Abstract
Introduction
Recently, genome-wide association studies (GWAS) in Caucasian populations have identified an association between single nucleotide polymorphisms (SNPs) in the CHRNA5-A3-B4 nicotinic acetylcholine receptor subunit gene cluster on chromosome 15q25, lung cancer risk and smoking behaviors. However, these SNPs are rare in Asians, and there is currently no consensus on whether SNPs in CHRNA5-A3-B4 have a direct or indirect carcinogenic effect through smoking behaviors on lung cancer risk. Though some studies confirmed rs6495308 polymorphisms to be associated with smoking behaviors and lung cancer, no research was conducted in China. Using a case-control study, we decided to investigate the associations between CHRNA3 rs6495308, CHRNB4 rs11072768, smoking behaviors and lung cancer risk, as well as explore whether the two SNPs have a direct or indirect carcinogenic effect on lung cancer.
Methods
A total of 1025 males were interviewed using a structured questionnaire (204 male lung cancer patients and 821 healthy men) to acquire socio-demographic status and smoking behaviors. Venous blood samples were collected to measure rs6495308 and rs11072768 gene polymorphisms. All subjects were divided into 3 groups: non-smokers, light smokers (1–15 cigarettes per day) and heavy smokers (>15 cigarettes per day).
Results
Compared to wild genotype, rs6495308 and rs11072768 variant genotypes reported smoking more cigarettes per day and a higher pack-years of smoking (P<0.05). More importantly, among smokers, both rs6495308 CT/TT and rs11072768 GT/GG had a higher risk of lung cancer compared to wild genotype without adjusting for potential confounding factors (OR = 1.36, 95%CI = 1.09–1.95; OR = 1.11, 95%CI = 1.07–1.58 respectively). Furthermore, heavy smokers with rs6495308 or rs11072768 variant genotypes have a positive interactive effect on lung cancer after adjustment for potential confounding factors (OR = 1.13, 95%CI = 1.01–3.09; OR = 1.09, 95%CI = 1.01–3.41 respectively). However, No significant associations were found between lung cancer risk and both rs6495308 and rs11072768 genotypes among non-smokers and smokers after adjusting for age, occupation, and education.
Conclusion
This study confirmed both rs6495308 and rs11072768 gene polymorphisms association with smoking behaviors and had an indirect link between gene polymorphisms and lung cancer risk.
Introduction
Lung cancer is the most common cancer worldwide and accounts for about 23% of the total cancer-related deaths [1]. In 2012 alone, about 0.42 million Chinese males died of lung cancer [2]. Smoking tobacco is a major risk factor for lung cancer; of the Smoking population, more than 80% are at risk for lung cancer [3, 4]. Cigarette smoke contains at least 250 harmful chemicals, more than 50 of which are carcinogens, including polycyclic aromatic hydrocarbons and nicotine metabolites such as 4-(methylnitrosamino)-1(3-pyridyl)-1-butanone (NNK) and N-nitrosonornicotine (NNN) [5]. These nitrosamines form DNA adducts that cause mutations resulting in lung cancer [6]. However, under the same environmental circumstances, only a small fraction of smokers (usually <20%) develop lung cancer; Inter-individual susceptibility to lung cancer may explain this outcome [7].
Nicotinic acetylcholine receptor subunits (nAChRs) belong to the super family of ligand-gated ion channels, and can be activated by nicotine and its metabolites such as NNK and NNN. Nicotine-mediated activation of nAChRs expressed in the key regions of brain can initiate nicotine addiction, thus making individuals susceptible to lung cancer[8,9]. In addition, nAChRs expressed in the alveolar epithelial cells can be activated by nicotine or its metabolites to cause cells’ loss of contact inhabitation and resistance to apoptosis[10–13]. Moreover, the variants in nAChRs may increase individuals’ vulnerability to nicotine and the harmful effects of tobacco smoke [14–16]. The above biologic mechanisms seem a plausible explanation of the associations between SNPs in CHRNA5-A3-B4 gene clusters, smoking behaviors, and lung cancer risk.
In 2008, three genome–wide association studies (GWAS) in Caucasian populations found three Single nucleotide polymorphisms (SNPs) (rs1051730, rs8034191 and rs16969968) in nicotinic acetylcholine receptor subunit gene cluster (CHRN5-A3-B4) on chromosome 15q25 to be associated with smoking behaviors[14,17,18]. Subsequently, a sea of case-control studies and meta-analyses have verified SNPs in CHRNA5-A3-B4 play an important role in susceptibility to lung cancer and smoking behaviors in European, American and Asian populations[17,19–21]. However, some SNPs, such as rs16969968, are extremely rare in Asians, and no association was found in relation to smoking behaviors and lung cancer in Chinese[19], these results reiterated underscored the differences in genetic markers among different ethnic populations[22,23]. In addition, there is currently no consensus on whether SNPs in CHRN5-A3-B4 have a direct or indirect carcinogenic effect through smoking behaviors on lung cancer risk [17,19–21,24].
