA) study. In this study, the diastolic blood pressure response of African Americans to the thiazide diuretic hydrochlorothiazide was associated with three SNPs located on chromosome 12q15 (a haplotype consisting of rs317689, rs315135 and rs7297610 near LYZ, FRS2 and YEATS4, with rs7297610 being the strongest signal)57. The PEAR study then replicated this association in an independent cohort of African Americans treated with hydrochlorothiazide58. A greater blood pressure lowering response was observed in African Americans with the rs7297610 CC genotype compared to the TT genotype (SBP −13 versus −9.6 mmHg and DBP −8 versus −5.2 mmHg for the CC and TT genotype, respectively). PEAR also explored YEATS4 and FRS2 gene expression before and after hydrochlorothiazide treatment and observed that patients with the FRS2 CC genotype had higher baseline gene expression than those with the CT genotype, providing a potential mechanistic explanation for the genetic association between the CC genotype and the greater blood pressure response to hydrochlorothiazide treatment58.
Protein kinase Cα
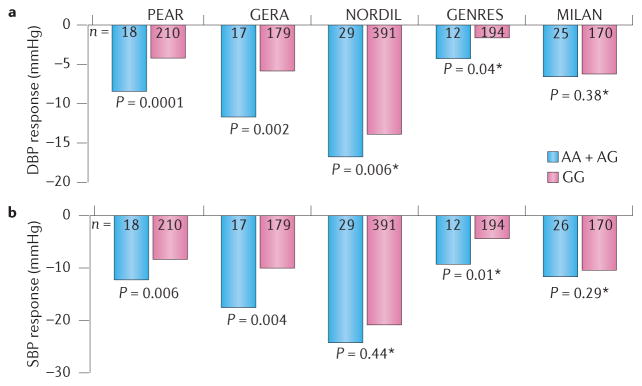
A 2013 GWAS of hydrochlorothiazide response59 included cohorts of white hypertensive patients from several pharmacogenomics studies. Cohorts from PEAR and GERA were included as the discovery cohorts and replication analyses were performed in three other cohorts, including the Genetics of Drug Responsiveness in Essential Hypertension (GENRES) study, the NORDIL Study and the Milan Hypertension Pharmacogenomics of hydrochlorothiazide (MIHYPHCTZ) study. An intronic SNP, rs16960228, in PRKCA (which encodes protein kinase Cα) on chromosome 17q24.3 was identified in the PEAR and GERA discovery cohorts in association with blood pressure response to hydrochlorothiazide. This signal was successfully replicated in the NORDIL and GENRES cohorts with the same direction of effect, but not in MIHYPHCTZ. The P value of the combined meta-analysis of the four studies was 3.3 × 10−8, meeting genome-wide significance. Systolic and diastolic blood pressure responses were consistently greater in carriers of the rs16960228A allele than in homozygote GG carriers (with a difference in both DBP and SBP >4 mmHg) across all study samples (FIG. 2). This association was further validated by other findings. First, in PEAR, the rs16960228 polymorphism was also associated with DBP response to the β-blocker, atenolol, with the opposite direction of effect. This finding is not surprising given the different pharmacologic profiles of thiazide diuretics and β-blockers. Second, associations between DBP and SNPs within PRKCA (P = 10−5) were also observed in individuals of African American ancestry59. Third, expression studies revealed that the expression level of PRKCA was significantly higher among carriers of the rs16960228 variant A allele than among those with the GG genotype59.
Figure 2. The effect of PRKCA genotype on blood pressure response to hydrochlorothiazide in participants from five independent studies (PEAR, GERA, NORDIL, GENRES and MILAN).
Participants were grouped according to the genotype of an intronic single nucleotide polymorphism, rs16960228, in the gene that encodes protein kinase Cα. The a | diastolic blood pressure (DBP) responses and b | systolic blood pressure (SBP) responses were greater in carriers of AA and AG alleles than in homozygous carriers of GG alleles in four of the five studies. The blood pressure responses are adjusted for pretreatment blood pressure levels, age and sex. P values are for contrast of adjusted means between genotype groups. Asterisks indicate replication cohorts. Permission obtained from the American Heart Association © Turner, S. T. et al. Hypertension 62, 391–397 (2013).
GNAS–EDN3
In the 2013 GWAS described above, the GNAS–EDN3 region variant rs2273359 was significantly associated with SBP response to hydrochlorothiazide in the PEAR, GERA and NORDIL studies. GENRES and MIHYPHCTZ did not have data available for this SNP. Among the three studies with available data, carriers of the CG genotype had a better blood pressure response to hydrochlorothiazide than those with the CC genotype. A meta-analysis of data from the three studies reported a P value of 5.5 × 10−8 for the association, which approached the level of genome-wide significance59.
TET2 and CSMD1
In a 2015 GWAS of two Italian cohorts, the Pharmacogenomics of Hydrochlorothiazide Sardinian Study (PHSS) and MIHYPHCTZ, TET2, an aldosterone-responsive mediator of αENaC transcription, and CSMD1, which encodes a transmembrane protein belonging to the vacuolar-protein-sorting-13 family and has been previously associated with hypertension, peripheral arterial disease and metabolic syndrome, were also associated with greater blood pressure lowering response to hydrochlorothiazide. Six SNPs — rs12505746, rs7387065, rs11993031, rs9285669, rs11189015, rs9915451 — were associated with SBP (β values ranging from −10.1 to −3.4 mmHg per variant allele). Five SNPs — rs4431329, rs7706429, rs9590353, rs113095083, rs77876672 — were associated with DBP (β values ranging from −6.5 to −2.6 mmHg). The CSMD1 signal was replicated in the GENRES cohort and showed the same direction of effect in NORDIL60.
G-protein β3 subunit
Candidate gene studies identified an association between a SNP, C825T, in GNB3 (which encodes the G-protein β3 subunit), with DBP and plasma renin, whereby carriers of the T allele had elevated DBP and low plasma renin61. On the basis of this finding we could hypothesize that the variant T allele might also be linked with volume expansion and sodium sensitivity, suggesting that this genotype might be associated with a thiazide diuretic-responsive form of hypertension. This hypothesis was tested in GERA, and indeed, participants with a T allele achieved greater lowering of SBP and DBP with hydrochlorothiazide than those who did not have a T allele (difference of −6 mmHg and −5 mmHg for SBP and DBP, respectively). This finding remained significant after adjusting for clinically appropriate covariates62. Although this finding was subsequently replicated by another study conducted in the Netherlands63, it has not been consistently replicated64.
