The anhydrous morpholinium salts of 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) with 4-aminobenzoic acid, 3,5-dinitrobenzoic acid and 3,5-dinitrosalicylic acid, provide one example of a three-dimensional hydrogen-bonded network polymer and two of weakly inter-associated hydrogen-bonded cation–anion units.
Keywords: crystal structure; 1,8-diazabicyclo[5.4.0]undec-7-ene; BDU; benzoate salts; hydrogen bonding
Abstract
The anhydrous salts of the Lewis base 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) with 4-aminobenzoic acid [1-aza-8-azoniabicyclo[5.4.0]undec-7-ene 4-aminobenzoate, C9H17N2 +·C7H6NO2 − (I)], 3,5-dinitrobenzoic acid [1-aza-8-azoniabicyclo[5.4.0]undec-7-ene 3,5-dinitrobenzoate, C9H17N2 +·C7H3N2O6 −, (II)] and 3,5-dinitrosalicylic acid (DNSA) [1-aza-8-azoniabicyclo[5.4.0]undec-7-ene 2-hydroxy-3,5-dinitrobenzoate, C9H17N2 +·C7H3N2O7 −, (III)] have been determined and their hydrogen-bonded structures are described. In both (II) and (III), the DBU cations have a common disorder in three of the C atoms of the six-membered ring moieties [site-occupancy factors (SOF) = 0.735 (3)/0.265 (3) and 0.686 (4)/0.314 (4), respectively], while in (III), there is additional rotational disorder in the DNSA anion, giving two sites (SOF = 0.72/0.28, values fixed) for the phenol group. In the crystals of (I) and (III), the cation–anion pairs are linked through a primary N—H⋯Ocarboxyl hydrogen bond [2.665 (2) and 2.869 (3) Å, respectively]. In (II), the ion pairs are linked through an asymmetric three-centre R 1 2(4), N—H⋯O,O′ chelate association. In (I), structure extension is through amine N—H⋯Ocarboxyl hydrogen bonds between the PABA anions, giving a three-dimensional structure. The crystal structures of (II) and (III) are very similar, the cation–anion pairs being associated only through weak C—H⋯O hydrogen bonds, giving in both overall two-dimensional layered structures lying parallel to (001). No π–π ring associations are present in any of the structures.
Chemical context and database survey
The Lewis base 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) is an alkaloid isolated from the sponge Niphates digitalis (Regalado et al., 2010 ▸) but is commonly synthesized. It finds use as a curing agent for epoxy resins, as a catalyst in organic syntheses, and as a counter-cation in metal complex chemistry, e.g. with the pentabromo(triphenylphosphane)platinum(IV) monoanion (Motevalli et al., 1989 ▸). It has also found use in binding organic liquids (BOLs), which usually comprise a mixture of amidines or guanidine and alcohol, and are used to reversibly capture and release gases such as CO2, CS2, SO2 or COS (Shannon et al., 2015 ▸; Pérez et al., 2004 ▸; Heldebrant et al., 2009 ▸). The structure of one of these formed from the absorption of CO2 is the bicarbonate (Pérez et al., 2004 ▸).
As a very strong base (pK a ca 14), protonation of the N8 group of the six-membered hetero-ring of DBU is readily achieved and results in the formation of salts with carboxylic acids and phenols. The Cambridge Structural Database (2015 version) (Groom & Allen, 2014 ▸) contains 35 examples of organic salts of DBU, among them the benzyl dithiocarbonate (Heldebrant et al., 2009 ▸) and the phenolate from 2,6-di(tert-butyl)-4-nitrophenol (Lynch & McClenaghan, 2003 ▸). However, of the total there are surprisingly few carboxylate salts, e.g. with Kemp’s triacid (1,3,5-trimethylcyclohexane-1,3,5-tricarboxylic acid) (a monoanionic acetonitrile salt) (Huczyński et al., 2008 ▸) and the dianionic salt of the tetra(3-carboxyphenyl)-substituted porphyrin (Lipstman & Goldberg, 2013 ▸).
No reported crystal structures of salts with simple substituted benzoic acids are found, so in order to examine the hydrogen-bonding in crystals of the DBU salts with some common ring-substituted benzoic acids, a number of these were prepared. Suitable crystals were obtained with 4-aminobenzoic acid (PABA), (3,5-dinitrobenzoic acid (DNBA) and (3,5-dinitrosalicylic acid (DNSA), giving the anhydrous salts, C9H17N2
+ C7H6NO2
− (I), C9H17N2
+ C7H3N2O6
− (II) and C9H17N2
+ C7H3N2O7
− (III), respectively and their structures and hydrogen-bonding modes are reported herein.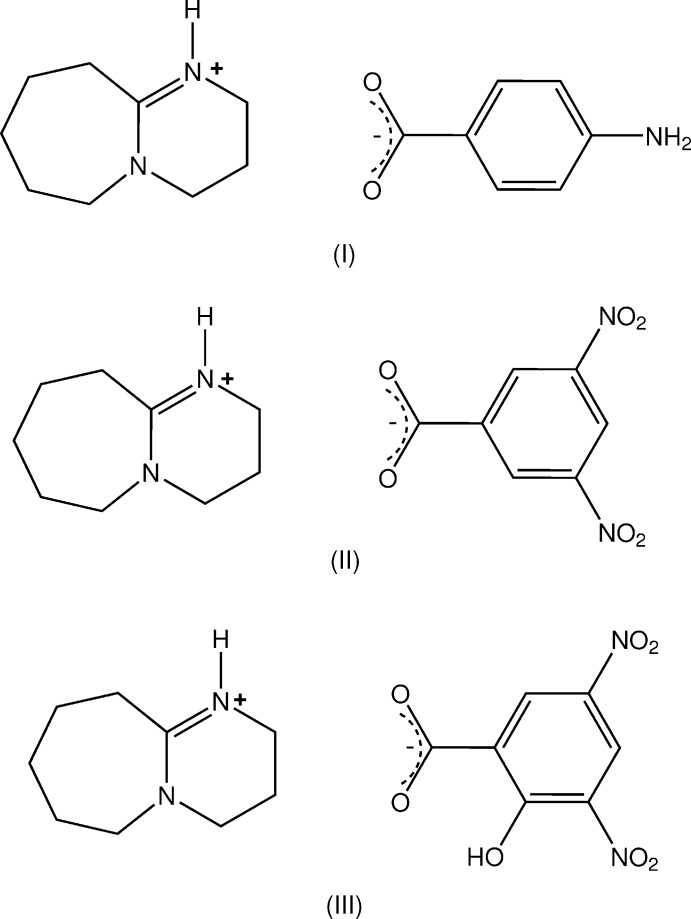
Structural commentary
The asymmetric units of (I)–(III) comprise a BDU cation (A) and a 4-aminobenzoate anion (B), (I) (Fig. 1 ▸), a 3,5-dinitrobenzoate anion (B), (II) (Fig. 2 ▸), and a 3,5-dinitrosalicylate anion (B), (III) (Fig. 3 ▸). The cation–anion pairs in (I) and (III) are linked through a primary N8A—H⋯Ocarboxyl hydrogen bond [2.665 (2) and 2.871 (3) Å, respectively; Tables 1 ▸ and 3 ▸]. In (II), the ion pairs are linked through an asymmetric three-centre  (4), N8A—H⋯O,O′ chelate association [2.777 (2), 3.117 (2) Å; Table 2 ▸]. With (III), the corresponding longer contact with the second carboxyl O12B atom is 3.222 (3) Å (Fig. 3 ▸).
(4), N8A—H⋯O,O′ chelate association [2.777 (2), 3.117 (2) Å; Table 2 ▸]. With (III), the corresponding longer contact with the second carboxyl O12B atom is 3.222 (3) Å (Fig. 3 ▸).
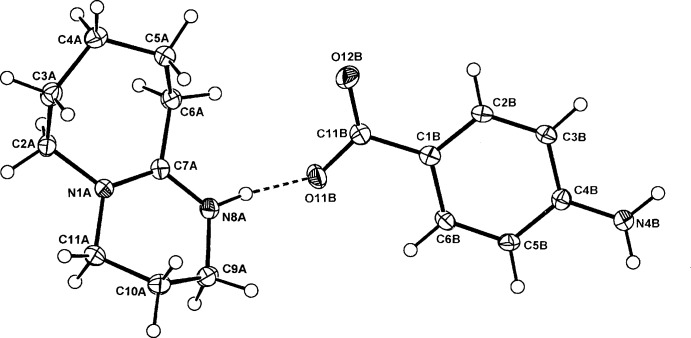
Figure 1.
The atom-numbering scheme and the molecular conformation of the DBU cation (A) and the PABA anion (B) in (I) with displacement ellipsoids drawn at the 40% probability level. The cation–anion hydrogen bond is shown as a dashed line.
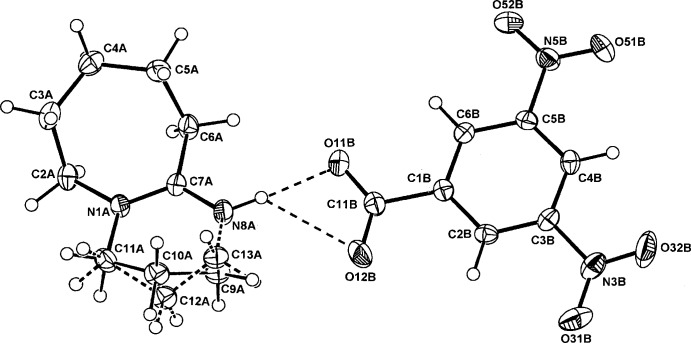
Figure 2.
The atom-numbering scheme and the molecular conformation of the DBU cation (A) and the DNBA anion (B) in (II) with displacement ellipsoids drawn at the 40% probability level. The bonds in the minor disordered section of the six-membered ring of the cation and the cation–anion hydrogen bonds are shown as dashed lines.
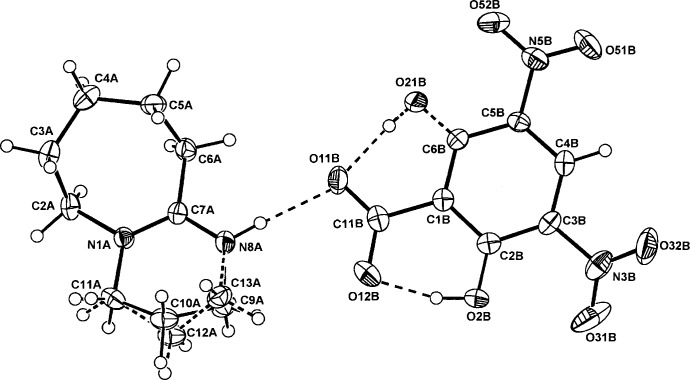
Figure 3.
The atom-numbering scheme and the molecular conformation of the DBU cation (A) and the DNSA anion (B) in (III) with displacement ellipsoids drawn at the 40% probability level. The bonds in the minor disordered section of the six-membered ring of the cation are shown as dashed lines.
Table 1. Hydrogen-bond geometry (Å, °) for (I) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N8A—H8A⋯O11B | 0.89 (2) | 1.78 (2) | 2.665 (2) | 170 (2) |
| N4B—H41B⋯O11B i | 0.89 (2) | 2.05 (2) | 2.939 (2) | 176 (2) |
| N4B—H42B⋯O12B ii | 0.92 (2) | 1.98 (2) | 2.891 (2) | 176 (2) |
Symmetry codes: (i) 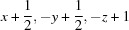 ; (ii)
; (ii) 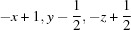 .
.
Table 3. Hydrogen-bond geometry (Å, °) for (III) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N8A—H8A⋯O11B | 0.88 (2) | 1.99 (2) | 2.871 (3) | 176 (2) |
| O2B—H2B⋯O12B | 0.84 | 1.72 | 2.473 (3) | 149 |
| C10A—H11A⋯O32B i | 0.99 | 2.45 | 3.251 (5) | 138 |
| C2A—H21A⋯O31B ii | 0.99 | 2.48 | 3.281 (3) | 138 |
Symmetry codes: (i) 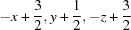 ; (ii)
; (ii) 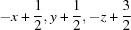 .