These previous studies encouraged us to investigate the associations between other SNPs in CHRNA5-A3-B4, smoking behaviors, and lung cancer risk in the Chinese population. Although some previous studies confirmed an association between rs6495308 polymorphisms, smoking behaviors, and lung cancer risk, no research was conducted in China. We, therefore, examined 10 SNPs including rs6495308 (MAF (Minor Allele Frequency) > 0.1 in Asians) in CHRNA5-A3-B4 gene clusters According to the HapMap data and previous studies[14,17,20,25–27], and, after our initial results, conducted a detailed analysis of rs6495308 and rs11072768 polymorphisms, and explored whether the two SNPs have a direct or indirect effect through smoking behaviors on lung cancer using a case-control study of 1025 patients: 204 with lung cancer and 821 healthy controls.
Methods
Study subjects
A community-based case-control study consisting of 204 male lung cancer patients and 821 healthy men was conducted. Patients who were newly diagnosed with cytological or histologically confirmed lung cancer were recruited from The First Affiliated Hospital of Guangzhou Medical University in China from May to October 2013. Controls consisted of healthy male subjects from a chronic disease epidemiology study conducted in Guangzhou and Zhuhai China from July 2006 to June 2007. All eligible subjects were of the Chinese Han population. In the present study, all subjects have no other cancers or occupational carcinogen exposure history. Ethics Committee of The First Affiliated Hospital of Guangzhou Medical University approved this study.
Data collection
Trained medical students used a structured questionnaire to acquire socio-demographic characteristics (e.g., age, income, marriage and occupation), complete clinical information such as family history of lung cancer and smoking behaviors via face to face interview. Data regarding histological classification and clinical stage of Lung cancer were obtained by our hospital case management system. 2 ml of venous blood was collected from all subjects for DNA extraction and genotyping, and stored at -80°C until use. Written informed consent was obtained from all patients and controls for the use of their DNA and clinical information.
Measures of cigarette smoking
A patient who has smoked more than 100 cigarettes in one’s life-time or smoked at least one cigarette per day for more than one year was referred as a ‘smoker’ [4]. ‘Cigarettes per day (CPD)’ was defined as average number of cigarettes smoked per day [4]. ‘Pack-years of smoking’ was counted by dividing 20 from daily cigarette consumption and multiplied duration of smoking among smokers [4]. All subjects were categorized into three groups according to their smoking quantity: never smokers (0 cigarettes/day), lighter smokers (1–15 cigarettes/day), and heavier smokers (>15 cigarettes/day).
DNA extraction and genotyping
Genomic DNA was extracted from venous blood using a commercial blood DNA kit according to the manufacturer’s instructions (TaKaRa, Dalian, CA, China).The selected SNPs were genotyped using the SNaPshot SNP parting technology (Life technology, Carlsbad, CA, USA). Primers for polymerase chain reaction (PCR) and single-base extension were designed using Assay Designers software version 3.1 (Sequenom, San Diego, CA, USA). Firstly the SNPs were amplified by Multiple PCR reaction using HotStarTaq DNA polymerase (TIANGEN, Beijing, CA, China). Next, genomic amplification products were amplified again using SNaPshot Multiplex extension reactions kit (ABI, Carlsbad, CA, USA) after Purification by shrimp alkali enzyme (Promega, Beijing, CA, China) and external enzyme (Epicentre, Beijing, CA, China). Finally genomic amplification products were assessed by 3730xl genetic analyzer (ABI, Carlsbad, CA, USA). SNP Genotypes were completed by GeneMapper4.1 (ABI, Carlsbad, CA, USA). Genotyping was performed by Commercial genetic testing company (Genesky, Shanghai, CA, China). CHRNA3 rs6495308 genotypes were categorized into homozygous wild-type (CC), hybrid variant type (CT) and homozygous variant (TT). rs11072768 in CHRNB4 was classified into homozygous wild-type (TT), hybrid variant type (GT) and homozygous variant (GG). For quality control, 5% of the samples were randomly selected and re-genotyped for all of the selected genes, and the results were 100% concordant.