Angiotensin-converting enzyme
ACE catalyses the formation of the active peptide angiotensin II from angiotensin I. Angiotensin II is a peptide hormone that increases blood pressure and affects fluid and electrolyte balance via vasoconstriction. Compelling data suggest a link between plasma ACE activity and the ACE insertion and deletion (I/D) polymorphism of intron 16. Those with the deletion polymorphism (D allele) have increased ACE activity65, making ACE an attractive target for candidate gene studies. Thiazide diuretics activate the renin–angiotensin system, particularly early in therapy, and the ACE I/D polymorphism has been investigated in terms of the blood pressure response to hydrochlorothiazide. Although results from these studies have been inconsistent, a meta-analysis of four studies including over 1,400 individuals found a small but significant lesser reduction in blood pressure response to hydrochlorothiazide in homozygous carriers of the insertion alleles (ACE II) compared with homozygous carriers of DD alleles (standard differences in means 0.256, 95% CI 0.109–0.403)66. Another study observed that following treatment with hydrochlorothiazide, carriers of the ACE II genotype were more likely to develop hypokalaemia than were carriers of the DD genotype67.
α-adducin
ADD1 encodes the protein α-adducin, which is one of three adducing subunits. α-adducin is expressed ubiquitously and might have a role in the modulation of ion transport. Through a candidate gene approach, the ADD1 polymorphism Gly460Trp, which is a nonsynonymous SNP, has been linked with salt sensitivity. Carriers of the 460Trp variant had reduced plasma renin activity before treatment with hydrochlorothiazide and a better blood pressure response to treatment than those with the Gly/Gly genotype (−15.9 versus −7.4 mmHg, P = 0.001)68. Subsequently, an in vitro study that utilized erythrocytes to model cell membrane transport found that the Gly460Trp variant affects renal sodium handling by altering cell membrane ion transport69. However, as has been the case with many of the candidate gene findings from pharmacogenomic studies, the Gly460Trp SNP findings have been replicated by some70,71, but not all, subsequently published studies59,64,72,73. Finally, in a meta-analysis of 1,001 individuals from four studies, carriers of the Gly/Gly genotype had a slightly, but significantly, better blood pressure lowering response to hydrochlorothiazide than did carriers of the Trp/Trp genotype66.
Pharmacogenomic signals for β-blocker response
G protein-coupled receptor kinase 4
GRK4, which encodes G protein-coupled receptor kinase 4, affects blood pressure when the G protein-coupled dopamine and β1 adrenergic receptors are inactivated by phosphorylation by G protein-coupled receptor kinases. The African American Study of Kidney Disease and Hypertension (AASK) used a candidate gene approach to assess the association between polymorphisms in GRK4 (Arg65Leu [rs2960306], Ala486Val [rs1801058], and Ala142Val [rs1024323]), with time to achieve a mean arterial blood pressure target of 107 mmHg following treatment with metoprolol. The AASK investigators found that among African American men, all of whom had early hypertension and nephrosclerosis, each Ala142 variant was associated with a 50% lower likelihood of achieving the 107 mmHg blood pressure target following treatment with metoprolol. Additionally, each Ala142 variant in men with the Leu65 variant was associated with a twofold reduction in the likelihood of achieving the target blood pressure. This link was not identified in the female AASK participants74. In the PEAR study, these variants were also found to be associated with blood pressure levels after treatment with atenolol, whereby carriers of variant alleles had or trended toward reduced blood pressure lowering following treatment. An analysis of the haplotype of the two more common variants revealed a significant association between increasing copies of the 65Leu–142Val haplotype and reduced DBP response to treatment with atenolol (−9.1 ± 6.8, −6.8 ± 7.1 and −5.3 ± 6.4 mmHg in participants with zero, one, and two copies of 65Leu–142Val, respectively; P = 0.0088). This effect was not observed after treatment with hydrochlorothiazide75. A 2014 study investigated the relationship between GRK4 SNPs and the number of antihypertensive agents required to achieve blood pressure control. Homozygous carriers of 65Leu and 142Val variants were significantly more likely to require two or more antihypertensive agents to achieve blood pressure control than those who were heterozygous carriers or expressed wild type alleles76. In a study of Japanese patients with hypertension exposed to multiple ARBs, carriers of one or two variant alleles at Ala486Val were less likely than those with no variant Ala486Val allele to respond adequately to an ARB, whereas individuals with at least one variant at each of the GRK4 polymorphic sites were significantly less likely to respond to an ARB and had less of a decrease in SBP and DBP than those with no variants77. Lastly, in an investigation of risk of adverse cardiovascular outcomes and GRK4 polymorphisms in INVEST, homozygous carriers of the 486Val allele had a significant twofold increased risk of adverse outcomes (OR 2.29, 95% CI 1.48–3.55; P = 0.0002), which was independent of antihypertensive treatment75.
Aminoacylase III
A GWAS of the GENRES study identified a signal (rs2514036) on chromosome 11 that achieved genome-wide significance for ambulatory blood pressure response to the β-blocker bisoprolol (β value −5.4 mmHg per variant allele, P = 2 × 10−8). These SNPs lie within the coding and regulatory regions of ACY3, which encodes aminoacylase III78. This chromosome 11 signal was not replicated within PEAR (which investigated the pharmacogenomic response to atenolol), and more research is necessary to validate this GWAS signal.
ACE inhibitor or ARB response
Angiotensinogen
Angiotensinogen (also known as the renin substrate) is a component of the renin–angiotensin system, a hormone system that regulates blood pressure and fluid balance. Angiotensinogen is catalytically cleaved by renin to produce angiotensin I in response to lowered blood pressure. ACE subsequently removes a dipeptide to produce angiotensin II, which is the physiologically active compound that functions to regulate fluid volume. Mutations in AGT, which encodes angiotensinogen, have been evaluated in pharmacogenomic candidate gene studies to assess the effects of AGT variants on response to ACE inhibitors and ARBs. In a 2014 study of a cohort of Chinese patients with hypertension, the AGT variant −6A>G resulted in significantly greater blood pressure lowering in response to treatment with an ACE inhibitor (SBP −22.5 mmHg in homozygous carriers of the GG variant compared to −13.7 mmHg in homozygous carriers of AA alleles), with similar trends for DBP, pulse pressure and mean arterial pressure79. Another study that investigated the same AGT variant in Swedish patients with hypertension, however, found that homozygous carriers of the AA alleles had the best blood pressure response to treatment with atenolol (−17 mmHg compared to −3 mmHg for homozygous carriers of the GG variant) with no difference observed by genotype following treatment with the ARB irbesartan80. Another AGT variant, rs699, which leads to the substitution Thr235Met has also been studied extensively with regard to blood pressure response to treatment. Although the rs699 variant is functional81, studies of blood pressure response associated with this variant have again resulted in mixed findings. The Thr/Thr genotype has been shown to be associated with a better blood pressure response to the ACE inhibitor enalapril than the Met/Thr or Met/Met variants (SBP −26mmHg, −3.0mmHg and −1.2 mmHg for the Thr/Thr, Met/Thr and Met/Met genotypes, respectively)82. Other studies, however, have shown no association with response to treatment with an ACE inhibitor83,84.
Collectively the data generated from these candidate gene studies are less compelling than those of other candidate genes since most of the available studies are small and many of the genetic signals lack substantial replication.