.
Table 2. Hydrogen-bond geometry (Å, °) for (II) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N8A—H8A⋯O11B | 0.90 (2) | 1.88 (2) | 2.777 (2) | 177 (2) |
| N8A—H8A⋯O12B | 0.90 (2) | 2.53 (2) | 3.117 (2) | 124 (1) |
| C10A—H11A⋯O32B i | 0.99 | 2.44 | 3.247 (3) | 138 |
| C2A—H21A⋯O31B ii | 0.99 | 2.56 | 3.309 (2) | 133 |
| C6A—H62A⋯O11B | 0.99 | 2.60 | 3.438 (2) | 143 |
Symmetry codes: (i) 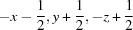 ; (ii)
; (ii) 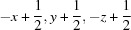 .
.
With the structures of (II) and (III), there is disorder in the six-membered ring system involving atoms C9A and C10A (with alternative minor occupancy sites C12A and C13A), giving similar site occupancy factors [SOF 0.735 (3)/0.265 (3) and 0.686 (4)/0.314 (4) for (II) and (III), respectively]. This feature is found in three other structures among the CSD set: the previously mentioned 2,6-di(tert-butyl)-4-nitrophenolate (SOF 0.60/0.40) (Lynch & McClenaghan, 2003 ▸); in the 8-bromoguanosine 8-bromoguanoside adduct salt (SOF = 0.63/0.37) (Saftić et al., 2012 ▸) and in the counter-cation of a bromocarbyne Mo complex (SOF = 0.83/0.17) (Cordiner et al., 2008 ▸).
With the PABA anion in (I), the carboxylate group is essentially coplanar with the benzene ring [torsion angle C2B—C1B—C11B— O11B = 179.25 (15)°, a feature similar to those found in the parent acid (Gracin & Fischer, 2005 ▸) and its co-crystals, e.g. with 4-nitrobenzoic acid (Bowers et al., 2005 ▸).
The carboxylate groups of the DNBA and DNSA anions in both (II) and (III) are also essentially coplanar with the benzene rings: torsion angles C2B—C1B—C11B—O11B = −176.60 (16) and −179.4 (2)°, respectively. The 5- and 3-substituted nitro groups are also either in-plane or out-of-plane [torsion angles C4B—C5B—N5B— O52B = 179.61 (16)° in (II) and −177.5 (2)° in (III) and C2B—C3B—N3B—O32B = −166.31 (17)° in (II) and −155.2 (2)° in (III)]. Also, in (III), the phenolic substituent group (O2B) is disordered by rotation about the C1B⋯C4B ring vector giving a minor site-occupancy factor for the O21B—H21B group of 0.28 (SOF fixed in the final refinement cycles). This is similar to the disorder in three examples among the DNSA proton-transfer salts with Lewis bases, e.g. with nicotinamide (SOF = 0.76/0.24) (Koman et al., 2003 ▸), with 2,6-diaminopyridine (0.90/0.10) (Smith et al., 2003 ▸) and with quinoline-2-carboxylic acid (0.51/0.49) (Smith et al., 2007 ▸). In (III), the usual short intramolecular phenol O—H⋯Ocarboxyl hydrogen bond is present (Table 3 ▸).
Supramolecular features
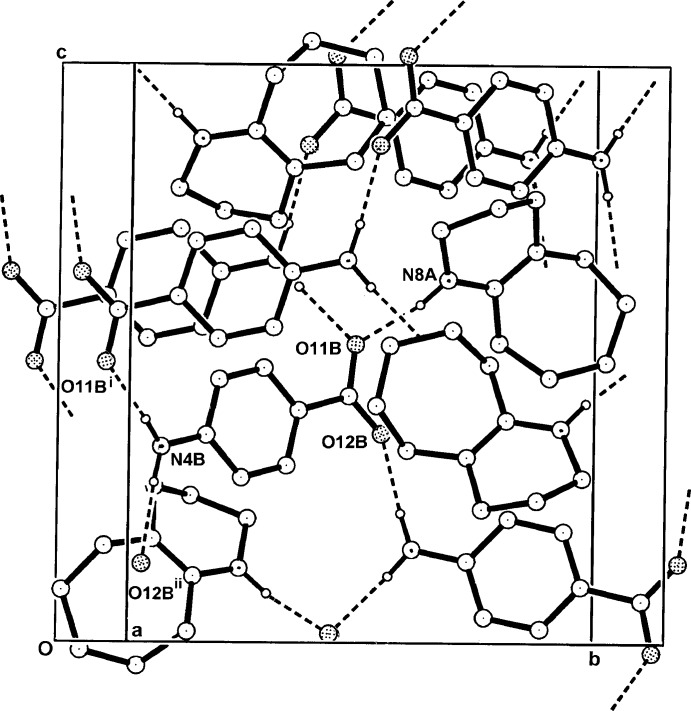
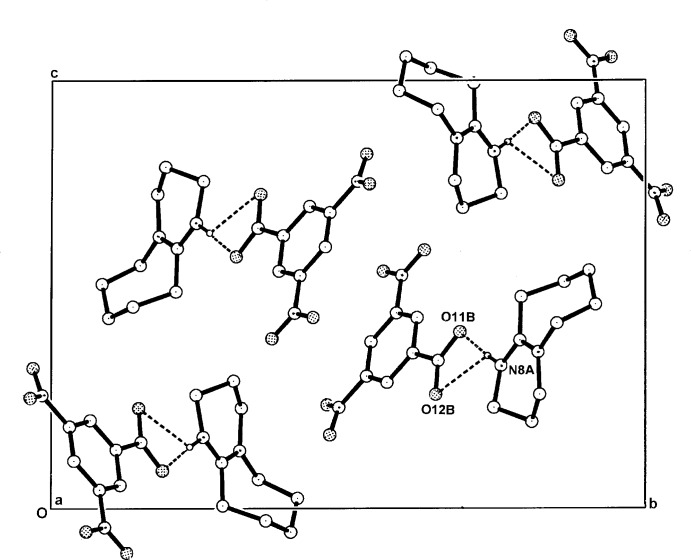
In the crystal of (I), the N8A—H⋯O11B hydrogen-bonded cation–anion pairs are extended through intermolecular N4B—H⋯ O11B i and ⋯N12B ii hydrogen-bonding extensions (Table 1 ▸), giving an overall three-dimensional network structure (Fig. 4 ▸). The structure contains no inter-ring π–π interactions or C—H⋯O hydrogen bonds.
Figure 4.
The three-dimensional hydrogen-bonded framework structure of (I) viewed approximately along a. For symmetry codes, see Table 1 ▸.
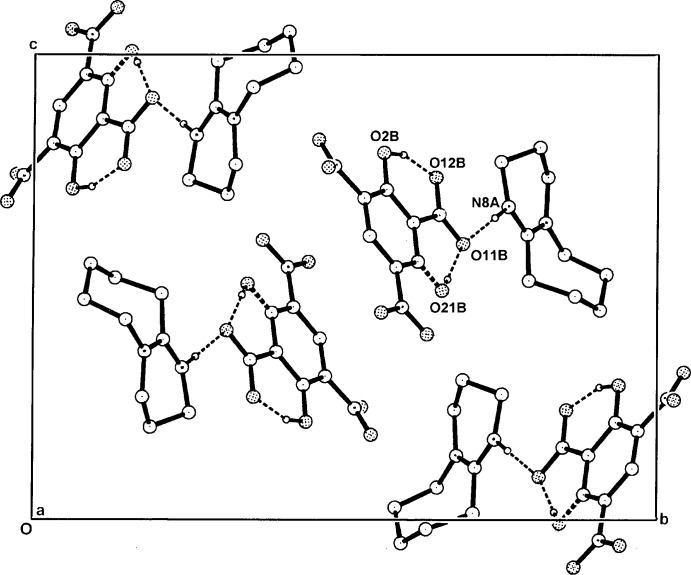
The unit-cell parameters, space group (Table 4 ▸), and the overall crystal packing of (II) and (III) are very similar (Figs. 5 ▸ and 6 ▸). Although no classical hydrogen-bonding interactions are present between the primary cation–anion pairs, with both structures there are two minor cation C—H⋯O hydrogen-bonding extensions to nitro O-atom acceptors, C2A—H⋯O31B ii [3.309 (2) Å in (II) and 3.281 (3) Å in (III)] and C10A—H⋯O32B i [3.247 (3) Å in (II) and 3.251 (5) Å in (III)] (Tables 2 ▸ and 3 ▸). These give two-dimensional layered structures lying parallel to (001). There are no inter-ring π–π interactions in either (II) or (III).
Table 4. Experimental details.
| (I) | (II) | (III) | |
|---|---|---|---|
| Crystal data | |||
| Chemical formula | C9H17N2 +·C7H6NO2 − | C9H17N2 +·C7H3N2O6 − | C9H17N2 +·C7H3N2O7 − |
| M r | 289.37 | 364.36 | 380.36 |
| Crystal system, space group | Orthorhombic, P212121 | Monoclinic, P21/n | Monoclinic, P21/n |
| Temperature (K) | 200 | 200 | 200 |
| a, b, c (Å) | 8.0986 (4), 12.9213 (6), 13.7344 (7) | 6.0197 (4), 19.6228 (13), 14.3866 (8) | 6.1537 (3), 19.1541 (14), 14.5527 (11) |
| α, β, γ (°) | 90, 90, 90 | 90, 98.078 (5), 90 | 90, 98.343 (6), 90 |
| V (Å3) | 1437.23 (12) | 1682.53 (18) | 1697.2 (2) |
| Z | 4 | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα |
| μ (mm−1) | 0.09 | 0.11 | 0.12 |
| Crystal size (mm) | 0.40 × 0.26 × 0.24 | 0.30 × 0.13 × 0.08 | 0.30 × 0.13 × 0.10 |
| Data collection | |||
| Diffractometer | Oxford Diffraction Gemini-S CCD-detector | Oxford Diffraction Gemini-S CCD-detector | Oxford Diffraction Gemini-S CCD-detector |
| Absorption correction | Multi-scan (CrysAlis PRO; Agilent, 2014 ▸) | Multi-scan (CrysAlis PRO; Agilent, 2014 ▸) | Multi-scan (CrysAlis PRO; Agilent, 2014 ▸) |
| T min, T max | 0.93, 0.99 | 0.90, 0.99 | 0.920, 0.990 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 7372, 3324, 2847 | 7082, 3311, 2561 | 7800, 3339, 2347 |
| R int | 0.031 | 0.024 | 0.034 |
| (sin θ/λ)max (Å−1) | 0.687 | 0.617 | 0.617 |
| Refinement | |||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.044, 0.098, 1.07 | 0.045, 0.109, 1.02 | 0.058, 0.123, 1.03 |
| No. of reflections | 3324 | 3311 | 3339 |
| No. of parameters | 199 | 245 | 263 |
| No. of restraints | 3 | 3 | 3 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.20, −0.25 | 0.18, −0.22 | 0.29, −0.29 |
Figure 5.
The packing of the hydrogen-bonded cation-anion pairs in the unit cell of (II), viewed along a. The minor-component disordered atoms and the non-associative H atoms have been omitted.
Figure 6.
The packing of the hydrogen-bonded cation-anion pairs in the unit cell of (III), viewed along a. The minor-component disordered atoms and the non-associative H atoms have been omitted.
Synthesis and crystallization
The title compounds (I)–(III) were prepared by first dissolving 100 mg of either PABA, DNBA, or DNSA in 5 mL of warm ethanol followed by the addition, with stirring, of 111 mg (I), 72 mg (II) or 67 mg (III) of BDU, respectively. Slow evaporation at room temperature gave colourless needles of (I), colourless prisms of (II), and fine yellow needles of (III), from which specimens were cleaved for the X-ray analyses.