Statistical analysis
In this study, either the Chi Square tests (χ2 tests) or the Fisher’s Exact test, whichever appropriate, were used to analyze the differences in the distribution of general demographic characteristics and CPD between cases and controls, Subsequently, Kruskal-Wallis test was conducted to assess the association of rs6495308 and rs11072768 polymorphisms with smoking behaviors. We adjusted the significance for multiple comparisons using the Bonferroni correction. Finally, a series of unconditional logistic regression binary logistic regression analyses was carried out to evaluate the associations between rs514743 genotypes, rs11072768 genotypes, smoking behaviors and lung cancer. Hardy–Weinberg distribution testing was performed for rs6495308 and rs11072768 among controls. For all tests, a two sided P<0.05 was considered statistically significant, and all statistical analysis were performed on SPSS version 19.0 software package (SPSS, Chicago, IL, USA).
Results
Comparison of demographic characteristics between case and control groups
There were statistically different distributions of age, daily cigarette consumption, occupation, education, marriage and the allele distribution of these two SNPs between cases and controls. More details are presented in Table 1.
Table 1. Comparison of demographic characteristics between case and control groups.
| Characteristics | Cases | Controls | χ2 | P value |
|---|---|---|---|---|
| (n = 204) (%) | (n = 821) (%) | |||
| Age group | 82.79 | <0.001 | ||
| 20–29 | 3(1.5) | 41(5) | ||
| 30–39 | 4(2) | 127(15.5) | ||
| 40–49 | 28(13.7) | 166(20.2) | ||
| 50–59 | 57(27.9) | 278(33.9) | ||
| 60–69 | 77(37.7) | 162(19.7) | ||
| 70–81 | 35(17.2) | 47(5.7) | ||
| Daily cigarette consumption | 150.54 | <0.001 | ||
| 0 cigarettes/day | 45(22.1) | 302(36.8) | ||
| 1–15 cigarettes/day | 18(8.8) | 316(38.5) | ||
| >15 cigarettes/day | 141(69.1) | 203(24.8) | ||
| Smoking pack years | 44.89±38.38 | 13.57±18.59 | 11.33 | <0.001 |
| Occupation | 76.46 | <0.001 | ||
| Worker | 24(11.8) | 229(27.9) | ||
| Farmer | 38(18.6) | 71(8.6) | ||
| Person in charge | 2(1) | 42(5.1) | ||
| Technician | 9(4.4) | 49(6) | ||
| Service personnel | 33(16.2) | 118(14.4) | ||
| Retired personnel | 83(40.7) | 171(20.8) | ||
| Jobless | 11(5.4) | 101(12.3) | ||
| Other | 4(2) | 40(4.9) | ||
| Education | 25.6 | <0.001 | ||
| Illiteracy | 7(3.4) | 52(6.3) | ||
| Elementary school | 50(24.5) | 105(12.8) | ||
| Junior middle school | 66(32.4) | 230(28) | ||
| Senior middle school | 56(27.5) | 264(32.2) | ||
| College or above | 25(12.3) | 170(20.7) | ||
| Familial history of cancer | ||||
| Yes | 30(14.7) | 78(9.5) | 4.697 | 0.030 |
| No | 174(85.3) | 706(90.5) | ||
| Marriage | 14.15 | <0.001 | ||
| Yes | 195(95.6) | 706(86.0) | ||