Treatment-related outcomes
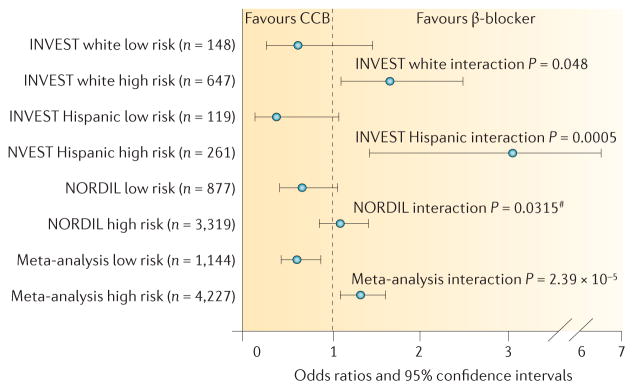
Clinical implementation of pharmacogenetics in patients with hypertension will probably require an approach that takes into account the potential of multiple genetic polymorphisms to guide one of several antihypertensive drug classes. An example of how such an approach might work was detailed in an analysis of INVEST, in which a genome-spanning array of genes related to cardiovascular and metabolic phenotypes were tested for their association with treatment-related outcomes. From this analysis, three nonsynonymous variants (rs16982743, rs893184 and rs4525, which are located in SIGLEC12, A1BG and F5, respectively) were significantly associated with differential outcomes with β-blocker versus calcium channel blocker treatment in white individuals and also, in a separate analysis, in individuals of Hispanic ancestry85. These SNPs have not been associated with blood pressure or hypertension in previous studies, although other SNPs in F5 have been associated with hypertension86. From these data a ‘risk score’ was then created for rs16982743, rs893184 and rs4525. Patients with a risk score of 0 or 1 had better cardiovascular outcomes if treated with a calcium channel blocker compared to a β-blocker (OR 0.60, 95% CI 0.42–0.86), and those with a risk score of 2 or 3 had worse outcomes if treated with a calcium channel blocker compared to a β-blocker (OR 1.31, 95% CI 1.08–1.59). These findings were then replicated in the NORDIL trial85 (FIG. 3).
Figure 3. Development of a genetic risk-score associated with treatment-related adverse cardiovascular outcomes on the basis of data from the INVEST and NORDIL trials.
A ‘risk score’ was developed on the basis of the association of nonsynonymous variants (rs16982743, rs893184 and rs4525, in SIGLEC12, A1BG and F5, respectively), with differential outcomes with β-blocker and calcium-channel blocker (CCB) treatment. One point was given for each genotype that conferred higher risk in the CCB group compared to the β-blocker group. Low risk was defined as a risk score of 0 or 1; high risk was defined as a risk score of 2 or 3. #One-sided P value based on a 1-sided hypothesis in the replication cohort. Permission obtained from the American Heart Association © McDonough, C. W. et al. Hypertension 62, 48–54 (2013).
These data not only identify genes of potential importance to define treatment-related outcomes, but this approach also highlights the manner in which genetic information might be used in the future to select therapy for hypertension.
A number of instances exist in the literature in which only one paper has been published describing the association of a particular gene variant with treatment-related cardiovascular outcomes. Although such studies do not provide sufficient evidence to guide treatment, the data are interesting, and many of the genes described have strong biologic plausibility. Thus future studies to independently replicate these findings are important. These genes include KCNMB1 (REF. 87), NR1H3 (REF. 88), MMP9 and MMP12 (REF. 89), NOS3 (REF. 90), NPPA91, RYR3 (REF. 92), and AGTR1 (REFS 93,94).
In contrast, ADD1, which might be associated with blood pressure response to hydrochlorothiazide as described above, has also been extensively studied with regard to cardiovascular outcomes. An observational case–control study found that carriers of the ADD1 Trp460 variant who used drugs of the thiazide class of diuretics had a significant 50% reduction in their risk of adverse outcomes, including myocardial infarction and stroke, compared to carriers of the ADD1 Trp460 variant who used other antihypertensive drug classes95. However, this association was also studied in large randomized trials of patients with hypertension that were designed to investigate cardiovascular outcomes. These studies, including the Genetics of Hypertension Associated Treatment (GenHAT)96, INVEST97, and Losartan Intervention for Endpoint Reduction in Hypertension (LIFE)98, as well as the population-based Pharmaco-Morbidity Record Linkage System (PHARMO)99, did not replicate the finding of the observational study. Collectively, the available evidence does not support a pharmacogenomic effect of the ADD1 Gly460Trp polymorphism on adverse outcomes in users of thiazide diuretics.
Future directions
The long-term goal of hypertension pharmacogenomics is to use genetic and clinical information to select the antihypertensive drug regimen that is most likely to provide the greatest efficacy with the lowest risk of adverse effects for an individual patient. However, many challenges have prevented this goal from being met. The biggest of these challenges is the relatively small sample sizes of the available studies with a well characterized blood pressure response and genetic data, which limits the power for detecting genetic associations at the GWAS level. Other challenges include the ability to replicate genomic signals in independent cohorts, particularly in datasets that include individuals from diverse ancestral backgrounds; identify the biological basis for the genetic association; and define the clinical utility of replicated signals for predicting blood pressure response. Previous examples of collaborative success in disease genetics studies, for example as seen with CHARGE, BPGEN and HYPERGEN, and in pharmacogenomics (for example, the International Warfarin Pharmacogenetics Consortium) clearly show that collaborative consortia represent the best approach to overcome these challenges.
ICAPS
The International Consortium for Antihypertensive Pharmacogenomics Studies (ICAPS) was formed in 2012 with the aim of increasing the opportunities to discover and replicate the genetic signatures of many different phenotypes related to antihypertensive treatment response. ICAPS promotes the discovery of genes that influence response to antihypertensive drugs with the primary goal of advancing pharmacogenomics discoveries that will lead to definitive evidence regarding the use of genetic information to guide antihypertensive treatment decisions. Through ICAPS collaborations, several cohorts have successfully identified pharmacogenomic associations59,60,78, demonstrating the importance of the consortial approach for successful pharmacogenomics discoveries.
ICAPS currently includes >350,000 participants from 31 cohorts, which are based in 10 countries on three continents. These cohorts include those from studies that tested blood pressure response (and in some cases other phenotypes including adverse metabolic responses) to various antihypertensive drugs, as well as large randomized controlled trials that tested clinical outcomes (such as death, myocardial infarction, stroke, and diabetes) following antihypertensive treatment. Additionally, a number of observational cohorts with blood pressure response, antihypertensive treatment and outcomes data are included in ICAPS and will be utilized for validation and replication of genetic findings. Through ICAPS, it will be possible to use meta-analytic approaches with genome-wide data for discovery, followed by replication in independent cohorts for many antihypertensive response phenotypes. These phenotypes include blood pressure response to first line antihypertensive drug classes as well as adverse cardiovascular outcomes with different antihypertensive treatment regimens utilized in clinical trials, antihypertensive treatment resistant hypertension (that is, patients with uncontrolled blood pressure despite three appropriately dosed medications, or requiring four or more medications to achieve blood pressure control), and adverse effects, including metabolic effects (for example hyperglycaemia, hypokalaemia, hyponatraemia and hyperuricaemia), new-onset diabetes and rare adverse events (for example angioedema) associated with antihypertensive medication treatment.