Refinement details
Crystal data, data collection and structure refinement details are given in Table 4 ▸. Hydrogen atoms were placed in calculated positions [C—Haromatic = 0.95 Å or C—Hmethylene = 0.99 Å] and were allowed to ride in the refinements, with U iso(H) = 1.2U eq(C). The amine and aminium H-atoms were located in difference-Fourier analyses and were allowed to refine with distance restraints [N—H = 0.90 (2) Å] and with U iso(H) = 1.2U eq(N). Disorder involving atoms C9A and C10A of the six-membered ring systems of both (II) and (III) gave refined minor occupancy sites C12A and C13A, with site occupancy factors of 0.735 (3)/0.265 (3) and 0.686 (4)/0.314 (4), respectively. Also in (III), the phenol group of the DNSA anion was found to be disordered with the minor occupancy site (O21B) having a SOF = 0.28, which was fixed in the final cycles of refinement. In the structure of (I), although of no relevance in the achiral molecule, the Flack parameter (Flack, 1983 ▸) was determined as −0.1 (13) for 1668 Friedel pairs, which serves to indicate the lack of any usable anomalous scattering signal, as expected for an all-light-atom structure determined with Mo Kα X-rays.
Supplementary Material
Crystal structure: contains datablock(s) global, I, II, III. DOI: 10.1107/S205698901600267X/pk2574sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901600267X/pk2574Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S205698901600267X/pk2574IIsup3.hkl
Structure factors: contains datablock(s) III. DOI: 10.1107/S205698901600267X/pk2574IIIsup4.hkl
Supporting information file. DOI: 10.1107/S205698901600267X/pk2574Isup5.cml
Supporting information file. DOI: 10.1107/S205698901600267X/pk2574IIsup6.cml
Supporting information file. DOI: 10.1107/S205698901600267X/pk2574IIIsup7.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
GS acknowledges financial support from the Science and Engineering Faculty, Queensland University of Technology.
supplementary crystallographic information
(I) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 4-aminobenzoate. Crystal data
| C9H17N2+·C7H6NO2− | F(000) = 624 |
| Mr = 289.37 | Dx = 1.337 Mg m−3 |
| Orthorhombic, P212121 | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: P 2ac 2ab | Cell parameters from 2097 reflections |
| a = 8.0986 (4) Å | θ = 3.5–28.4° |
| b = 12.9213 (6) Å | µ = 0.09 mm−1 |
| c = 13.7344 (7) Å | T = 200 K |
| V = 1437.23 (12) Å3 | Prism, colourless |
| Z = 4 | 0.40 × 0.26 × 0.24 mm |
(I) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 4-aminobenzoate. Data collection
| Oxford Diffraction Gemini-S CCD-detector diffractometer | 3324 independent reflections |
| Radiation source: Enhance (Mo) X-ray source | 2847 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.031 |
| Detector resolution: 16.067 pixels mm-1 | θmax = 29.2°, θmin = 3.3° |
| ω scans | h = −10→10 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2014) | k = −16→15 |
| Tmin = 0.93, Tmax = 0.99 | l = −17→18 |
| 7372 measured reflections |
(I) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 4-aminobenzoate. Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.044 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.098 | w = 1/[σ2(Fo2) + (0.0438P)2 + 0.0476P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.07 | (Δ/σ)max < 0.001 |
| 3324 reflections | Δρmax = 0.20 e Å−3 |
| 199 parameters | Δρmin = −0.25 e Å−3 |
| 3 restraints | Absolute structure: Flack (1983), 1668 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Absolute structure parameter: −0.1 (13) |
(I) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 4-aminobenzoate. Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell esds are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
(I) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 4-aminobenzoate. Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1A | 0.32105 (18) | 0.84571 (11) | 0.67893 (11) | 0.0229 (4) | |
| N8A | 0.36282 (18) | 0.67732 (12) | 0.62864 (11) | 0.0241 (5) | |
| C2A | 0.2390 (2) | 0.94651 (14) | 0.66676 (13) | 0.0256 (5) | |
| C3A | 0.3174 (2) | 1.01454 (14) | 0.58999 (14) | 0.0291 (6) | |
| C4A | 0.2728 (2) | 0.98456 (14) | 0.48576 (14) | 0.0288 (6) | |
| C5A | 0.3145 (2) | 0.87339 (14) | 0.45932 (13) | 0.0271 (5) | |
| C6A | 0.2207 (2) | 0.79201 (14) | 0.51882 (13) | 0.0262 (5) | |
| C7A | 0.3028 (2) | 0.77086 (13) | 0.61456 (13) | 0.0209 (5) | |
| C9A | 0.4591 (2) | 0.64922 (14) | 0.71447 (13) | 0.0262 (5) | |
| C10A | 0.5429 (2) | 0.74497 (13) | 0.75333 (13) | 0.0280 (6) | |
| C11A | 0.4170 (2) | 0.82988 (15) | 0.76868 (13) | 0.0302 (6) | |
| O11B | 0.28719 (17) | 0.51621 (9) | 0.51597 (9) | 0.0320 (4) | |
| O12B | 0.29529 (19) | 0.56473 (11) | 0.36120 (11) | 0.0428 (5) | |
| N4B | 0.6170 (2) | 0.11141 (13) | 0.33808 (12) | 0.0296 (5) | |
| C1B | 0.3958 (2) | 0.39741 (13) | 0.40190 (12) | 0.0206 (5) | |
| C2B | 0.43611 (19) | 0.36990 (13) | 0.30648 (12) | 0.0212 (5) | |
| C3B | 0.5089 (2) | 0.27615 (13) | 0.28504 (12) | 0.0220 (5) | |
| C4B | 0.5475 (2) | 0.20495 (13) | 0.35867 (13) | 0.0220 (5) | |
| C5B | 0.5100 (2) | 0.23325 (13) | 0.45489 (12) | 0.0243 (5) | |
| C6B | 0.4347 (2) | 0.32664 (13) | 0.47496 (13) | 0.0227 (5) | |
| C11B | 0.3204 (2) | 0.50006 (14) | 0.42672 (14) | 0.0238 (5) | |
| H8A | 0.342 (2) | 0.6279 (14) | 0.5850 (13) | 0.0290* | |
| H10A | 0.59810 | 0.72880 | 0.81580 | 0.0340* | |
| H11A | 0.34180 | 0.81070 | 0.82260 | 0.0360* | |
| H12A | 0.47390 | 0.89490 | 0.78670 | 0.0360* | |
| H13A | 0.62810 | 0.76860 | 0.70660 | 0.0340* | |
| H21A | 0.24080 | 0.98360 | 0.72980 | 0.0310* | |
| H22A | 0.12200 | 0.93460 | 0.64920 | 0.0310* | |
| H31A | 0.43890 | 1.01140 | 0.59740 | 0.0350* | |
| H32A | 0.28290 | 1.08700 | 0.60140 | 0.0350* | |
| H41A | 0.15290 | 0.99540 | 0.47620 | 0.0350* | |
| H42A | 0.33170 | 1.03140 | 0.44050 | 0.0350* | |
| H51A | 0.43450 | 0.86260 | 0.46870 | 0.0320* | |
| H52A | 0.29000 | 0.86260 | 0.38940 | 0.0320* | |
| H61A | 0.10660 | 0.81660 | 0.53060 | 0.0310* | |
| H62A | 0.21410 | 0.72690 | 0.48100 | 0.0310* | |
| H91A | 0.54280 | 0.59660 | 0.69700 | 0.0310* | |
| H92A | 0.38570 | 0.61950 | 0.76490 | 0.0310* | |
| H2B | 0.41290 | 0.41690 | 0.25510 | 0.0250* | |
| H3B | 0.53320 | 0.25950 | 0.21920 | 0.0260* | |
| H5B | 0.53680 | 0.18760 | 0.50670 | 0.0290* | |
| H6B | 0.40870 | 0.34320 | 0.54060 | 0.0270* | |
| H41B | 0.666 (2) | 0.0742 (16) | 0.3845 (13) | 0.0360* | |
| H42B | 0.647 (2) | 0.0939 (16) | 0.2759 (12) | 0.0360* |
(I) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 4-aminobenzoate. Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1A | 0.0263 (7) | 0.0220 (8) | 0.0205 (7) | 0.0030 (6) | −0.0033 (6) | −0.0022 (6) |
| N8A | 0.0294 (8) | 0.0197 (8) | 0.0233 (8) | −0.0004 (6) | −0.0008 (6) | −0.0024 (7) |
| C2A | 0.0276 (9) | 0.0223 (9) | 0.0269 (10) | 0.0053 (7) | −0.0018 (7) | −0.0053 (8) |
| C3A | 0.0311 (10) | 0.0218 (9) | 0.0343 (11) | −0.0005 (8) | −0.0031 (8) | −0.0024 (8) |
| C4A | 0.0314 (10) | 0.0263 (9) | 0.0287 (10) | 0.0011 (8) | −0.0008 (8) | 0.0041 (8) |
| C5A | 0.0295 (9) | 0.0292 (10) | 0.0225 (9) | 0.0039 (8) | −0.0010 (7) | −0.0007 (8) |
| C6A | 0.0312 (9) | 0.0221 (9) | 0.0253 (9) | −0.0010 (7) | −0.0062 (8) | −0.0035 (8) |
| C7A | 0.0202 (8) | 0.0203 (9) | 0.0223 (9) | −0.0013 (7) | 0.0013 (7) | −0.0012 (7) |