| No | 9(4.4) | 115(14.0) | ||
| rs6495308 | ||||
| C/C | 87(42.6) | 415(50.5) | 4.152 | 0.125 |
| C/T | 98(48.0) | 336(40.9) | ||
| T/T | 19(9.3) | 70(8.5) | ||
| rs11072768 | ||||
| T/T | 110(53.9) | 484(59.0) | 2.014 | 0.365 |
| G/T | 85(41.7) | 298(36.3) | ||
| G/G | 9(4.4) | 39(4.8) |
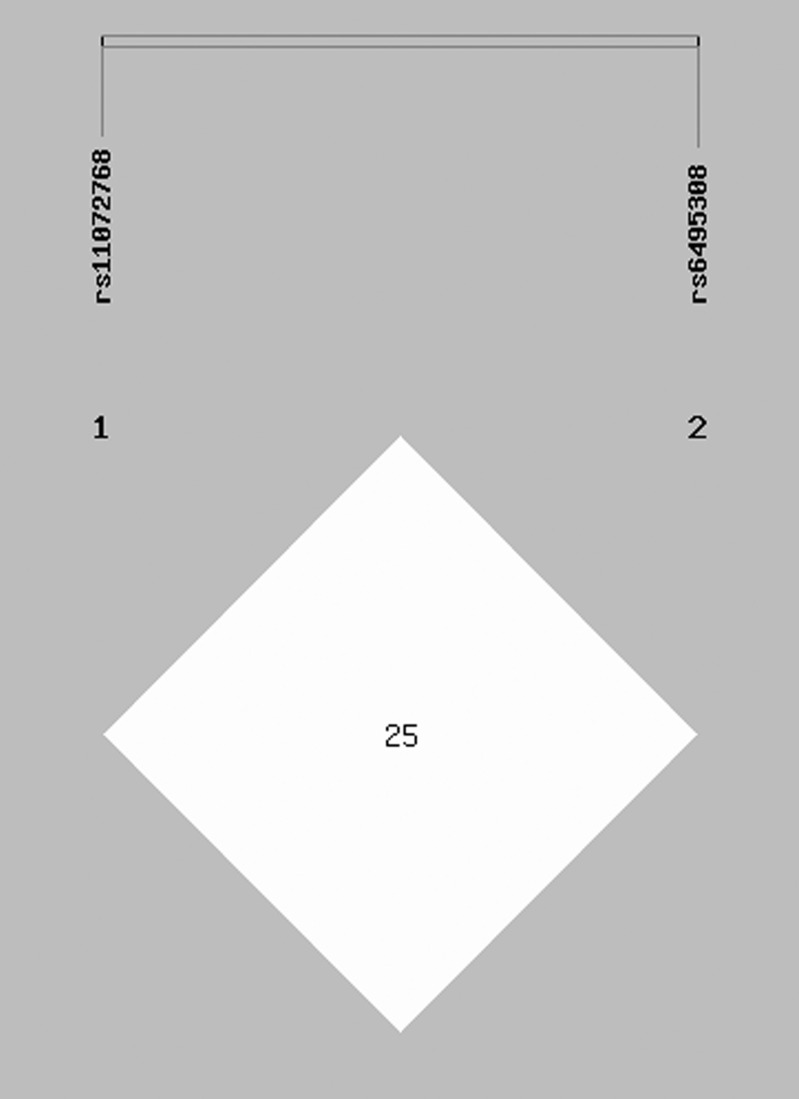
At the same time, we examined the linkage disequilibrium (LD) between rs6495308 and rs11072768 in our data. The LD structure (rs6495308/rs11072768) showed low correlation (r2 = 0.251) (Fig 1).
Fig 1. Linkage disequilibrium (LD) patterns of the two SNPs in 15q25.1.
Numbers inside the boxes represent r2 values for LD. Colors indicate the strength of LD between pair-wise combinations of SNPs (white, low LD; red, high LD).
Association between gene polymorphisms and smoking behaviors among smokers of lung cancer cases and controls
All subjects were genotyped for two SNPs (rs6495308 C/T, rs11072768 T/G) in 15q25. The SNPs were in Hardy-Weinberg equilibrium (HWE) both in controls and in cases (p>0.05). The associations of rs6495308 and rs11072768 polymorphisms with smoking behaviors were analyzed in 159 smokers with lung cancer and 519 smokers in the control group. The values of cigarettes per day and pack-years were regularly used as the measurements of smoking behaviors for this analysis and can be seen in Table 2. Compared with the wild genotype, rs6495308 and rs11072768 variant genotypes reported smoking more cigarettes per day and had a higher pack-years of smoking among smokers in both groups (P<0.05).
Table 2. Association between gene polymorphisms and smoking behaviors among smokers of lung cancer cases and controls.
Data were expressed as mean±standard deviation.
| Genotype | n(%) | case | control | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CPD | P | Pack-years | P | CPD | P | Pack-years | P | ||
| rs6495308 | 0.039 | 0.027 | 0.060 | 0.047 | |||||
| CC | 25.31±8.26 | 52.75±20.79 | 13.25±8.06 | 20.52±17.28 | |||||
| CT | 28.92±15.54* | 57.02±35.38* | 14.19±8.45 | 21.17±20.00 | |||||
| TT | 29.62±16.21* | 59.11±35.23* | 15.02±10.33* | 22.05±19.29* | |||||
| rs11072768 | 0.044 | 0.045 | 0.037 | 0.050 | |||||
| TT | 22.14±7.56 | 48.04±22.52 | 14.39±9.99 | 20.75±16.43 | |||||
| GT | 28.75±15.25# | 56.05±33.95# | 14.47±8.29 | 21.68±21.18 | |||||
| GG | 29.71±15.80# | 60.70±35.16# | 14.81±6.98# | 24.47±19.80# | |||||
*: Compared with CC group, p<0.05.
#: Compared with TT group, p<0.05.
Association between gene polymorphisms and lung cancer risk in non-smokers
Table 3. presents the results of associations between rs6495308 and rs11072768 genotypes and lung cancer risks in non-smokers. No significant associations were found between rs6495308 and rs11072768 genotypes and lung cancer risk after adjusting for age, occupation, and education.