By including many different types of cohorts in ICAPS, including shorter term clinical trials with surrogate end points (such as blood pressure and glucose); longer term randomized trial data with overall blood pressure response and hard clinical outcomes (such as myocardial infarction, stroke and death); epidemiological studies with rich data sets that include medication history, medical history with physical exam and laboratory data, and in many cases cardiovascular outcomes data; and cohorts from diverse ancestral backgrounds, analyses conducted within ICAPS will be able to overcome many of the existing shortcomings of antihypertensive pharmacogenomics research.
Other ‘omics’ data as biomarkers
Over the past decade, metabolomics, a global biochemical approach to identify metabolic pathways and networks associated with human disease, has been used to assess drug responses. In 2013 we reported a differential metabolomics signature of response to atenolol treatment in white Americans and African Americans100. When the signals of the metabolomics signature were combined with genetic signals involved in relevant metabolite pathways, we observed atenolol-induced changes in the metabolome that were dependent on ancestry and genotype, which could help to explain the observed differential response in blood pressure22,100. In addition, a well replicated branched chain amino acid signal that is associated with insulin resistance and diabetes is also associated with atenolol-induced hyperglycaemia and impaired fasting glucose, which is an important phenotype in the diabetes continuum101.
Studies of pharmacometabolics and blood pressure response are currently underway and will shed further light on important biochemical pathways involved in the response to antihypertensive drugs. In the UK, the Ancestry and Biological Informative Markers for Stratification of Hypertension (AIM HY) study102 will investigate pharmacogenomic and pharmacometabolomic signatures of antihypertensive drug response, and use this information to prospectively identify and test a personalized antihypertensive treatment approach. Lastly, transcriptomics — the study of the complete set of RNA transcripts that are produced by the genome — is another ‘omics’ approach that could be useful to elucidate drug response mechanisms103. Tools for studying genome-wide microRNAs that may affect expression, methylation, and other genetic modifiers are also gaining interest. These approaches are likely to be used to help define biomarkers of response to antihypertensive drugs and, therefore, increase our understanding of the causes of interindividual differences in drug responses.
Conclusions
Great hope exists for personalized treatment approaches — or precision medicine — in the near future. A number of approaches might be used to achieve a more personalized approach to the treatment of hypertension than is currently used, including the consideration of relatively simple factors, such as age and ancestry, or measurement of plasma renin activity. Great hope has also been placed on the potential to use genetic markers, or other ‘omics’ level data to refine treatment strategies. The available data are promising in terms of defining genetic determinants of responses to antihypertensive drugs, although no studies to date have been sufficiently replicated, or of large enough effect size to warrant changes in clinical practice. For the first time, a new Scientific Statement from the American College of Cardiology, the American Heart Association and the American Society of Hypertension on the treatment of hypertension in patients with coronary artery disease includes discussion of genetic polymorphisms that could affect blood pressure response to antihypertensive agents, and suggests that determination of genetic variants could be useful in the future to help select appropriate antihypertensive agents to reduce both blood pressure and the risk of adverse cardiovascular outcomes104. The question remains, however, of how to realize the promise of pharmacogenetics to guide antihypertensive treatment. In the field of disease genetics, and particularly hypertension genetics, any discoveries that are made from GWAS by extending well beyond the >100,000 participants that have been included in studies to date will only identify genetic variants with clinically irrelevant effect sizes. However, the same cannot be said for hypertension pharmacogenomics, where sample sizes have typically been in the hundreds (for studies of blood pressure responses) or a few thousand (for studies of adverse outcomes). Thus, use of larger sample sizes, facilitated through ICAPS, should enable the identification of replicable genetic associations. These sample sizes will enable the continued detection of moderate effect size associations (for example, those with an OR >1.5 or leading to a lowering of blood pressure in the range of 2–5 mmHg per allele), which are also sufficiently large to have potential clinical relevance if considered collectively.
Any future clinical utility in hypertension pharmacogenomics clearly needs to include a cadre of genetic markers, perhaps along with clinical factors and/or other biomarkers. The precise approach by which such information could be used remains to be defined, but some excellent examples of the possibilities exist. One potential approach is the development of a mathematical algorithm, such as that used for warfarin pharmacogenetic dosing105, which incorporates genetic and clinical factors with different weights assigned to the different genotypes included in the algorithm. This approach could be used to predict blood pressure response to a given antihypertensive drug. Prediction of the best drug to avoid long-term cardiovascular outcomes in a particular patient is more likely to occur through a genetic risk scoring approach, as has been suggested for numerous cardiovascular phenotypes, including cardiovascular events prediction, stroke and atrial fibrillation106–110. Using these studies as examples, the risk score could include genetic markers or genetics plus other factors, and the genetic data might be weighted or unweighted depending on the level of evidence in each case. Additional work is required to advance hypertension pharmacogenomics to the level of understanding currently occupied by most cardiovascular disease phenotypes. Full use of genetic risk score approaches, even if they improve predictability, is clearly only going be possible once large amounts of genetic data are readily available for individual patients (for example, genome-wide SNP or whole genome data), and recorded in an accessible manner as part of an electronic health record.
Other ‘omics’ approaches might also prove useful as predictors of response. Although use of metabolomics and transcriptomics data is in its nascent stages, promise exists for the potential of additional insights to be provided through these tools.
Work over the next 5–10 years should define the potential for genetically-guided antihypertensive drug therapy. Even if a clinically relevant pharmacogenomics signal does not come to fruition in the near term, the work in the field is nonetheless likely to provide important insights into mechanisms of drug action, particularly for thiazide diuretics, for which a detailed, mechanistic understanding of long-term blood pressure lowering remains limited. In addition, discoveries in the field could potentially lead to identification of novel drug targets as has happened in other areas, such as PCSK9 inhibition for cholesterol lowering, with two PCSK9 inhibitors now FDA approved111.
Increasing focus has been placed on personalized treatment approaches — or precision medicine — in health care, and it is important to define whether effective methods exist to achieve a personalized approach to selecting therapy for patients with hypertension. The best tools currently available for such personalization of treatment are consideration of a patient’s age and ancestry, and perhaps measurement of plasma renin activity. The next decade will provide clarity as to whether additional ‘omics’ biomarkers can be added to the available tools to enable a more precise selection of suitable antihypertensive therapy for an individual patient.
Acknowledgments
R.M.C.-D. has received grant funding from NIH, NIGMS U01 GM074492 and U01 GM092586; and NIH, NHGRI U01 HG007269. J.A.J. has received grant funding from NIH, NIGMS U01 GM074492; NIH, NHGRI U01 HG007269, and NIH, NINDS R01 NS073346 and FDA U01 FD005235.