| C9A | 0.0268 (9) | 0.0251 (9) | 0.0267 (10) | 0.0030 (8) | −0.0003 (7) | 0.0033 (8) |
| C10A | 0.0269 (9) | 0.0301 (10) | 0.0269 (10) | 0.0034 (8) | −0.0065 (8) | −0.0010 (8) |
| C11A | 0.0365 (11) | 0.0306 (10) | 0.0235 (9) | 0.0053 (8) | −0.0094 (8) | −0.0056 (8) |
| O11B | 0.0520 (8) | 0.0207 (7) | 0.0233 (7) | −0.0025 (6) | 0.0074 (6) | −0.0035 (5) |
| O12B | 0.0643 (9) | 0.0337 (8) | 0.0305 (8) | 0.0187 (7) | 0.0123 (7) | 0.0099 (7) |
| N4B | 0.0406 (9) | 0.0260 (9) | 0.0223 (9) | 0.0072 (7) | 0.0000 (7) | −0.0008 (7) |
| C1B | 0.0194 (8) | 0.0215 (9) | 0.0209 (9) | −0.0045 (6) | −0.0004 (7) | 0.0005 (7) |
| C2B | 0.0225 (9) | 0.0237 (9) | 0.0174 (8) | −0.0015 (7) | −0.0013 (6) | 0.0033 (7) |
| C3B | 0.0240 (8) | 0.0241 (8) | 0.0179 (8) | −0.0032 (7) | 0.0001 (7) | 0.0010 (7) |
| C4B | 0.0216 (8) | 0.0195 (9) | 0.0250 (9) | −0.0025 (7) | −0.0011 (7) | −0.0019 (7) |
| C5B | 0.0322 (9) | 0.0221 (9) | 0.0185 (8) | 0.0004 (8) | −0.0011 (7) | 0.0046 (7) |
| C6B | 0.0288 (9) | 0.0228 (9) | 0.0166 (8) | −0.0043 (7) | 0.0018 (7) | −0.0017 (7) |
| C11B | 0.0255 (9) | 0.0221 (9) | 0.0237 (9) | −0.0043 (7) | 0.0028 (7) | 0.0001 (7) |
(I) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 4-aminobenzoate. Geometric parameters (Å, º)
| O11B—C11B | 1.272 (2) | C4A—H41A | 0.9900 |
| O12B—C11B | 1.245 (2) | C5A—H52A | 0.9900 |
| N1A—C11A | 1.471 (2) | C5A—H51A | 0.9900 |
| N1A—C7A | 1.319 (2) | C6A—H61A | 0.9900 |
| N1A—C2A | 1.472 (2) | C6A—H62A | 0.9900 |
| N8A—C7A | 1.317 (2) | C9A—H91A | 0.9900 |
| N8A—C9A | 1.459 (2) | C9A—H92A | 0.9900 |
| N8A—H8A | 0.892 (18) | C10A—H10A | 0.9900 |
| N4B—C4B | 1.363 (2) | C10A—H13A | 0.9900 |
| N4B—H41B | 0.892 (18) | C11A—H12A | 0.9900 |
| N4B—H42B | 0.916 (17) | C11A—H11A | 0.9900 |
| C2A—C3A | 1.513 (3) | C1B—C11B | 1.499 (2) |
| C3A—C4A | 1.526 (3) | C1B—C2B | 1.397 (2) |
| C4A—C5A | 1.520 (3) | C1B—C6B | 1.394 (2) |
| C5A—C6A | 1.533 (2) | C2B—C3B | 1.379 (2) |
| C6A—C7A | 1.499 (2) | C3B—C4B | 1.402 (2) |
| C9A—C10A | 1.509 (2) | C4B—C5B | 1.404 (2) |
| C10A—C11A | 1.513 (2) | C5B—C6B | 1.380 (2) |
| C2A—H21A | 0.9900 | C2B—H2B | 0.9500 |
| C2A—H22A | 0.9900 | C3B—H3B | 0.9500 |
| C3A—H31A | 0.9900 | C5B—H5B | 0.9500 |
| C3A—H32A | 0.9900 | C6B—H6B | 0.9500 |
| C4A—H42A | 0.9900 | ||
| C2A—N1A—C7A | 121.49 (15) | C5A—C6A—H62A | 109.00 |
| C2A—N1A—C11A | 117.17 (14) | H61A—C6A—H62A | 108.00 |
| C7A—N1A—C11A | 121.26 (15) | C7A—C6A—H62A | 109.00 |
| C7A—N8A—C9A | 122.97 (15) | C7A—C6A—H61A | 109.00 |
| C7A—N8A—H8A | 119.3 (11) | C10A—C9A—H91A | 110.00 |
| C9A—N8A—H8A | 117.8 (12) | N8A—C9A—H92A | 110.00 |
| C4B—N4B—H41B | 120.9 (13) | N8A—C9A—H91A | 110.00 |
| H41B—N4B—H42B | 114.5 (17) | C10A—C9A—H92A | 110.00 |
| C4B—N4B—H42B | 121.5 (13) | H91A—C9A—H92A | 108.00 |
| N1A—C2A—C3A | 113.83 (14) | C9A—C10A—H10A | 110.00 |
| C2A—C3A—C4A | 114.02 (15) | H10A—C10A—H13A | 108.00 |
| C3A—C4A—C5A | 114.29 (15) | C9A—C10A—H13A | 110.00 |
| C4A—C5A—C6A | 114.26 (14) | C11A—C10A—H10A | 110.00 |
| C5A—C6A—C7A | 111.90 (14) | C11A—C10A—H13A | 110.00 |
| N8A—C7A—C6A | 117.38 (15) | N1A—C11A—H11A | 110.00 |
| N1A—C7A—N8A | 122.23 (16) | N1A—C11A—H12A | 110.00 |
| N1A—C7A—C6A | 120.28 (15) | C10A—C11A—H11A | 110.00 |
| N8A—C9A—C10A | 108.79 (14) | H11A—C11A—H12A | 108.00 |
| C9A—C10A—C11A | 109.93 (14) | C10A—C11A—H12A | 110.00 |
| N1A—C11A—C10A | 109.88 (14) | C6B—C1B—C11B | 120.58 (15) |
| C3A—C2A—H22A | 109.00 | C2B—C1B—C11B | 122.25 (15) |
| H21A—C2A—H22A | 108.00 | C2B—C1B—C6B | 117.12 (15) |
| N1A—C2A—H22A | 109.00 | C1B—C2B—C3B | 121.59 (15) |
| C3A—C2A—H21A | 109.00 | C2B—C3B—C4B | 121.17 (15) |
| N1A—C2A—H21A | 109.00 | N4B—C4B—C3B | 121.61 (16) |
| C2A—C3A—H31A | 109.00 | N4B—C4B—C5B | 121.03 (16) |
| C2A—C3A—H32A | 109.00 | C3B—C4B—C5B | 117.36 (15) |
| H31A—C3A—H32A | 108.00 | C4B—C5B—C6B | 120.75 (16) |
| C4A—C3A—H31A | 109.00 | C1B—C6B—C5B | 122.00 (16) |
| C4A—C3A—H32A | 109.00 | O11B—C11B—O12B | 123.50 (17) |
| C3A—C4A—H42A | 109.00 | O11B—C11B—C1B | 116.76 (16) |
| H41A—C4A—H42A | 108.00 | O12B—C11B—C1B | 119.74 (17) |
| C5A—C4A—H41A | 109.00 | C1B—C2B—H2B | 119.00 |
| C5A—C4A—H42A | 109.00 | C3B—C2B—H2B | 119.00 |
| C3A—C4A—H41A | 109.00 | C2B—C3B—H3B | 119.00 |
| C6A—C5A—H52A | 109.00 | C4B—C3B—H3B | 119.00 |
| H51A—C5A—H52A | 108.00 | C4B—C5B—H5B | 120.00 |
| C4A—C5A—H52A | 109.00 | C6B—C5B—H5B | 120.00 |
| C6A—C5A—H51A | 109.00 | C1B—C6B—H6B | 119.00 |
| C4A—C5A—H51A | 109.00 | C5B—C6B—H6B | 119.00 |
| C5A—C6A—H61A | 109.00 | ||
| C7A—N1A—C2A—C3A | −74.8 (2) | N8A—C9A—C10A—C11A | 52.82 (18) |
| C11A—N1A—C2A—C3A | 108.53 (17) | C9A—C10A—C11A—N1A | −52.69 (19) |
| C2A—N1A—C7A—N8A | −173.67 (15) | C6B—C1B—C2B—C3B | 1.0 (2) |
| C2A—N1A—C7A—C6A | 10.2 (2) | C11B—C1B—C2B—C3B | 178.48 (15) |
| C11A—N1A—C7A—N8A | 2.9 (3) | C2B—C1B—C6B—C5B | 0.1 (2) |
| C11A—N1A—C7A—C6A | −173.23 (15) | C11B—C1B—C6B—C5B | −177.44 (15) |
| C2A—N1A—C11A—C10A | −157.91 (14) | C2B—C1B—C11B—O11B | 179.25 (15) |
| C7A—N1A—C11A—C10A | 25.4 (2) | C2B—C1B—C11B—O12B | −1.6 (3) |
| C9A—N8A—C7A—N1A | −2.2 (3) | C6B—C1B—C11B—O11B | −3.4 (2) |
| C9A—N8A—C7A—C6A | 174.06 (15) | C6B—C1B—C11B—O12B | 175.83 (16) |
| C7A—N8A—C9A—C10A | −26.7 (2) | C1B—C2B—C3B—C4B | −0.9 (2) |
| N1A—C2A—C3A—C4A | 77.87 (18) | C2B—C3B—C4B—N4B | 178.92 (16) |
| C2A—C3A—C4A—C5A | −56.71 (19) | C2B—C3B—C4B—C5B | −0.4 (2) |
| C3A—C4A—C5A—C6A | 62.97 (19) | N4B—C4B—C5B—C6B | −177.86 (16) |
| C4A—C5A—C6A—C7A | −83.76 (18) | C3B—C4B—C5B—C6B | 1.4 (2) |
| C5A—C6A—C7A—N1A | 60.9 (2) | C4B—C5B—C6B—C1B | −1.3 (3) |
| C5A—C6A—C7A—N8A | −115.39 (17) |
(I) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 4-aminobenzoate. Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N8A—H8A···O11B | 0.89 (2) | 1.78 (2) | 2.665 (2) | 170 (2) |
| N4B—H41B···O11Bi | 0.89 (2) | 2.05 (2) | 2.939 (2) | 176 (2) |
| N4B—H42B···O12Bii | 0.92 (2) | 1.98 (2) | 2.891 (2) | 176 (2) |
Symmetry codes: (i) x+1/2, −y+1/2, −z+1; (ii) −x+1, y−1/2, −z+1/2.
(II) Aza-8-azoniabicyclo[5.4.0]undec-7-ene 3,5-dinitrobenzoate. Crystal data
| C9H17N2+·C7H3N2O6− | F(000) = 768 |
| Mr = 364.36 | Dx = 1.438 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 1784 reflections |
| a = 6.0197 (4) Å | θ = 4.0–28.0° |
| b = 19.6228 (13) Å | µ = 0.11 mm−1 |
| c = 14.3866 (8) Å | T = 200 K |
| β = 98.078 (5)° | Needle, colourless |
| V = 1682.53 (18) Å3 | 0.30 × 0.13 × 0.08 mm |
| Z = 4 |
(II) Aza-8-azoniabicyclo[5.4.0]undec-7-ene 3,5-dinitrobenzoate. Data collection
| Oxford Diffraction Gemini-S CCD-detector diffractometer | 3311 independent reflections |
| Radiation source: fine-focus sealed tube | 2561 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.024 |
| Detector resolution: 16.077 pixels mm-1 | θmax = 26.0°, θmin = 3.4° |
| ω scans | h = −7→7 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2014) | k = −14→24 |
| Tmin = 0.90, Tmax = 0.99 | l = −9→17 |
| 7082 measured reflections |
(II) Aza-8-azoniabicyclo[5.4.0]undec-7-ene 3,5-dinitrobenzoate. Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.045 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.109 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.01 | w = 1/[σ2(Fo2) + (0.0435P)2 + 0.5615P] where P = (Fo2 + 2Fc2)/3 |
| 3311 reflections | (Δ/σ)max < 0.001 |
| 245 parameters | Δρmax = 0.18 e Å−3 |
| 3 restraints | Δρmin = −0.22 e Å−3 |