Table 3. Association between gene polymorphisms and lung cancer risk in non-smokers.
| Variables | Cases | Controls | OR (95%CI)a | P-value | OR (95%CI) b | P-value |
|---|---|---|---|---|---|---|
| (n = 45) (%) | (n = 302) (%) | |||||
| rs6495308 | ||||||
| CC | 19(42.2%) | 152(50.3%) | 1 | 0.727 | 1 | 0.368 |
| CT | 21(46.7%) | 120(39.7%) | 1.30(0.68–2.49) | 0.427 | 1.93(0.89–3.95) | 0.238 |
| TT | 5(11.1%) | 30(9.9%) | 1.20(0.33–4.41) | 0.780 | 1.54(0.42–6.56) | 0.632 |
| CT/TT | 26(57.8%) | 150(49.7%) | 1.29(0.69–2.41) | 0.131 | 1.35(0.58–2.50) | 0.512 |
| rs11072768 | ||||||
| TT | 25(55.6%) | 173(57.2%) | 1 | 0.367 | 1 | 0.723 |
| GT | 17(37.8%) | 114(37.8%) | 1.59(0.84–3.04) | 0.157 | 1.65(0.69–3.24) | 0.318 |
| GG | 3(6.7%) | 15(4.8%) | 1.25(0.27–5.90) | 0.777 | 1.39(0.18–6.58) | 0.894 |
| GT/GG | 20(33.3%) | 129(42.7%) | 1.56(0.83–2.92) | 0.169 | 1.54(0.70–3.12) | 0.325 |
aOR value unajust for any confoundings
bOR value adjustment for age, education, education and marriage and familial history of cancer.
Association between gene polymorphisms and lung cancer risk in smokers
Associations between gene polymorphisms and lung cancer risk in smokers are shown in Table 4. Among smokers both rs6495308 CT/TT and rs11072768 GT/GG had a higher risk of lung cancer compared to wild genotype without adjusting for potential confounding factors (OR = 1.36, 95%CI = 1.09–1.95; OR = 1.11, 95%CI = 1.07–1.58 respectively). However, no significant associations were confirmed between rs6495308 and rs11072768 genotypes, and lung cancer risk after adjusting for CPD, age, occupation, and education.
Table 4. Association between gene polymorphisms and lung cancer risk in smokers.
| Variables | Cases | Controls | OR (95%CI) a | P-value | OR (95%CI) b | P-value | OR (95%CI)c | P-value |
|---|---|---|---|---|---|---|---|---|
| (n = 159) | (n = 519) (%) | |||||||
| rs6495308 | ||||||||
| CC | 68(42.8%) | 263(50.7%) | 1 | 0.021 | 1 | 0.297 | 1 | 0.812 |
| CT | 77(48.4%) | 216(41.6%) | 1.40(1.02–2.03) | 0.050 | 1.39(0.92–2.10) | 0.120 | 1.19(0.68–1.95 | 0.628 |
| TT | 14(8.8%) | 40(7.7%) | 1.21(0.65–2.26) | 0.545 | 1.22(0.61–2.42) | 0.577 | 1.30(0.50–2.85) | 0.596 |
| CT/TT | 91(57.2%) | 256(49.3%) | 1.36(1.09–1.95) | 0.049 | 1.20(0.77–1.86) | 0.420 | 1.25(0.68–1.88) | 0.583 |
| rs11072768 | ||||||||
| TT | 85(53.4%) | 311(60.0%) | 1 | 0.047 | 1 | 0.799 | 1 | 0.983 |
| GT | 68(42.7%) | 185(35.6%) | 1.13(0.78–1.64) | 0.056 | 1.14(0.76–1.71) | 0.527 | 1.19(0.67–1.85) | 0.716 |
| GG | 6(3.8%) | 24(4.6%) | 0.92(0.39–2.19) | 0.849 | 0.95(0.37–2.46) | 0.914 | 1.00(0.25–3.01) | 0.998 |
| GT/GG | 74(46.5%) | 213(40.0%) | 1.11(1.07–1.58) | 0.027 | 1.12(0.75–1.66) | 0.579 | 1.16(0.65–1.82) | 0.710 |
aOR value unajustment for any confoundings
bOR value adjustment for age, education, education and marriage.
cOR value adjustment for age, education, education, marriage daily cigarettes consumption, familial history of cancer and smoking pack years.
Interaction between gene polymorphisms and smoking behaviors on lung cancer risk
After adjustment for potential confounding factors, we found that the interactions between CPD and both rs6495308 and rs11072768 genotypes were statistically significant: heavy smokers with rs6495308 or rs11072768 variant genotypes were more likely to have lung cancer than respective light smokers with wild genotypes (OR = 1.08, 95%CI = 1.02–3.26; OR = 1.05, 95%CI = 1.01–3.68 respectively), More details are presented in Table 5.