Footnotes
Author contributions
Both authors researched data for the article, made substantial contributions to planning the review, shared in the writing of the article and reviewed or edited the manuscript before and after submission.
Competing interests statement
The authors declare no competing interests.
References
- 1.Mozaffarian D, et al. Heart disease and stroke statistics — 2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–e322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 2.Egan BM, Zhao Y, Axon RN. US trends in prevalence, awareness, treatment, and control of hypertension, 1988–2008. JAMA. 2010;303:2043–2050. doi: 10.1001/jama.2010.650. [DOI] [PubMed] [Google Scholar]
- 3.Kearney PM, et al. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 4.Lim SS, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2224–2260. doi: 10.1016/S0140-6736(12)61766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO. Adherence to long-term therapies: evidence for action [online] Press Release. http://www.who.int/chp/knowledge/publications/adherence_report/en/
- 6.Sundstrom J, et al. Effects of blood pressure reduction in mild hypertension: a systematic review and meta-analysis. Ann Intern Med. 2014;162:184–191. doi: 10.7326/M14-0773. [DOI] [PubMed] [Google Scholar]
- 7.Sundstrom J, et al. Blood pressure-lowering treatment based on cardiovascular risk: a meta-analysis of individual patient data. Lancet. 2014;384:591–598. doi: 10.1016/S0140-6736(14)61212-5. [DOI] [PubMed] [Google Scholar]
- 8.Blood Pressure Lowering Treatment Trialists’ Collaboration. Effects of ACE inhibitors calcium antagonists and other blood-pressure-lowering drugs: results of prospectively designed overviews of randomised trials. Lancet. 2000;356:1955–1964. doi: 10.1016/s0140-6736(00)03307-9. [DOI] [PubMed] [Google Scholar]
- 9.SHEP Cooperative Research Group. Prevention of stroke by antihypertensive drug treatment in older persons with isolated systolic hypertension. JAMA. 1991;265:3255–3264. [PubMed] [Google Scholar]
- 10.Beckett NS, et al. Treatment of hypertension in patients 80 years of age or older. N Engl J Med. 2008;358:1887–1898. doi: 10.1056/NEJMoa0801369. [DOI] [PubMed] [Google Scholar]
- 11.Lewington S, Clarke R, Qizilbash N, Peto R, Collins R. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360:1903–1913. doi: 10.1016/s0140-6736(02)11911-8. [DOI] [PubMed] [Google Scholar]
- 12.Hsu CY, McCulloch CE, Darbinian J, Go AS, Iribarren C. Elevated blood pressure and risk of end-stage renal disease in subjects without baseline kidney disease. Arch Intern Med. 2005;165:923–928. doi: 10.1001/archinte.165.8.923. [DOI] [PubMed] [Google Scholar]
- 13.Lawes CM, Vander Hoorn S, Rodgers A. Global burden of blood-pressure-related disease, 2001. Lancet. 2008;371:1513–1518. doi: 10.1016/S0140-6736(08)60655-8. [DOI] [PubMed] [Google Scholar]
- 14.Gu Q, Burt VL, Dillon CF, Yoon S. Trends in antihypertensive medication use and blood pressure control among United States adults with hypertension: the National Health And Nutrition Examination Survey, 2001 to 2010. Circulation. 2012;126:2105–2114. doi: 10.1161/CIRCULATIONAHA.112.096156. [DOI] [PubMed] [Google Scholar]
- 15.Hajjar I, Kotchen TA. Trends in prevalence, awareness, treatment, and control of hypertension in the United States, 1988–2000. JAMA. 2003;290:199–206. doi: 10.1001/jama.290.2.199. [DOI] [PubMed] [Google Scholar]
- 16.James PA, et al. 2014 evidence-based guideline for the management of high blood pressure in adults: report from the panel members appointed to the Eighth Joint National Committee (JNC 8) JAMA. 2014;311:507–520. doi: 10.1001/jama.2013.284427. [DOI] [PubMed] [Google Scholar]
- 17.Chobanian AV, et al. The seventh report of the Joint National Committee on prevention, detection, evaluation, and treatment of high blood pressure. JAMA. 2003;289:2560–2572. doi: 10.1001/jama.289.19.2560. [DOI] [PubMed] [Google Scholar]
- 18.Mancia G, et al. 2013 ESH/ESC guidelines for the management of arterial hypertension: the task force for the management of arterial hypertension of the European Society of Hypertension (ESH) and of the European Society of Cardiology (ESC) Eur Heart J. 2013;34:2159–2219. doi: 10.1093/eurheartj/eht151. [DOI] [PubMed] [Google Scholar]
- 19.American Diabetes Association. Standards of medical care in diabetes — 2014. Diabetes Care. 2014;37:S14–S80. doi: 10.2337/dc14-S014. [DOI] [PubMed] [Google Scholar]
- 20.Materson BJ, et al. Single-drug therapy for hypertension in men — a comparison of six antihypertensive agents with placebo. N Engl J Med. 1993;328:914–921. doi: 10.1056/NEJM199304013281303. [DOI] [PubMed] [Google Scholar]
- 21.Mahmud A, Feely J. Choice of first antihypertensive: simple as ABCD? Am J Hypertens. 2007;20:923–927. doi: 10.1016/j.amjhyper.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 22.Johnson JA, et al. Hydrochlorothiazide and atenolol combination antihypertensive therapy: effects of drug initiation order. Clin Pharmacol Ther. 2009;86:533–539. doi: 10.1038/clpt.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz GL, Bailey K, Chapman AB, Boerwinkle E, Turner ST. The role of plasma renin activity, age, and race in selecting effective initial drug therapy for hypertension. Am J Hypertens. 2013;26:957–964. doi: 10.1093/ajh/hpt047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laragh JH, et al. Renin, angiotensin and aldosterone system in pathogenesis and management of hypertensive vascular disease. Am J Med. 1972;52:633–652. doi: 10.1016/0002-9343(72)90054-x. [DOI] [PubMed] [Google Scholar]
- 25.Turner ST, et al. Plasma renin activity predicts blood pressure responses to β-blocker and thiazide diuretic as monotherapy and add-on therapy for hypertension. Am J Hypertens. 2010;23:1014–1022. doi: 10.1038/ajh.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenetics Research Network. PharmGKB. doi: 10.1038/clpt.2010.279. [online], https://www.pharmgkb.org/page/cpic. [DOI] [PMC free article] [PubMed]
- 27.Scott SA, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94:317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson JA, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011;90:625–629. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramsey LB, et al. The Clinical Pharmacogenetics Implementation Consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin Pharmacol Ther. 2014;96:423–428. doi: 10.1038/clpt.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owusu-Obeng A, et al. Emerging roles for pharmacists in clinical implementation of pharmacogenomics. Pharmacotherapy. 2014;34:1102–1112. doi: 10.1002/phar.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weitzel KW, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet. 2014;166:56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El Rouby N, Cooper-DeHoff RM. Genetics of resistant hypertension: a novel pharmacogenomics phenotype. Curr Hypertens Rep. 2015;17:583. doi: 10.1007/s11906-015-0583-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rizzo F, Staub O. NEDD4-2 and salt-sensitive hypertension. Curr Opin Nephrol Hypertens. 2015;24:111–116. doi: 10.1097/MNH.0000000000000097. [DOI] [PubMed] [Google Scholar]