(II) Aza-8-azoniabicyclo[5.4.0]undec-7-ene 3,5-dinitrobenzoate. Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell esds are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
(II) Aza-8-azoniabicyclo[5.4.0]undec-7-ene 3,5-dinitrobenzoate. Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O11B | −0.0061 (2) | 0.68797 (7) | 0.41185 (9) | 0.0408 (4) | |
| O12B | −0.0380 (2) | 0.64765 (8) | 0.26602 (10) | 0.0556 (5) | |
| O31B | −0.5921 (3) | 0.46963 (8) | 0.17213 (10) | 0.0567 (5) | |
| O32B | −0.8865 (3) | 0.46500 (9) | 0.24178 (11) | 0.0703 (6) | |
| O51B | −0.8471 (2) | 0.55899 (8) | 0.55381 (10) | 0.0514 (5) | |
| O52B | −0.5787 (3) | 0.62813 (8) | 0.60351 (10) | 0.0576 (6) | |
| N3B | −0.6966 (3) | 0.48464 (8) | 0.23584 (11) | 0.0416 (5) | |
| N5B | −0.6770 (3) | 0.59011 (8) | 0.54409 (10) | 0.0363 (5) | |
| C1B | −0.2967 (3) | 0.60944 (8) | 0.36270 (11) | 0.0264 (5) | |
| C2B | −0.3972 (3) | 0.56537 (8) | 0.29419 (11) | 0.0288 (5) | |
| C3B | −0.5892 (3) | 0.53100 (8) | 0.30888 (11) | 0.0289 (5) | |
| C4B | −0.6880 (3) | 0.53844 (8) | 0.38905 (12) | 0.0293 (5) | |
| C5B | −0.5807 (3) | 0.58130 (8) | 0.45649 (11) | 0.0265 (5) | |
| C6B | −0.3873 (3) | 0.61637 (8) | 0.44556 (11) | 0.0269 (5) | |
| C11B | −0.0952 (3) | 0.65174 (9) | 0.34516 (13) | 0.0327 (5) | |
| N1A | 0.6514 (2) | 0.81846 (7) | 0.36517 (9) | 0.0288 (4) | |
| N8A | 0.3270 (3) | 0.75625 (8) | 0.33371 (10) | 0.0364 (5) | |
| C2A | 0.8281 (3) | 0.85009 (9) | 0.43221 (13) | 0.0350 (6) | |
| C3A | 0.7557 (3) | 0.91508 (9) | 0.47621 (13) | 0.0378 (6) | |
| C4A | 0.6172 (3) | 0.90383 (10) | 0.55531 (13) | 0.0390 (6) | |
| C5A | 0.4046 (3) | 0.86226 (10) | 0.52884 (13) | 0.0383 (6) | |
| C6A | 0.4433 (3) | 0.78996 (9) | 0.49381 (11) | 0.0334 (5) | |
| C7A | 0.4773 (3) | 0.78797 (8) | 0.39270 (11) | 0.0262 (5) | |
| C9A | 0.3565 (6) | 0.74500 (17) | 0.2353 (2) | 0.0357 (10) | 0.735 (3) |
| C10A | 0.4681 (5) | 0.80757 (15) | 0.20241 (17) | 0.0364 (8) | 0.735 (3) |
| C11A | 0.6839 (3) | 0.82115 (10) | 0.26593 (12) | 0.0364 (6) | |
| C13A | 0.3000 (16) | 0.7705 (5) | 0.2305 (8) | 0.0357 (10) | 0.265 (3) |
| C12A | 0.5368 (12) | 0.7669 (4) | 0.2074 (5) | 0.0364 (8) | 0.265 (3) |
| H2B | −0.33470 | 0.55890 | 0.23780 | 0.0350* | |
| H4B | −0.82260 | 0.51530 | 0.39720 | 0.0350* | |
| H6B | −0.31700 | 0.64500 | 0.49430 | 0.0320* | |
| H8A | 0.217 (3) | 0.7342 (9) | 0.3570 (12) | 0.0440* | |
| H10A | 0.49920 | 0.80070 | 0.13730 | 0.0440* | 0.735 (3) |
| H21A | 0.95830 | 0.86030 | 0.39950 | 0.0420* | |
| H22A | 0.87810 | 0.81700 | 0.48270 | 0.0420* | |
| H31A | 0.66700 | 0.94280 | 0.42680 | 0.0450* | |
| H32A | 0.89130 | 0.94160 | 0.50080 | 0.0450* | |
| H41A | 0.57520 | 0.94880 | 0.57870 | 0.0470* | |
| H42A | 0.71200 | 0.88050 | 0.60760 | 0.0470* | |
| H51A | 0.30550 | 0.88690 | 0.47920 | 0.0460* | |
| H52A | 0.32480 | 0.85900 | 0.58440 | 0.0460* | |
| H61A | 0.57690 | 0.77030 | 0.53250 | 0.0400* | |
| H62A | 0.31250 | 0.76120 | 0.50230 | 0.0400* | |
| H91A | 0.20920 | 0.73760 | 0.19650 | 0.0430* | 0.735 (3) |
| H92A | 0.45120 | 0.70430 | 0.22990 | 0.0430* | 0.735 (3) |
| H11A | 0.36680 | 0.84730 | 0.20280 | 0.0440* | 0.735 (3) |
| H12A | 0.79710 | 0.78680 | 0.25410 | 0.0440* | 0.735 (3) |
| H13A | 0.74150 | 0.86670 | 0.25170 | 0.0440* | 0.735 (3) |
| H14A | 0.53670 | 0.77550 | 0.13960 | 0.0440* | 0.265 (3) |
| H15A | 0.59940 | 0.72090 | 0.22210 | 0.0440* | 0.265 (3) |
| H16A | 0.23430 | 0.81620 | 0.21620 | 0.0430* | 0.265 (3) |
| H17A | 0.20330 | 0.73590 | 0.19490 | 0.0430* | 0.265 (3) |
| H18A | 0.84390 | 0.81290 | 0.26060 | 0.0440* | 0.265 (3) |
| H19A | 0.64380 | 0.86710 | 0.24050 | 0.0440* | 0.265 (3) |
(II) Aza-8-azoniabicyclo[5.4.0]undec-7-ene 3,5-dinitrobenzoate. Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O11B | 0.0376 (7) | 0.0403 (8) | 0.0454 (7) | −0.0139 (6) | 0.0090 (6) | −0.0054 (6) |
| O12B | 0.0558 (9) | 0.0740 (11) | 0.0419 (8) | −0.0258 (8) | 0.0238 (7) | −0.0043 (7) |
| O31B | 0.0690 (10) | 0.0566 (10) | 0.0423 (8) | 0.0063 (8) | 0.0003 (7) | −0.0213 (7) |
| O32B | 0.0733 (11) | 0.0811 (12) | 0.0539 (9) | −0.0503 (10) | −0.0006 (8) | −0.0091 (9) |
| O51B | 0.0472 (8) | 0.0586 (9) | 0.0543 (8) | −0.0128 (7) | 0.0276 (7) | 0.0036 (7) |
| O52B | 0.0663 (10) | 0.0729 (11) | 0.0380 (8) | −0.0175 (8) | 0.0228 (7) | −0.0229 (8) |
| N3B | 0.0534 (11) | 0.0353 (9) | 0.0329 (8) | −0.0074 (8) | −0.0055 (8) | −0.0016 (7) |
| N5B | 0.0392 (9) | 0.0378 (9) | 0.0343 (8) | −0.0006 (7) | 0.0132 (7) | 0.0021 (7) |
| C1B | 0.0250 (8) | 0.0242 (8) | 0.0298 (8) | 0.0011 (7) | 0.0034 (7) | 0.0032 (7) |
| C2B | 0.0342 (9) | 0.0280 (9) | 0.0251 (8) | 0.0037 (7) | 0.0070 (7) | 0.0017 (7) |
| C3B | 0.0338 (9) | 0.0244 (9) | 0.0268 (8) | −0.0015 (7) | −0.0021 (7) | −0.0001 (7) |
| C4B | 0.0263 (8) | 0.0258 (9) | 0.0352 (9) | −0.0016 (7) | 0.0021 (7) | 0.0055 (8) |
| C5B | 0.0284 (8) | 0.0259 (9) | 0.0264 (8) | 0.0035 (7) | 0.0078 (7) | 0.0013 (7) |
| C6B | 0.0275 (8) | 0.0247 (9) | 0.0279 (8) | −0.0004 (7) | 0.0020 (7) | −0.0018 (7) |
| C11B | 0.0297 (9) | 0.0309 (9) | 0.0382 (10) | −0.0006 (8) | 0.0072 (8) | 0.0031 (8) |
| N1A | 0.0255 (7) | 0.0325 (8) | 0.0285 (7) | −0.0044 (6) | 0.0039 (6) | 0.0020 (6) |
| N8A | 0.0358 (8) | 0.0484 (10) | 0.0250 (7) | −0.0178 (7) | 0.0045 (6) | −0.0008 (7) |
| C2A | 0.0236 (9) | 0.0388 (10) | 0.0410 (10) | −0.0059 (8) | −0.0007 (8) | 0.0001 (8) |
| C3A | 0.0347 (10) | 0.0330 (10) | 0.0434 (10) | −0.0076 (8) | −0.0020 (8) | 0.0000 (9) |
| C4A | 0.0405 (10) | 0.0399 (11) | 0.0343 (9) | −0.0025 (8) | −0.0032 (8) | −0.0064 (9) |
| C5A | 0.0357 (10) | 0.0495 (12) | 0.0302 (9) | −0.0046 (9) | 0.0062 (8) | −0.0083 (8) |
| C6A | 0.0357 (10) | 0.0403 (10) | 0.0237 (8) | −0.0113 (8) | 0.0024 (7) | 0.0050 (8) |
| C7A | 0.0254 (8) | 0.0250 (8) | 0.0272 (8) | −0.0015 (7) | 0.0006 (7) | 0.0028 (7) |
| C9A | 0.0418 (19) | 0.041 (2) | 0.0236 (10) | −0.0039 (14) | 0.0026 (12) | −0.0018 (16) |
| C10A | 0.0443 (15) | 0.0390 (16) | 0.0263 (10) | −0.0024 (12) | 0.0061 (10) | 0.0036 (12) |
| C11A | 0.0348 (10) | 0.0427 (11) | 0.0339 (9) | −0.0043 (8) | 0.0126 (8) | 0.0047 (8) |
| C13A | 0.0418 (19) | 0.041 (2) | 0.0236 (10) | −0.0039 (14) | 0.0026 (12) | −0.0018 (16) |
| C12A | 0.0443 (15) | 0.0390 (16) | 0.0263 (10) | −0.0024 (12) | 0.0061 (10) | 0.0036 (12) |
(II) Aza-8-azoniabicyclo[5.4.0]undec-7-ene 3,5-dinitrobenzoate. Geometric parameters (Å, º)
| O11B—C11B | 1.253 (2) | C5A—C6A | 1.534 (3) |
| O12B—C11B | 1.238 (2) | C6A—C7A | 1.498 (2) |
| O31B—N3B | 1.218 (2) | C9A—C10A | 1.507 (4) |
| O32B—N3B | 1.221 (3) | C10A—C11A | 1.503 (3) |
| O51B—N5B | 1.217 (2) | C12A—C13A | 1.510 (12) |
| O52B—N5B | 1.223 (2) | C12A—C11A | 1.555 (8) |
| N3B—C3B | 1.469 (2) | C2A—H21A | 0.9900 |
| N5B—C5B | 1.470 (2) | C2A—H22A | 0.9900 |
| N1A—C2A | 1.469 (2) | C3A—H31A | 0.9900 |
| N1A—C7A | 1.315 (2) | C3A—H32A | 0.9900 |
| N1A—C11A | 1.469 (2) | C4A—H41A | 0.9900 |
| N8A—C13A | 1.497 (11) | C4A—H42A | 0.9900 |
| N8A—C9A | 1.468 (3) | C5A—H51A | 0.9900 |
| N8A—C7A | 1.308 (2) | C5A—H52A | 0.9900 |
| N8A—H8A | 0.895 (18) | C6A—H61A | 0.9900 |
| C1B—C11B | 1.520 (3) | C6A—H62A | 0.9900 |
| C1B—C2B | 1.385 (2) | C9A—H91A | 0.9900 |
| C1B—C6B | 1.386 (2) | C9A—H92A | 0.9900 |
| C2B—C3B | 1.380 (2) | C10A—H10A | 0.9900 |
| C3B—C4B | 1.378 (2) | C10A—H11A | 0.9900 |
| C4B—C5B | 1.375 (2) | C11A—H12A | 0.9900 |
| C5B—C6B | 1.380 (2) | C11A—H13A | 0.9900 |