Table 5. Interaction between gene polymorphisms and smoking behaviors on lung cancer risk.
| Variables | Cases | Controls | OR (95%CI) a | P-value |
|---|---|---|---|---|
| (n = 204) (%) | (n = 821) (%) | |||
| CPD | ||||
| 1–15 | 1 | 0.008 | ||
| >15 | 9.28(5.86–19.12) | |||
| rs6495308 | ||||
| CC | 87(42.6%) | 415(50.5%) | 1 | 0.798 |
| CT | 98(48.0%) | 336(40.9%) | 1.08(0.70–1.96) | 0.632 |
| TT | 19(9.3%) | 70(8.5%) | 1.23(0.54–2.88) | 0.594 |
| CT+TT | 117(57.3%) | 406(49.4%) | 1.25(0.64–1.95) | 0.562 |
| rs11072768 | ||||
| TT | 110(53.9%) | 484(59.0%) | 1 | 0.961 |
| GT | 85(41.7%) | 298(36.2%) | 1.19(0.67–1.81) | 0.683 |
| GG | 9(4.4%) | 39(4.8%) | 1.04(0.36–2.93) | 0.999 |
| GT+GG | 94(46.1%) | 337(41.0%) | 1.08(0.73–1.97) | 0.717 |
| interaction(rs6495308×CPD) | ||||
| CT/TT×>15 | 1.08(1.02–3.26) | 0.038 | ||
| interaction(rs11072768×CPD) | ||||
| GT/GG×>15 | 1.05(1.01–3.68) | 0.045 |
aOR value adjustment for age, education, education, marriage, familial history of cancer and smoking pack years.
Discussion
To our knowledge, this is the first time an investigation on the association between rs6495308, rs11072768 genotypes, smoking behaviors and lung cancer risk in a Chinese male population has been performed. We found rs6495308 and rs11072768 variant genotypes reported smoking more cigarettes over a long period. More importantly, rs6495308 and rs11072768 variant genotypes have a higher risk of lung cancer without adjustment for any potential confounding factors, and heavy smoking has a positive interactive effect with rs6495308 and rs11072768 variant genotypes on lung cancer among smokers. However, no significant associations were found between rs6495308, rs11072768 genotypes and lung cancer risk among nonsmokers and smokers after adjusting for CPD and other potential confounding factors. In this study, a series of results have provided strong evidence that rs6495308 and rs11072768 gene polymorphisms have an indirect impact on lung cancer through smoking behaviors, and there is a correlation between variants in 15q25 and smoking on lung cancer.
A fair amount of research supports the above results. A GWAS meta-analysis in a total sample of 41,150 individuals identified rs6495308 variants in CHRNA3 increased smoking quantity[28], subsequently, a cohort study confirmed carriers of the rs6495308 TT genotypes have approximately two fold greater odds for ND defined using CPD in European-American smokers[29], these findings suggest that rs6495308 variants lead indirectly to lung cancer via smoking behavior. Furthermore, large GWAS meta-analyses confirmed that the strongest genetic contribution to smoking-related traits comes from variation in CHRNA5-A3-B4[10,15,17,30,31], as first revealed on a genome-wide significant level by Thorgeirsson et al. in a study of over 13,000 smokers from Iceland[15]. In addition, a GWAS study in the Chinese population speculated that one non-synonymous mutation in the rs6495308c risk allele results in a higher CHRNA3 receptor production which may make individuals more sensitive to nicotine and thus more susceptible to nicotine dependency, a well-established etiological factor for lung cancer [19]. Li et al found that the G allele of rs11072768 in CHRNB4 was significantly associated with smoking initiation (SI) (P = 0.001; OR = 1.22;95%CI: 1.08, 1.37), smoking quantity (SQ) (P = 0.016; OR = 1.16; 95%CI: 1.03, 1.31), and smoking cessation (SC) (P = 0.01; OR = 1.18; 95%CI: 1.04, 1.34) in a sample of Korean males [32]. Other studies have affirmed some SNPs (rs12914385, rs8042374 and rs588765) to be associated with smoking behaviors and are in complete agreement with an indirect link between genotypes and lung cancer[24,27].Nevertheless, there is much assertion that, rather than being an agent for smoking, CHRNA5-A3-B4 variants have a direct impact on lung cancer risk. Lips et al. calculated a 1.2 difference in CPD due to the fact that rs16969968 homozygotes in CHRNA5 only contribute a 9% increase in lung cancer risk, which cannot account for the association between the variants, smoking behavior and lung cancer[31]. Hung et al. also indicated an increased risk for lung cancer, even among non-smokers, due to rs16969968 variants, in addition to other evidence suggesting that rs16969968 variant genotypes are not directly associated with an increase in the risk for other smoking-related cancers such as head and neck cancer [33]. Moreover, other studies confirmed an over-representation of CHRNA5-A3-B4 variants in familial lung cancer cases indicating a direct impact of variants on lung cancer [34]. These discrepancies in the previous studies can most probably be attributed to genetic, racial and environmental differences [14,18,24].