- 34.Ronzaud C, et al. Renal tubular NEDD4-2 deficiency causes NCC-mediated salt-dependent hypertension. J Clin Invest. 2013;123:657–665. doi: 10.1172/JCI61110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russo CJ, et al. Association of NEDD4L ubiquitin ligase with essential hypertension. Hypertension. 2005;46:488–491. doi: 10.1161/01.HYP.0000178594.63193.c0. [DOI] [PubMed] [Google Scholar]
- 36.Luo F, et al. A functional variant of NEDD4L is associated with hypertension, antihypertensive response, and orthostatic hypotension. Hypertension. 2009;54:796–801. doi: 10.1161/HYPERTENSIONAHA.109.135103. [DOI] [PubMed] [Google Scholar]
- 37.Dahlberg J, Sjögren M, Hedblad B, Engström G, Melander O. Genetic variation in NEDD4L, an epithelial sodium channel regulator, is associated with cardiovascular disease and cardiovascular death. J Hypertens. 2014;32:294–299. doi: 10.1097/HJH.0000000000000044. [DOI] [PubMed] [Google Scholar]
- 38.Dahlberg J, Nilsson LO, von Wowern F, Melander O. Polymorphism in NEDD4L is associated with increased salt sensitivity, reduced levels of P-renin and increased levels of Nt-proANP. PLoS ONE. 2007;2:e432. doi: 10.1371/journal.pone.0000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svensson-Farbom P, et al. A functional variant of the NEDD4L gene is associated with beneficial treatment response with β-blockers and diuretics in hypertensive patients. J Hypertens. 2011;29:388–395. doi: 10.1097/HJH.0b013e3283410390. [DOI] [PubMed] [Google Scholar]
- 40.McDonough CW, et al. Association of variants in NEDD4L with blood pressure response and adverse cardiovascular outcomes in hypertensive patients treated with thiazide diuretics. J Hypertens. 2013;31:698–704. doi: 10.1097/HJH.0b013e32835e2a71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson JA, Liggett SB. Cardiovascular pharmacogenomics of adrenergic receptor signaling: clinical implications and future directions. Clin Pharmacol Ther. 2011;89:366–378. doi: 10.1038/clpt.2010.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang F, Steinberg SF. S49G and R389G polymorphisms of the β1-adrenergic receptor influence signaling via the cAMP-PKA and ERK pathways. Physiol Genom. 2013;45:1186–1192. doi: 10.1152/physiolgenomics.00087.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson AD, et al. Association of hypertension drug target genes with blood pressure and hypertension in 86,588 individuals. Hypertension. 2011;57:903–910. doi: 10.1161/HYPERTENSIONAHA.110.158667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wain LV, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43:1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ganesh SK, et al. Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum Mol Genet. 2013;22:1663–1678. doi: 10.1093/hmg/dds555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson JA, et al. β1-adrenergic receptor polymorphisms and antihypertensive response to metoprolol. Clin Pharmacol Ther. 2003;74:44–52. doi: 10.1016/S0009-9236(03)00068-7. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, et al. β1-adrenergic receptor polymorphisms influence the response to metoprolol monotherapy in patients with essential hypertension. Clin Pharmacol Ther. 2006;80:23–32. doi: 10.1016/j.clpt.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 48.Wu D, et al. Associations between ADRB1 and CYP2D6 gene polymorphisms and the response to β-blocker therapy in hypertension. J Int Med Res. 2015;43:424–434. doi: 10.1177/0300060514563151. [DOI] [PubMed] [Google Scholar]
- 49.Si D, et al. Association of common polymorphisms in β1-adrenergic receptor with antihypertensive response to carvedilol. J Cardiovasc Pharmacol. 2014;64:306–309. doi: 10.1097/FJC.0000000000000119. [DOI] [PubMed] [Google Scholar]
- 50.O’Shaughnessy KM, Fu B, Dickerson C, Thurston D, Brown MJ. The gain-of-function G389R variant of the β1-adrenoceptor does not influence blood pressure or heart rate response to β-blockade in hypertensive subjects. Clin Sci (Lond) 2000;99:233–238. [PubMed] [Google Scholar]
- 51.Karlsson J, et al. β1-adrenergic receptor gene polymorphisms and response to β1-adrenergic receptor blockade in patients with essential hypertension. Clin Cardiol. 2004;27:347–350. doi: 10.1002/clc.4960270610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pacanowski MA, et al. β-adrenergic receptor gene polymorphisms and β-blocker treatment outcomes in hypertension. Clin Pharmacol Ther. 2008;84:715–721. doi: 10.1038/clpt.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lemaitre RN, et al. β1- and β2-adrenergic receptor gene variation, β-blocker use and risk of myocardial infarction and stroke. Am J Hypertens. 2008;21:290–296. doi: 10.1038/ajh.2007.71. [DOI] [PubMed] [Google Scholar]
- 54.Aleong RG, et al. Prevention of atrial fibrillation by bucindolol is dependent on the β1389 Arg/Gly adrenergic receptor polymorphism. JACC Heart Fail. 2013;1:338–344. doi: 10.1016/j.jchf.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aleong RG, Sauer WH, Robertson AD, Liggett SB, Bristow MR. Adrenergic receptor polymorphisms and prevention of ventricular arrhythmias with bucindolol in patients with chronic heart failure. Circ Arrhythm Electrophysiol. 2013;6:137–143. doi: 10.1161/CIRCEP.111.969618. [DOI] [PubMed] [Google Scholar]
- 56.Liggett SB, et al. A polymorphism within a conserved β1-adrenergic receptor motif alters cardiac function and β-blocker response in human heart failure. Proc Natl Acad Sci USA. 2006;103:11288–11293. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turner ST, et al. Genomic association analysis suggests chromosome 12 locus influencing antihypertensive response to thiazide diuretic. Hypertension. 2008;52:359–365. doi: 10.1161/HYPERTENSIONAHA.107.104273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duarte JD, et al. Association of chromosome 12 locus with antihypertensive response to hydrochlorothiazide may involve differential YEATS4 expression. Pharmacogenom J. 2013;13:257–263. doi: 10.1038/tpj.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turner ST, et al. Genomic association analysis of common variants influencing antihypertensive response to hydrochlorothiazide. Hypertension. 2013;62:391–397. doi: 10.1161/HYPERTENSIONAHA.111.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chittani M, et al. TET2 and CSMD1 genes affect SBP response to hydrochlorothiazide in never-treated essential hypertensives. J Hypertens. 2015;33:1301–1309. doi: 10.1097/HJH.0000000000000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schunkert H, Hense HW, Döring A, Riegger GA, Siffert W. Association between a polymorphism in the G protein β3 subunit gene and lower renin and elevated diastolic blood pressure levels. Hypertension. 1998;32:510–513. doi: 10.1161/01.hyp.32.3.510. [DOI] [PubMed] [Google Scholar]