| C2B—H2B | 0.9500 | C12A—H14A | 0.9900 |
| C4B—H4B | 0.9500 | C12A—H15A | 0.9900 |
| C6B—H6B | 0.9500 | C13A—H16A | 0.9900 |
| C2A—C3A | 1.515 (3) | C13A—H17A | 0.9900 |
| C3A—C4A | 1.518 (3) | C11A—H18A | 0.9900 |
| C4A—C5A | 1.520 (3) | C11A—H19A | 0.9900 |
| O31B—N3B—O32B | 124.33 (17) | C3A—C2A—H22A | 109.00 |
| O31B—N3B—C3B | 117.77 (17) | H21A—C2A—H22A | 108.00 |
| O32B—N3B—C3B | 117.89 (16) | C2A—C3A—H31A | 109.00 |
| O51B—N5B—O52B | 123.92 (16) | C2A—C3A—H32A | 109.00 |
| O51B—N5B—C5B | 118.63 (15) | C4A—C3A—H31A | 109.00 |
| O52B—N5B—C5B | 117.44 (16) | C4A—C3A—H32A | 109.00 |
| C2A—N1A—C11A | 116.09 (13) | H31A—C3A—H32A | 108.00 |
| C7A—N1A—C11A | 121.98 (14) | C3A—C4A—H41A | 109.00 |
| C2A—N1A—C7A | 121.91 (14) | C3A—C4A—H42A | 109.00 |
| C7A—N8A—C9A | 122.1 (2) | C5A—C4A—H41A | 108.00 |
| C7A—N8A—C13A | 121.6 (4) | C5A—C4A—H42A | 108.00 |
| C13A—N8A—H8A | 118.6 (12) | H41A—C4A—H42A | 108.00 |
| C9A—N8A—H8A | 119.0 (11) | C4A—C5A—H51A | 109.00 |
| C7A—N8A—H8A | 117.9 (11) | C4A—C5A—H52A | 109.00 |
| C2B—C1B—C6B | 119.23 (16) | C6A—C5A—H51A | 109.00 |
| C2B—C1B—C11B | 120.13 (15) | C6A—C5A—H52A | 109.00 |
| C6B—C1B—C11B | 120.60 (15) | H51A—C5A—H52A | 108.00 |
| C1B—C2B—C3B | 119.19 (15) | C5A—C6A—H61A | 109.00 |
| N3B—C3B—C2B | 119.14 (15) | C5A—C6A—H62A | 109.00 |
| N3B—C3B—C4B | 117.78 (16) | C7A—C6A—H61A | 109.00 |
| C2B—C3B—C4B | 123.08 (15) | C7A—C6A—H62A | 109.00 |
| C3B—C4B—C5B | 116.13 (16) | H61A—C6A—H62A | 108.00 |
| C4B—C5B—C6B | 123.01 (16) | N8A—C9A—H91A | 110.00 |
| N5B—C5B—C4B | 118.32 (16) | N8A—C9A—H92A | 110.00 |
| N5B—C5B—C6B | 118.67 (14) | C10A—C9A—H91A | 110.00 |
| C1B—C6B—C5B | 119.30 (15) | C10A—C9A—H92A | 110.00 |
| O11B—C11B—C1B | 116.66 (16) | H91A—C9A—H92A | 108.00 |
| O11B—C11B—O12B | 126.65 (17) | C9A—C10A—H10A | 110.00 |
| O12B—C11B—C1B | 116.68 (16) | C9A—C10A—H11A | 110.00 |
| C3B—C2B—H2B | 120.00 | C11A—C10A—H10A | 110.00 |
| C1B—C2B—H2B | 120.00 | C11A—C10A—H11A | 110.00 |
| C3B—C4B—H4B | 122.00 | H10A—C10A—H11A | 108.00 |
| C5B—C4B—H4B | 122.00 | N1A—C11A—H12A | 109.00 |
| C5B—C6B—H6B | 120.00 | N1A—C11A—H13A | 109.00 |
| C1B—C6B—H6B | 120.00 | C10A—C11A—H12A | 109.00 |
| N1A—C2A—C3A | 113.94 (15) | C10A—C11A—H13A | 109.00 |
| C2A—C3A—C4A | 114.29 (15) | H12A—C11A—H13A | 108.00 |
| C3A—C4A—C5A | 114.95 (15) | C13A—C12A—H14A | 110.00 |
| C4A—C5A—C6A | 114.65 (15) | C13A—C12A—H15A | 110.00 |
| C5A—C6A—C7A | 113.01 (14) | H14A—C12A—H15A | 108.00 |
| N1A—C7A—N8A | 121.94 (15) | N8A—C13A—H16A | 111.00 |
| N1A—C7A—C6A | 120.27 (15) | N8A—C13A—H17A | 111.00 |
| N8A—C7A—C6A | 117.79 (16) | C12A—C13A—H16A | 111.00 |
| N8A—C9A—C10A | 107.5 (2) | C12A—C13A—H17A | 111.00 |
| C9A—C10A—C11A | 109.8 (2) | H16A—C13A—H17A | 109.00 |
| N1A—C11A—C10A | 111.27 (16) | N1A—C11A—H18A | 109.00 |
| N8A—C13A—C12A | 103.6 (7) | N1A—C11A—H19A | 109.00 |
| N1A—C2A—H21A | 109.00 | C12A—C11A—H18A | 109.00 |
| N1A—C2A—H22A | 109.00 | C12A—C11A—H19A | 109.00 |
| C3A—C2A—H21A | 109.00 | H18A—C11A—H19A | 108.00 |
| O31B—N3B—C3B—C2B | 12.0 (2) | C6B—C1B—C11B—O11B | 5.9 (2) |
| O31B—N3B—C3B—C4B | −168.62 (16) | C6B—C1B—C11B—O12B | −173.03 (16) |
| O32B—N3B—C3B—C2B | −166.31 (17) | C11B—C1B—C2B—C3B | −175.52 (15) |
| O32B—N3B—C3B—C4B | 13.1 (2) | C2B—C1B—C6B—C5B | −2.6 (2) |
| O51B—N5B—C5B—C4B | 0.3 (2) | C11B—C1B—C6B—C5B | 174.94 (15) |
| O51B—N5B—C5B—C6B | −179.75 (16) | C1B—C2B—C3B—N3B | 179.56 (15) |
| O52B—N5B—C5B—C4B | 179.61 (16) | C1B—C2B—C3B—C4B | 0.2 (3) |
| O52B—N5B—C5B—C6B | −0.5 (2) | C2B—C3B—C4B—C5B | −1.7 (2) |
| C2A—N1A—C11A—C10A | −162.56 (17) | N3B—C3B—C4B—C5B | 178.91 (15) |
| C7A—N1A—C2A—C3A | −71.6 (2) | C3B—C4B—C5B—C6B | 1.1 (2) |
| C11A—N1A—C2A—C3A | 110.16 (17) | C3B—C4B—C5B—N5B | −178.96 (15) |
| C2A—N1A—C7A—N8A | −175.79 (16) | N5B—C5B—C6B—C1B | −178.93 (15) |
| C2A—N1A—C7A—C6A | 5.5 (2) | C4B—C5B—C6B—C1B | 1.0 (3) |
| C11A—N1A—C7A—N8A | 2.4 (3) | N1A—C2A—C3A—C4A | 78.97 (19) |
| C11A—N1A—C7A—C6A | −176.35 (15) | C2A—C3A—C4A—C5A | −57.1 (2) |
| C7A—N1A—C11A—C10A | 19.2 (2) | C3A—C4A—C5A—C6A | 60.0 (2) |
| C9A—N8A—C7A—C6A | −173.3 (2) | C4A—C5A—C6A—C7A | −81.00 (19) |
| C9A—N8A—C7A—N1A | 7.9 (3) | C5A—C6A—C7A—N1A | 63.5 (2) |
| C7A—N8A—C9A—C10A | −37.5 (3) | C5A—C6A—C7A—N8A | −115.29 (18) |
| C6B—C1B—C2B—C3B | 2.0 (2) | N8A—C9A—C10A—C11A | 55.9 (3) |
| C2B—C1B—C11B—O11B | −176.60 (16) | C9A—C10A—C11A—N1A | −48.3 (3) |
| C2B—C1B—C11B—O12B | 4.4 (2) |
(II) Aza-8-azoniabicyclo[5.4.0]undec-7-ene 3,5-dinitrobenzoate. Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N8A—H8A···O11B | 0.90 (2) | 1.88 (2) | 2.777 (2) | 177 (2) |
| N8A—H8A···O12B | 0.90 (2) | 2.53 (2) | 3.117 (2) | 124 (1) |
| C10A—H11A···O32Bi | 0.99 | 2.44 | 3.247 (3) | 138 |
| C11A—H13A···O52Bii | 0.99 | 2.52 | 3.071 (2) | 115 |
| C2A—H21A···O31Biii | 0.99 | 2.56 | 3.309 (2) | 133 |
| C6A—H62A···O11B | 0.99 | 2.60 | 3.438 (2) | 143 |
| C9A—H91A···O12B | 0.99 | 2.60 | 3.127 (4) | 114 |
Symmetry codes: (i) −x−1/2, y+1/2, −z+1/2; (ii) x+3/2, −y+3/2, z−1/2; (iii) −x+1/2, y+1/2, −z+1/2.
(III) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 2-hydroxy-3,5-dinitrobenzoate. Crystal data
| C9H17N2+·C7H3N2O7− | F(000) = 800 |
| Mr = 380.36 | Dx = 1.489 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 1891 reflections |
| a = 6.1537 (3) Å | θ = 3.5–26.6° |
| b = 19.1541 (14) Å | µ = 0.12 mm−1 |
| c = 14.5527 (11) Å | T = 200 K |
| β = 98.343 (6)° | Needle, yellow |
| V = 1697.2 (2) Å3 | 0.30 × 0.13 × 0.10 mm |
| Z = 4 |
(III) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 2-hydroxy-3,5-dinitrobenzoate. Data collection
| Oxford Diffraction Gemini-S CCD-detector diffractometer | 3339 independent reflections |
| Radiation source: Enhance (Mo) X-ray source | 2347 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.034 |
| Detector resolution: 16.077 pixels mm-1 | θmax = 26.0°, θmin = 3.4° |
| ω scans | h = −7→7 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2014) | k = −23→23 |
| Tmin = 0.920, Tmax = 0.990 | l = −17→17 |
| 7800 measured reflections |
(III) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 2-hydroxy-3,5-dinitrobenzoate. Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.058 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.123 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0374P)2 + 0.7569P] where P = (Fo2 + 2Fc2)/3 |
| 3339 reflections | (Δ/σ)max < 0.001 |
| 263 parameters | Δρmax = 0.29 e Å−3 |
| 3 restraints | Δρmin = −0.29 e Å−3 |
(III) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 2-hydroxy-3,5-dinitrobenzoate. Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell esds are taken into account in the estimation of distances, angles and torsion angles |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
(III) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 2-hydroxy-3,5-dinitrobenzoate. Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O2B | 0.8426 (4) | 0.56153 (13) | 0.78929 (15) | 0.0433 (8) | 0.720 |
| O11B | 0.5084 (3) | 0.68879 (9) | 0.59293 (13) | 0.0433 (6) | |
| O12B | 0.5450 (3) | 0.64525 (10) | 0.73596 (13) | 0.0522 (7) | |
| O31B | 1.1116 (4) | 0.45700 (12) | 0.81707 (17) | 0.0819 (10) | |
| O32B | 1.4080 (4) | 0.47261 (13) | 0.75765 (15) | 0.0761 (9) | |
| O51B | 1.3286 (3) | 0.55867 (11) | 0.44585 (14) | 0.0594 (7) | |
| O52B | 1.0707 (4) | 0.63206 (11) | 0.39870 (14) | 0.0670 (8) | |
| N3B | 1.2118 (4) | 0.48306 (12) | 0.76028 (16) | 0.0467 (8) | |