The specific mechanisms of the genetic variations in CHRNA5-A3-B4 that influenced smoking behaviors were unclear, but there was a potentially plausible biologic hypothesis underlying the observed results. The variants in CHRNA5-A3-B4 give rise to the over expression of the nAChRs in in the key regions of brain which may make individuals sensitive to nicotine, which includes the pharmacological effects of nicotine along with higher activation of the medial habenula and reduced activation of dopaminergic neurons after acute nicotine administration[35,36], and thus lead to smoking more cigarettes over a long period, a well-established etiological factor for lung cancer. The mechanism indicates the presence of a smoking-by-variant interaction, and such effect is only observed in smokers.
Several limitations of this study should be mentioned. Firstly, self-reported smoking behaviors may be prone to recall bias which influences the authenticity of associations between SNPs, smoking behavior and lung cancer. Secondly, the relatively small sample size could occasionally affect the results of this study. Next, the average age of the case group was higher than that of the control group. In the case group, the largest proportion were retirees, retirees may be higher risk of lung cancer due to their older age; in addition, those with higher education in the case group was lower than that of the control group, those with a higher degree may pay more attention to and be more knowledgeable in regards to their own health condition and, as a result, have relatively fewer adverse health behaviors. Finally, the functional study furtherly cannot be implemented because rs6495308 and rs11072768 are located on intron in 15q25, so there may be other causal SNPs in highly linkage disequilibrium (LD) with the two SNPs within 15q25 region to affect the function of nAchRs. Therefore, a large population-based study is still required to confirm the present findings.
In conclusion, the results of our study confirmed two SNPs (rs6495308 and rs11072768) in CHRNA5-A3-B4 have a indirect effect on lung cancer through smoking behaviors, and a positive correlation between CPD and both rs6495308 and rs11072768 on lung cancer among smokers. These findings extend our understanding of the possible mechanism of cigarette smoking on lung cancer and may have applications in healthcare for tailoring strategies of smoking cessation
Supporting Information
(XLS)
(DOCX)
(SAV)
Acknowledgments
This work was supported by Science and Technology Planning Project of Guangdong Province, P.R.China (No. 2011B061300111).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Science and Technology Planning Project of Guangdong Province, P. R. China (No. 2011B061300111). http://www.gdstc.gov.cn/. The funding institution is Guangdong Provincial Department of Science and Technology, Mei Jiang received the funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.ACS. Cancer Facts And Figures 2009.: World Health Organization; 2009. [Google Scholar]
- 2.IARC. Lung Cancer Incidence, Mortality and Prevalence Worldwide in2012. International Agency for Research on Cancer; 2012(2012):1–2. [Google Scholar]
- 3.WHO. Tobacco and health: a global status report Geneva: World Health Organization; 1997. [Google Scholar]
- 4.WHO. WHO Report on the Global Tobacco Epidemic, 2009:Implementing Smoke-Free Environments. World Health Organization; 2009;1(1):1–2. [Google Scholar]
- 5.Proctor R. Tobacco and the global lung cancer epidemic. Nat Rev Cancer. 2001;1(1):82–86. [DOI] [PubMed] [Google Scholar]
- 6.Lee PN, Forey BA, Coombs KJ. Systematic review with meta-analysis of the epidemiological evidence in the 1900s relating smoking to lung cancer. BMC Cancer. 2012;12:385 10.1186/1471-2407-12-385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang JB, Jiang Y, Wei WQ, Yang GH, Qiao YL, Boffetta P. Estimation of cancer incidence and mortality attributable to smoking in China. Cancer Causes Control. 2010;21(6):959–965. 10.1007/s10552-010-9523-8 [DOI] [PubMed] [Google Scholar]
- 8.Di Chiara G. Role of dopamine in the behavioural actions of nicotine related to addiction. Eur J Pharmacol. 2000;393(1–3):295–314. [DOI] [PubMed] [Google Scholar]
- 9.Corrigall WA, Franklin KB, Coen KM, Clarke PB. The mesolimbic dopaminergic system is implicated in the reinforcing effects of nicotine. Psychopharmacology (Berl). 1992;107(2–3):285–289. [DOI] [PubMed] [Google Scholar]
- 10.Le Novere N, Changeux JP. Molecular evolution of the nicotinic acetylcholine receptor: an example of multigene family in excitable cells. J Mol Evol. 1995;40(2):155–172. [DOI] [PubMed] [Google Scholar]