- 62.Turner ST, Schwartz GL, Chapman AB, Boerwinkle E. C825T polymorphism of the G protein β3-subunit and antihypertensive response to a thiazide diuretic. Hypertension. 2001;37:739–743. doi: 10.1161/01.hyp.37.2.739. [DOI] [PubMed] [Google Scholar]
- 63.Schelleman H, et al. Interactions between five candidate genes and antihypertensive drug therapy on blood pressure. Pharmacogenom J. 2006;6:22–26. doi: 10.1038/sj.tpj.6500339. [DOI] [PubMed] [Google Scholar]
- 64.Matayoshi T, et al. The thiazide-sensitive Na+-Cl− cotransporter gene, C1784T, and adrenergic receptor-β3 gene, T727C, may be gene polymorphisms susceptible to the antihypertensive effect of thiazide diuretics. Hypertens Res. 2004;27:821–833. doi: 10.1291/hypres.27.821. [DOI] [PubMed] [Google Scholar]
- 65.Rigat B, et al. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Choi HD, et al. Effects of ACE and ADD1 gene polymorphisms on blood pressure response to hydrochlorothiazide: a meta-analysis. Int J Clin Pharmacol Ther. 2013;51:718–724. doi: 10.5414/CP201899. [DOI] [PubMed] [Google Scholar]
- 67.Vormfelde SV, et al. Hydrochlorothiazide efficacy and polymorphisms in ACE, ADD1 and GNB3 in healthy, male volunteers. Eur J Clin Pharmacol. 2006;62:195–201. doi: 10.1007/s00228-005-0081-z. [DOI] [PubMed] [Google Scholar]
- 68.Cusi D, et al. Polymorphisms of α-adducin and salt sensitivity in patients with essential hypertension. Lancet. 1997;349:1353–1357. doi: 10.1016/S0140-6736(97)01029-5. [DOI] [PubMed] [Google Scholar]
- 69.Glorioso N, et al. α-adducin 460Trp allele is associated with erythrocyte Na transport rate in North Sardinian primary hypertensives. Hypertension. 2002;39:357–362. doi: 10.1161/hy0202.103065. [DOI] [PubMed] [Google Scholar]
- 70.Glorioso N, et al. The role of α-adducin polymorphism in blood pressure and sodium handling regulation may not be excluded by a negative association study. Hypertension. 1999;34:649–654. doi: 10.1161/01.hyp.34.4.649. [DOI] [PubMed] [Google Scholar]
- 71.Sciarrone MT, et al. ACE and α-adducin polymorphism as markers of individual response to diuretic therapy. Hypertension. 2003;41:398–403. doi: 10.1161/01.HYP.0000057010.27011.2C. [DOI] [PubMed] [Google Scholar]
- 72.Turner ST, Chapman AB, Schwartz GL, Boerwinkle E. Effects of endothelial nitric oxide synthase, α-adducin, and other candidate gene polymorphisms on blood pressure response to hydrochlorothiazide. Am J Hypertens. 2003;16:834–839. doi: 10.1016/s0895-7061(03)01011-2. [DOI] [PubMed] [Google Scholar]
- 73.Schelleman H, et al. The influence of the α-adducin G460W polymorphism and angiotensinogen M235T polymorphism on antihypertensive medication and blood pressure. Eur J Hum Genet. 2006;14:860–866. doi: 10.1038/sj.ejhg.5201632. [DOI] [PubMed] [Google Scholar]
- 74.Bhatnagar V, et al. G-protein-coupled receptor kinase 4 polymorphisms and blood pressure response to metoprolol among African Americans: sex-specificity and interactions. Am J Hypertens. 2009;22:332–338. doi: 10.1038/ajh.2008.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vandell AG, et al. G protein receptor kinase 4 polymorphisms: β-blocker pharmacogenetics and treatment-related outcomes in hypertension. Hypertension. 2012;60:957–964. doi: 10.1161/HYPERTENSIONAHA.112.198721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muskalla AM, et al. G-protein receptor kinase 4 polymorphism and response to antihypertensive therapy. Clin Chem. 2014;60:1543–1548. doi: 10.1373/clinchem.2014.226605. [DOI] [PubMed] [Google Scholar]
- 77.Sanada H, et al. Common variants of the G protein-coupled receptor type 4 are associated with human essential hypertension and predict the blood pressure response to angiotensin receptor blockade. Pharmacogenom J. doi: 10.1038/tpj.2015.6. http://dx.doi.org/10.1038/tpj.2015.6. [DOI] [PMC free article] [PubMed]
- 78.Hiltunen TP, et al. Pharmacogenomics of hypertension: a genome-wide, placebo-controlled cross-over study, using four classes of antihypertensive drugs. J Am Heart Assoc. 2015;4:e001521. doi: 10.1161/JAHA.114.001521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu H, et al. A core promoter variant of angiotensinogen gene and interindividual variation in response to angiotensin-converting enzyme inhibitors. J Renin Angiotensin Aldosterone Syst. 2014;15:540–546. doi: 10.1177/1470320313506481. [DOI] [PubMed] [Google Scholar]
- 80.Kurland L, et al. Angiotensinogen gene polymorphisms: relationship to blood pressure response to antihypertensive treatment: results from the Swedish Irbesartan Left Ventricular Hypertrophy Investigation vs Atenolol (SILVHIA) trial. Am J Hypertens. 2004;17:8–13. doi: 10.1016/j.amjhyper.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 81.Jeunemaitre X, et al. Molecular basis of human hypertension: role of angiotensinogen. Cell. 1992;71:169–180. doi: 10.1016/0092-8674(92)90275-h. [DOI] [PubMed] [Google Scholar]
- 82.Srivastava K, Chandra S, Bhatia J, Narang R, Saluja D. Association of angiotensinogen (M235T) gene polymorphism with blood pressure lowering response to angiotensin converting enzyme inhibitor (Enalapril) J Pharm Pharm Sci. 2012;15:399–406. doi: 10.18433/j3kw3b. [DOI] [PubMed] [Google Scholar]
- 83.Mondorf UF, et al. Contribution of angiotensin I converting enzyme gene polymorphism and angiotensinogen gene polymorphism to blood pressure regulation in essential hypertension. Am J Hypertens. 1998;11:174–183. doi: 10.1016/s0895-7061(97)00402-0. [DOI] [PubMed] [Google Scholar]
- 84.Hannila-Handelberg T, et al. Common genetic variations of the renin–angiotensin–aldosterone system and response to acute angiotensin I-converting enzyme inhibition in essential hypertension. J Hypertens. 2010;28:771–779. doi: 10.1097/HJH.0b013e328335c368. [DOI] [PubMed] [Google Scholar]