| N5B | 1.1654 (3) | 0.59169 (11) | 0.45698 (15) | 0.0407 (7) | |
| C1B | 0.8002 (3) | 0.60950 (11) | 0.63899 (16) | 0.0268 (7) | |
| C2B | 0.9062 (3) | 0.56600 (12) | 0.70947 (16) | 0.0297 (7) | |
| C3B | 1.0956 (4) | 0.53052 (12) | 0.69146 (16) | 0.0310 (7) | |
| C4B | 1.1816 (3) | 0.53943 (11) | 0.61041 (16) | 0.0308 (7) | |
| C5B | 1.0735 (3) | 0.58226 (11) | 0.54278 (15) | 0.0276 (7) | |
| C6B | 0.8810 (3) | 0.61671 (11) | 0.55531 (15) | 0.0263 (7) | |
| C11B | 0.6029 (4) | 0.65080 (12) | 0.65595 (19) | 0.0346 (8) | |
| O21B | 0.7762 (10) | 0.6571 (3) | 0.4915 (5) | 0.052 (3) | 0.280 |
| N1A | −0.1524 (3) | 0.82026 (10) | 0.63820 (13) | 0.0293 (6) | |
| N8A | 0.1714 (3) | 0.76040 (11) | 0.67301 (14) | 0.0369 (7) | |
| C2A | −0.3262 (3) | 0.85087 (13) | 0.56984 (17) | 0.0357 (8) | |
| C3A | −0.2606 (4) | 0.91805 (13) | 0.52684 (18) | 0.0397 (8) | |
| C4A | −0.1188 (4) | 0.90797 (14) | 0.45044 (17) | 0.0409 (8) | |
| C5A | 0.0934 (4) | 0.86814 (13) | 0.48033 (17) | 0.0393 (9) | |
| C6A | 0.0612 (4) | 0.79368 (13) | 0.51409 (16) | 0.0340 (8) | |
| C7A | 0.0226 (3) | 0.79083 (11) | 0.61294 (15) | 0.0265 (7) | |
| C9A | 0.1399 (9) | 0.7478 (2) | 0.7696 (4) | 0.0366 (18) | 0.686 (4) |
| C10A | 0.0234 (6) | 0.8111 (2) | 0.8005 (3) | 0.0379 (11) | 0.686 (4) |
| C11A | −0.1871 (4) | 0.82349 (13) | 0.73612 (16) | 0.0363 (8) | |
| C13A | 0.189 (2) | 0.7738 (7) | 0.7752 (11) | 0.0366 (18) | 0.314 (4) |
| C12A | −0.0464 (13) | 0.7704 (5) | 0.7958 (6) | 0.0379 (11) | 0.314 (4) |
| H4B | 1.31350 | 0.51650 | 0.60110 | 0.0370* | |
| H6B | 0.80240 | 0.64380 | 0.50700 | 0.0320* | 0.720 |
| H2B | 0.73870 | 0.58950 | 0.79190 | 0.0650* | 0.720 |
| H21B | 0.66080 | 0.67200 | 0.50930 | 0.0770* | 0.280 |
| H61B | 0.85460 | 0.56120 | 0.76770 | 0.0360* | 0.280 |
| H8A | 0.280 (3) | 0.7394 (11) | 0.6508 (15) | 0.0320* | |
| H10A | −0.00890 | 0.80380 | 0.86450 | 0.0460* | 0.686 (4) |
| H21A | −0.45670 | 0.85990 | 0.60060 | 0.0430* | |
| H22A | −0.36910 | 0.81640 | 0.51980 | 0.0430* | |
| H31A | −0.17940 | 0.94750 | 0.57630 | 0.0480* | |
| H32A | −0.39530 | 0.94360 | 0.50080 | 0.0480* | |
| H41A | −0.20570 | 0.88270 | 0.39820 | 0.0490* | |
| H42A | −0.08210 | 0.95440 | 0.42720 | 0.0490* | |
| H51A | 0.18300 | 0.89440 | 0.53080 | 0.0470* | |
| H52A | 0.17720 | 0.86610 | 0.42730 | 0.0470* | |
| H61A | −0.06570 | 0.77230 | 0.47440 | 0.0410* | |
| H62A | 0.19310 | 0.76570 | 0.50720 | 0.0410* | |
| H91A | 0.28350 | 0.74140 | 0.80920 | 0.0440* | 0.686 (4) |
| H92A | 0.05040 | 0.70540 | 0.77390 | 0.0440* | 0.686 (4) |
| H11A | 0.11950 | 0.85260 | 0.80070 | 0.0460* | 0.686 (4) |
| H12A | −0.29640 | 0.78780 | 0.74760 | 0.0440* | 0.686 (4) |
| H13A | −0.24650 | 0.86990 | 0.74910 | 0.0440* | 0.686 (4) |
| H14A | −0.10610 | 0.72290 | 0.78230 | 0.0460* | 0.314 (4) |
| H15A | −0.04950 | 0.78040 | 0.86230 | 0.0460* | 0.314 (4) |
| H16A | 0.25340 | 0.82040 | 0.79120 | 0.0440* | 0.314 (4) |
| H17A | 0.28080 | 0.73790 | 0.81100 | 0.0440* | 0.314 (4) |
| H18A | −0.34390 | 0.81460 | 0.74010 | 0.0440* | 0.314 (4) |
| H19A | −0.15090 | 0.87100 | 0.76060 | 0.0440* | 0.314 (4) |
(III) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 2-hydroxy-3,5-dinitrobenzoate. Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O2B | 0.0472 (13) | 0.0563 (16) | 0.0297 (14) | 0.0095 (12) | 0.0168 (11) | 0.0083 (12) |
| O11B | 0.0356 (9) | 0.0369 (10) | 0.0579 (12) | 0.0124 (8) | 0.0087 (8) | 0.0063 (9) |
| O12B | 0.0492 (11) | 0.0640 (13) | 0.0484 (12) | 0.0080 (10) | 0.0245 (9) | −0.0040 (10) |
| O31B | 0.0774 (15) | 0.0847 (18) | 0.0749 (17) | −0.0226 (13) | −0.0181 (12) | 0.0525 (14) |
| O32B | 0.0708 (15) | 0.0894 (18) | 0.0629 (15) | 0.0492 (13) | −0.0082 (11) | 0.0045 (12) |
| O51B | 0.0542 (11) | 0.0636 (13) | 0.0680 (14) | 0.0066 (10) | 0.0346 (10) | −0.0115 (11) |
| O52B | 0.0918 (15) | 0.0703 (15) | 0.0456 (13) | 0.0168 (13) | 0.0323 (11) | 0.0224 (11) |
| N3B | 0.0592 (15) | 0.0338 (13) | 0.0411 (14) | 0.0003 (11) | −0.0127 (12) | 0.0001 (11) |
| N5B | 0.0464 (12) | 0.0388 (13) | 0.0405 (13) | −0.0040 (10) | 0.0189 (10) | −0.0056 (11) |
| C1B | 0.0259 (11) | 0.0216 (12) | 0.0323 (13) | −0.0038 (9) | 0.0021 (9) | −0.0035 (10) |
| C2B | 0.0341 (12) | 0.0270 (13) | 0.0280 (13) | −0.0057 (10) | 0.0044 (10) | −0.0019 (10) |
| C3B | 0.0361 (12) | 0.0241 (12) | 0.0300 (14) | −0.0006 (10) | −0.0049 (10) | 0.0015 (10) |
| C4B | 0.0256 (11) | 0.0245 (12) | 0.0406 (15) | −0.0010 (10) | −0.0006 (10) | −0.0054 (11) |
| C5B | 0.0297 (11) | 0.0257 (12) | 0.0285 (13) | −0.0056 (10) | 0.0077 (10) | −0.0023 (10) |
| C6B | 0.0288 (11) | 0.0214 (12) | 0.0275 (13) | −0.0017 (9) | −0.0001 (9) | 0.0021 (10) |
| C11B | 0.0292 (12) | 0.0284 (13) | 0.0466 (16) | −0.0028 (10) | 0.0068 (11) | −0.0055 (12) |
| O21B | 0.043 (4) | 0.059 (5) | 0.053 (4) | 0.008 (3) | 0.009 (3) | 0.025 (4) |
| N1A | 0.0254 (9) | 0.0319 (11) | 0.0304 (11) | 0.0014 (8) | 0.0038 (8) | −0.0007 (9) |
| N8A | 0.0333 (11) | 0.0476 (13) | 0.0299 (12) | 0.0157 (10) | 0.0051 (9) | 0.0034 (10) |
| C2A | 0.0254 (11) | 0.0378 (14) | 0.0427 (15) | 0.0059 (10) | 0.0006 (10) | −0.0026 (12) |
| C3A | 0.0358 (13) | 0.0339 (14) | 0.0464 (16) | 0.0066 (11) | −0.0044 (11) | 0.0003 (12) |
| C4A | 0.0442 (14) | 0.0384 (15) | 0.0365 (15) | −0.0024 (12) | −0.0061 (11) | 0.0074 (12) |
| C5A | 0.0370 (13) | 0.0508 (17) | 0.0308 (14) | −0.0005 (12) | 0.0075 (10) | 0.0080 (12) |
| C6A | 0.0340 (12) | 0.0413 (15) | 0.0262 (13) | 0.0081 (11) | 0.0029 (10) | −0.0052 (11) |
| C7A | 0.0270 (11) | 0.0226 (12) | 0.0291 (13) | −0.0006 (9) | 0.0014 (9) | −0.0035 (10) |
| C9A | 0.042 (3) | 0.037 (4) | 0.0292 (18) | 0.001 (2) | 0.000 (2) | 0.005 (3) |
| C10A | 0.047 (2) | 0.041 (2) | 0.0263 (17) | −0.0052 (17) | 0.0070 (16) | −0.0033 (19) |
| C11A | 0.0363 (13) | 0.0419 (15) | 0.0335 (14) | −0.0014 (11) | 0.0150 (11) | −0.0058 (12) |
| C13A | 0.042 (3) | 0.037 (4) | 0.0292 (18) | 0.001 (2) | 0.000 (2) | 0.005 (3) |
| C12A | 0.047 (2) | 0.041 (2) | 0.0263 (17) | −0.0052 (17) | 0.0070 (16) | −0.0033 (19) |
(III) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 2-hydroxy-3,5-dinitrobenzoate. Geometric parameters (Å, º)
| O2B—C2B | 1.281 (3) | C3A—C4A | 1.522 (4) |
| O11B—C11B | 1.247 (3) | C4A—C5A | 1.520 (4) |
| O12B—C11B | 1.271 (3) | C5A—C6A | 1.531 (4) |
| O21B—C6B | 1.305 (7) | C6A—C7A | 1.493 (3) |
| O31B—N3B | 1.208 (3) | C9A—C10A | 1.510 (6) |
| O32B—N3B | 1.230 (4) | C10A—C11A | 1.503 (5) |
| O51B—N5B | 1.217 (3) | C12A—C13A | 1.523 (15) |
| O52B—N5B | 1.230 (3) | C2A—H21A | 0.9900 |
| O2B—H2B | 0.8400 | C2A—H22A | 0.9900 |
| O21B—H21B | 0.8400 | C3A—H31A | 0.9900 |
| N3B—C3B | 1.460 (3) | C3A—H32A | 0.9900 |
| N5B—C5B | 1.455 (3) | C4A—H41A | 0.9900 |
| N1A—C2A | 1.473 (3) | C4A—H42A | 0.9900 |
| N1A—C11A | 1.473 (3) | C5A—H51A | 0.9900 |
| N1A—C7A | 1.314 (3) | C5A—H52A | 0.9900 |
| N8A—C9A | 1.467 (6) | C6A—H61A | 0.9900 |
| N8A—C13A | 1.498 (16) | C6A—H62A | 0.9900 |
| N8A—C7A | 1.308 (3) | C9A—H91A | 0.9900 |
| N8A—H8A | 0.88 (2) | C9A—H92A | 0.9900 |
| C1B—C11B | 1.499 (3) | C10A—H10A | 0.9900 |
| C1B—C6B | 1.387 (3) | C10A—H11A | 0.9900 |
| C1B—C2B | 1.406 (3) | C11A—H12A | 0.9900 |
| C2B—C3B | 1.406 (3) | C11A—H13A | 0.9900 |
| C3B—C4B | 1.372 (3) | C12A—H14A | 0.9900 |
| C4B—C5B | 1.376 (3) | C12A—H15A | 0.9900 |
| C5B—C6B | 1.391 (3) | C13A—H16A | 0.9900 |
| C2B—H61B | 0.9500 | C13A—H17A | 0.9900 |
| C4B—H4B | 0.9500 | C11A—H18A | 0.9900 |
| C6B—H6B | 0.9500 | C11A—H19A | 0.9900 |
| C2A—C3A | 1.511 (3) | ||
| C2B—O2B—H2B | 109.00 | C9A—C10A—C11A | 110.2 (3) |
| C6B—O21B—H21B | 110.00 | N1A—C11A—C10A | 111.3 (2) |
| O32B—N3B—C3B | 117.7 (2) | N8A—C13A—C12A | 104.7 (9) |
| O31B—N3B—O32B | 123.7 (2) | N1A—C2A—H21A | 109.00 |
| O31B—N3B—C3B | 118.6 (2) | N1A—C2A—H22A | 109.00 |
| O52B—N5B—C5B | 117.7 (2) | C3A—C2A—H21A | 109.00 |
| O51B—N5B—C5B | 118.7 (2) | C3A—C2A—H22A | 109.00 |
| O51B—N5B—O52B | 123.5 (2) | H21A—C2A—H22A | 108.00 |
| C2A—N1A—C11A | 116.36 (18) | C2A—C3A—H31A | 109.00 |
| C7A—N1A—C11A | 121.90 (19) | C2A—C3A—H32A | 109.00 |
| C2A—N1A—C7A | 121.74 (19) | C4A—C3A—H31A | 109.00 |
| C7A—N8A—C13A | 122.0 (5) | C4A—C3A—H32A | 109.00 |
| C7A—N8A—C9A | 122.5 (3) | H31A—C3A—H32A | 108.00 |
| C9A—N8A—H8A | 119.3 (14) | C3A—C4A—H41A | 109.00 |
| C13A—N8A—H8A | 119.6 (15) | C3A—C4A—H42A | 109.00 |
| C7A—N8A—H8A | 117.0 (14) | C5A—C4A—H41A | 109.00 |
| C2B—C1B—C6B | 120.78 (18) | C5A—C4A—H42A | 109.00 |
| C2B—C1B—C11B | 119.6 (2) | H41A—C4A—H42A | 108.00 |
| C6B—C1B—C11B | 119.6 (2) | C4A—C5A—H51A | 109.00 |
| O2B—C2B—C1B | 122.0 (2) | C4A—C5A—H52A | 109.00 |
| C1B—C2B—C3B | 117.4 (2) | C6A—C5A—H51A | 109.00 |
| O2B—C2B—C3B | 120.5 (2) | C6A—C5A—H52A | 109.00 |
| N3B—C3B—C4B | 117.1 (2) | H51A—C5A—H52A | 108.00 |
| C2B—C3B—C4B | 122.3 (2) | C5A—C6A—H61A | 109.00 |
| N3B—C3B—C2B | 120.6 (2) | C5A—C6A—H62A | 109.00 |
| C3B—C4B—C5B | 118.84 (19) | C7A—C6A—H61A | 109.00 |
| C4B—C5B—C6B | 121.43 (19) | C7A—C6A—H62A | 109.00 |
| N5B—C5B—C4B | 118.69 (18) | H61A—C6A—H62A | 108.00 |
| N5B—C5B—C6B | 119.88 (19) | N8A—C9A—H91A | 110.00 |
| O21B—C6B—C1B | 118.7 (3) | N8A—C9A—H92A | 110.00 |
| O21B—C6B—C5B | 122.0 (3) | C10A—C9A—H91A | 110.00 |
| C1B—C6B—C5B | 119.23 (19) | C10A—C9A—H92A | 110.00 |
| O12B—C11B—C1B | 116.6 (2) | H91A—C9A—H92A | 109.00 |
| O11B—C11B—C1B | 119.3 (2) | C9A—C10A—H10A | 110.00 |
| O11B—C11B—O12B | 124.1 (2) | C9A—C10A—H11A | 110.00 |
| C3B—C2B—H61B | 121.00 | C11A—C10A—H10A | 110.00 |
| C1B—C2B—H61B | 122.00 | C11A—C10A—H11A | 110.00 |
| C5B—C4B—H4B | 121.00 | H10A—C10A—H11A | 108.00 |
| C3B—C4B—H4B | 121.00 | N1A—C11A—H12A | 109.00 |
| C1B—C6B—H6B | 120.00 | N1A—C11A—H13A | 109.00 |
| C5B—C6B—H6B | 121.00 | C10A—C11A—H12A | 109.00 |
| N1A—C2A—C3A | 114.04 (18) | C10A—C11A—H13A | 109.00 |
| C2A—C3A—C4A | 114.2 (2) | H12A—C11A—H13A | 108.00 |
| C3A—C4A—C5A | 114.5 (2) | C13A—C12A—H15A | 110.00 |
| C4A—C5A—C6A | 114.4 (2) | H14A—C12A—H15A | 108.00 |
| C5A—C6A—C7A | 112.9 (2) | N8A—C13A—H16A | 111.00 |
| N1A—C7A—N8A | 121.8 (2) | N8A—C13A—H17A | 111.00 |
| N1A—C7A—C6A | 120.35 (19) | C12A—C13A—H16A | 111.00 |
| N8A—C7A—C6A | 117.82 (19) | C12A—C13A—H17A | 111.00 |
| N8A—C9A—C10A | 106.7 (3) | H16A—C13A—H17A | 109.00 |
| O31B—N3B—C3B—C2B | 23.9 (3) | C6B—C1B—C11B—O11B | 3.1 (3) |
| O31B—N3B—C3B—C4B | −157.2 (2) | C6B—C1B—C11B—O12B | −175.8 (2) |
| O32B—N3B—C3B—C2B | −155.2 (2) | C2B—C1B—C11B—O11B | −179.4 (2) |
| O32B—N3B—C3B—C4B | 23.8 (3) | C2B—C1B—C11B—O12B | 1.8 (3) |
| O51B—N5B—C5B—C4B | 3.7 (3) | C11B—C1B—C6B—C5B | 175.2 (2) |
| O51B—N5B—C5B—C6B | −176.8 (2) | O2B—C2B—C3B—N3B | 5.6 (4) |
| O52B—N5B—C5B—C4B | −177.5 (2) | O2B—C2B—C3B—C4B | −173.3 (2) |
| O52B—N5B—C5B—C6B | 2.0 (3) | C1B—C2B—C3B—N3B | −178.5 (2) |
| C2A—N1A—C7A—N8A | −176.4 (2) | C1B—C2B—C3B—C4B | 2.7 (3) |
| C2A—N1A—C7A—C6A | 6.0 (3) | C2B—C3B—C4B—C5B | −2.9 (3) |
| C11A—N1A—C7A—N8A | 2.7 (3) | N3B—C3B—C4B—C5B | 178.3 (2) |
| C11A—N1A—C7A—C6A | −175.0 (2) | C3B—C4B—C5B—C6B | 0.4 (3) |
| C2A—N1A—C11A—C10A | −163.0 (2) | C3B—C4B—C5B—N5B | 179.9 (2) |
| C7A—N1A—C2A—C3A | −71.7 (3) | N5B—C5B—C6B—C1B | −177.36 (19) |
| C11A—N1A—C2A—C3A | 109.2 (2) | C4B—C5B—C6B—C1B | 2.1 (3) |
| C7A—N1A—C11A—C10A | 17.9 (3) | N1A—C2A—C3A—C4A | 78.8 (3) |
| C7A—N8A—C9A—C10A | −38.8 (4) | C2A—C3A—C4A—C5A | −57.5 (3) |
| C9A—N8A—C7A—C6A | −173.2 (3) | C3A—C4A—C5A—C6A | 61.0 (3) |
| C9A—N8A—C7A—N1A | 9.1 (4) | C4A—C5A—C6A—C7A | −82.0 (3) |
| C6B—C1B—C2B—C3B | 0.0 (3) | C5A—C6A—C7A—N1A | 63.3 (3) |
| C11B—C1B—C2B—O2B | −1.6 (3) | C5A—C6A—C7A—N8A | −114.5 (2) |
| C11B—C1B—C2B—C3B | −177.5 (2) | N8A—C9A—C10A—C11A | 56.2 (4) |
| C2B—C1B—C6B—C5B | −2.3 (3) | C9A—C10A—C11A—N1A | −47.7 (4) |
| C6B—C1B—C2B—O2B | 175.9 (2) |
(III) 1-Aza-8-azoniabicyclo[5.4.0]undec-7-ene 2-hydroxy-3,5-dinitrobenzoate. Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N8A—H8A···O11B | 0.88 (2) | 1.99 (2) | 2.871 (3) | 176 (2) |
| O2B—H2B···O12B | 0.84 | 1.72 | 2.473 (3) | 149 |
| C10A—H11A···O32Bi | 0.99 | 2.45 | 3.251 (5) | 138 |
| C11A—H13A···O52Bii | 0.99 | 2.59 | 3.093 (3) | 111 |
| C2A—H21A···O31Biii | 0.99 | 2.48 | 3.281 (3) | 138 |
Symmetry codes: (i) −x+3/2, y+1/2, −z+3/2; (ii) x−3/2, −y+3/2, z+1/2; (iii) −x+1/2, y+1/2, −z+3/2.
References
- Agilent (2014). CrysAlis PRO. Agilent Technologies Ltd, Yarnton, England.
- Altomare, A., Cascarano, G., Giacovazzo, C. & Guagliardi, A. (1993). J. Appl. Cryst. 26, 343–350.
- Bowers, J. R., Hopkins, G. W., Yap, G. P. A. & Wheeler, K. A. (2005). Cryst. Growth Des. 5, 727–736.
- Cordiner, R. L., Hill, A. F. & Wagler, J. (2008). Organometallics, 27, 4532–4540.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Gracin, S. & Fischer, A. (2005). Acta Cryst. E61, o1242–o1244.
- Groom, C. R. & Allen, F. H. (2014). Angew. Chem. Int. Ed. 53, 662–671. [DOI] [PubMed]
- Heldebrant, D. J., Yonker, C. R., Jessop, P. G. & Phan, L. (2009). Chem. Eur. J. 15, 7619–7627. [DOI] [PubMed]
- Huczyński, A., Ratajczak-Sitarz, M., Katrusiak, A. & Brzezinski, B. (2008). J. Mol. Struct. 889, 64–71.
- Koman, M., Martiška, L., Valigura, D. & Glowiak, T. (2003). Acta Cryst. E59, o441–o442.
- Lipstman, S. & Goldberg, I. (2013). Cryst. Growth Des. 13, 942–952.
- Lynch, D. E. & McClenaghan, I. (2003). Cryst. Eng. 6, 99–107.
- Motevalli, M., Hursthouse, M. B., Kelly, P. F. & Woollins, J. D. (1989). Polyhedron, 8, 893–896.
- Pérez, E. R., Santos, R. H. A., Gambardella, M. T. P., de Macedo, L. G. M., Rodrigues-Filho, U. P., Launay, J. & Franco, D. W. (2004). J. Org. Chem. 69, 8005–8011. [DOI] [PubMed]
- Regalado, E. L., Mendiola, J., Laguna, A., Nogueiras, C. & Thomas, O. P. (2010). Nat. Prod. Commun. 5, 1187–1190. [PubMed]
- Saftić, D., Žinić, B. & Višnjevac, A. (2012). Tetrahedron, 68, 1062–1070.
- Shannon, M. S., Irvin, A. C., Liu, H., Moon, J. D., Hindman, M. S., Turner, C. H. & Bara, J. E. (2015). Ind. Eng. Chem. Res. 54, 462–471.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Smith, G., Wermuth, U. D., Healy, P. C. & White, J. M. (2003). Aust. J. Chem. 56, 707–713.
- Smith, G., Wermuth, U. D., Healy, P. C. & White, J. M. (2007). Aust. J. Chem. 60, 264–277.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I, II, III. DOI: 10.1107/S205698901600267X/pk2574sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901600267X/pk2574Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S205698901600267X/pk2574IIsup3.hkl
Structure factors: contains datablock(s) III. DOI: 10.1107/S205698901600267X/pk2574IIIsup4.hkl
Supporting information file. DOI: 10.1107/S205698901600267X/pk2574Isup5.cml
Supporting information file. DOI: 10.1107/S205698901600267X/pk2574IIsup6.cml
Supporting information file. DOI: 10.1107/S205698901600267X/pk2574IIIsup7.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report