- 85.McDonough CW, et al. Pharmacogenomic association of nonsynonymous SNPs in SIGLEC12, A1BG, and the selectin region and cardiovascular outcomes. Hypertension. 2013;62:48–54. doi: 10.1161/HYPERTENSIONAHA.111.00823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ehret GB, O’Connor AA, Weder A, Cooper RS, Chakravarti A. Follow-up of a major linkage peak on chromosome 1 reveals suggestive QTLs associated with essential hypertension: GenNet study. Eur J Hum Genet. 2009;17:1650–1657. doi: 10.1038/ejhg.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Beitelshees AL, et al. KCNMB1 genotype influences response to verapamil SR and adverse outcomes in the INternational VErapamil SR/Trandolapril STudy (INVEST) Pharmacogenet Genom. 2007;17:719–729. doi: 10.1097/FPC.0b013e32810f2e3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Price ET, et al. Liver X receptor α gene polymorphisms and variable cardiovascular outcomes in patients treated with antihypertensive therapy: results from the INVEST-GENES study. Pharmacogenet Genom. 2011;21:333–340. doi: 10.1097/FPC.0b013e3283452fec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tanner RM, et al. Pharmacogenetic associations of MMP9 and MMP12 variants with cardiovascular disease in patients with hypertension. PLoS ONE. 2011;6:e23609. doi: 10.1371/journal.pone.0023609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang X, et al. Pharmacogenetic association of NOS3 variants with cardiovascular disease in patients with hypertension: the GenHAT study. PLoS ONE. 2012;7:e34217. doi: 10.1371/journal.pone.0034217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lynch AI, et al. Pharmacogenetic association of the NPPA T2238C genetic variant with cardiovascular disease outcomes in patients with hypertension. JAMA. 2008;299:296–307. doi: 10.1001/jama.299.3.296. [DOI] [PubMed] [Google Scholar]
- 92.Lynch AI, et al. Gene panels to help identify subgroups at high and low risk of coronary heart disease among those randomized to antihypertensive treatment: the GenHAT study. Pharmacogenet Genom. 2012;22:355–366. doi: 10.1097/FPC.0b013e3283516ff8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schelleman H, et al. Interaction between polymorphisms in the renin–angiotensin-system and angiotensin-converting enzyme inhibitor or β-blocker use and the risk of myocardial infarction and stroke. Pharmacogenom J. 2008;8:400–407. doi: 10.1038/sj.tpj.6500493. [DOI] [PubMed] [Google Scholar]
- 94.Maitland-van der Zee AH, et al. Genetic variation in the renin–angiotensin system, use of renin–angiotensin system inhibitors and the risk of myocardial infarction. J Renin Angiotensin Aldosterone Syst. 2011;12:208–214. doi: 10.1177/1470320310391834. [DOI] [PubMed] [Google Scholar]
- 95.Psaty BM, et al. Diuretic therapy, the α-adducin gene variant, and the risk of myocardial infarction or stroke in persons with treated hypertension. JAMA. 2002;287:1680–1689. doi: 10.1001/jama.287.13.1680. [DOI] [PubMed] [Google Scholar]
- 96.Davis BR, et al. Antihypertensive therapy, the α-adducin polymorphism, and cardiovascular disease in high-risk hypertensive persons: the Genetics of Hypertension-Associated Treatment Study. Pharmacogenom J. 2007;7:112–122. doi: 10.1038/sj.tpj.6500395. [DOI] [PubMed] [Google Scholar]
- 97.Gerhard T, et al. α-adducin polymorphism associated with increased risk of adverse cardiovascular outcomes: results from GENEtic Substudy of the INternational VErapamil SR-trandolapril STudy (INVEST-GENES) Am Heart J. 2008;156:397–404. doi: 10.1016/j.ahj.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nordestgaard BG, et al. Effect of ACE insertion/deletion and 12 other polymorphisms on clinical outcomes and response to treatment in the LIFE study. Pharmacogenet Genom. 2010;20:77–85. doi: 10.1097/FPC.0b013e328333f70b. [DOI] [PubMed] [Google Scholar]
- 99.van Wieren-de Wijer DB, et al. Interaction between the Gly460Trp α-adducin gene variant and diuretics on the risk of myocardial infarction. J Hypertens. 2009;27:61–68. doi: 10.1097/hjh.0b013e328317a74d. [DOI] [PubMed] [Google Scholar]
- 100.Wikoff WR, et al. Pharmacometabolomics reveals racial differences in response to atenolol treatment. PLoS ONE. 2013;8:e57639. doi: 10.1371/journal.pone.0057639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cooper-Dehoff RM, et al. Is diabetes mellitus-linked amino acid signature associated with β-blocker-induced impaired fasting glucose? Circ Cardiovasc Genet. 2014;7:199–205. doi: 10.1161/CIRCGENETICS.113.000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.King’s College London. Ancestry and biological Informative Markers for stratification of HYpertension: the AIM HY study. Research Councils UK. 2015 [online], http://gtr.rcuk.ac.uk/project/8ED525DF-F1B0-4254-800B-85B4357EACAE.
- 103.Barrie ES, Smith RM, Sanford JC, Sadee W. mRNA transcript diversity creates new opportunities for pharmacological intervention. Mol Pharmacol. 2012;81:620–630. doi: 10.1124/mol.111.076604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rosendorff C, et al. Treatment of hypertension in patients with coronary artery disease: a scientific statement from the American Heart Association, American College of Cardiology, and American Society of Hypertension. Circulation. 2015;60:753–764. doi: 10.1161/CIR.0000000000000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Klein TE, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lluis-Ganella C, et al. Assessment of the value of a genetic risk score in improving the estimation of coronary risk. Atherosclerosis. 2012;222:456–463. doi: 10.1016/j.atherosclerosis.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Havulinna AS, et al. A blood pressure genetic risk score is a significant predictor of incident cardiovascular events in 32,669 individuals. Hypertension. 2013;61:987–994. doi: 10.1161/HYPERTENSIONAHA.111.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ganna A, et al. Multilocus genetic risk scores for coronary heart disease prediction. Arterioscler Thromb Vasc Biol. 2013;33:2267–2272. doi: 10.1161/ATVBAHA.113.301218. [DOI] [PubMed] [Google Scholar]
- 109.Malik R, et al. Multilocus genetic risk score associates with ischemic stroke in case-control and prospective cohort studies. Stroke. 2014;45:394–402. doi: 10.1161/STROKEAHA.113.002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tada H, et al. Twelve-single nucleotide polymorphism genetic risk score identifies individuals at increased risk for future atrial fibrillation and stroke. Stroke. 2014;45:2856–2862. doi: 10.1161/STROKEAHA.114.006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mabuchi H, Nohara A. Therapy: PCSK9 inhibitors for treating familial hypercholesterolaemia. Nat Rev Endocrinol. 2015;11:8–9. doi: 10.1038/nrendo.2014.205. [DOI] [PubMed] [Google Scholar]