- 11.Conklin BS, Zhao W, Zhong DS, Chen C. Nicotine and cotinine up-regulate vascular endothelial growth factor expression in endothelial cells. Am J Pathol. 2002;160(2):413–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng Y, Ritzenthaler JD, Roman J, Han S. Nicotine stimulates human lung cancer cell growth by inducing fibronectin expression. Am J Respir Cell Mol Biol. 2007;37(6):681–690. [DOI] [PubMed] [Google Scholar]
- 13.Sher E, Codignola A, Passafaro M, Tarroni P, Magnelli V, Carbone E, et al. Nicotinic receptors and calcium channels in small cell lung carcinoma. Functional role, modulation, and autoimmunity. Ann N Y Acad Sci. 1998;841:606–624. [DOI] [PubMed] [Google Scholar]
- 14.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–642. 10.1038/nature06846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorgeirsson TE, Stefansson K. Commentary: gene-environment interactions and smoking-related cancers. Int J Epidemiol. 2010;39(2):577–579. 10.1093/ije/dyp385 [DOI] [PubMed] [Google Scholar]
- 16.Le Marchand L, Derby KS, Murphy SE, Hecht SS, Hatsukami D, Carmella SG, et al. Smokers with the CHRNA lung cancer-associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine. Cancer Res. 2008;68(22):9137–9140. 10.1158/0008-5472.CAN-08-2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T,et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–622. 10.1038/ng.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. 10.1038/nature06885 [DOI] [PubMed] [Google Scholar]
- 19.Wu C, Hu Z, Yu D, Huang L, Jin G, Liang J, et al. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Res. 2009;69(12):5065–5072. 10.1158/0008-5472.CAN-09-0081 [DOI] [PubMed] [Google Scholar]
- 20.Amos CI, Gorlov IP, Dong Q, Wu X, Zhang H, Lu EY, et al. Nicotinic acetylcholine receptor region on chromosome 15q25 and lung cancer risk among African Americans: a case-control study. J Natl Cancer Inst. 2010;102(15):1199–1205. 10.1093/jnci/djq232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broderick P, Wang Y, Vijayakrishnan J, Matakidou A, Spitz MR, Eisen T, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69(16):6633–6641. 10.1158/0008-5472.CAN-09-0680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ware JJ, van den Bree M, Munafo MR. From men to mice: CHRNA5/CHRNA3, smoking behavior and disease. Nicotine Tob Res. 2012;14(11):1291–1299. 10.1093/ntr/nts106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Improgo MR, Scofield MD, Tapper AR, Gardner PD. From smoking to lung cancer: the CHRNA5/A3/B4 connection. Oncogene. 2010;29(35):4874–4884. 10.1038/onc.2010.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Broderick P, Matakidou A, Eisen T, Houlston RS. Chromosome 15q25 (CHRNA3-CHRNA5) variation impacts indirectly on lung cancer risk. PLoS One. 2011;6(4):e19085 10.1371/journal.pone.0019085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Bao S, Xu X, Bao Y, Zhang Y. Polymorphisms of CHRNA5-CHRNA3-CHRNB4 Gene Cluster and NSCLC Risk in Chinese Population. Transl Oncol. 2012;5(6):448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tekpli X, Landvik NE, Skaug V, Gulsvik A, Haugen A, Zienolddiny S. Functional effect of polymorphisms in 15q25 locus on CHRNA5 mRNA, bulky DNA adducts and TP53 mutations. Int J Cancer. 2013;132(8):1811–1820. 10.1002/ijc.27870 [DOI] [PubMed] [Google Scholar]
- 27.Shen B, Zhu Q, Zheng MQ, Chen J, Shi MQ, Feng JF. CHRNA5 polymorphism and susceptibility to lung cancer in a Chinese population. Braz J Med Biol Res. 2013;46(1):79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet.2010;42(5):436–440. 10.1038/ng.572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bousman CA, Rivard C, Haese JD, Ambrosone C, Hyland A. Alpha-5 and -3 nicotinic receptor gene variants predict nicotine dependence but not cessation: findings from the COMMIT cohort. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(2):227–235. 10.1002/ajmg.b.32019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lips EH, Gaborieau V, McKay JD, Chabrier A, Hung RJ, Boffetta P, et al. Association between a 15q25 gene variant, smoking quantity and tobacco-related cancers among 17 000 individuals. Int J Epidemiol. 2010;39(2):563–577. 10.1093/ije/dyp288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li MD, Yoon D, Lee JY, Han BG, Niu T, Payne TJ, et al. Associations of variants in CHRNA5/A3/B4 gene cluster with smoking behaviors in a Korean population. PLoS One. 2010. August 16;5(8):e12183 10.1371/journal.pone.0012183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. 10.1038/nature06885 [DOI] [PubMed] [Google Scholar]
- 34.Liu P, Vikis HG, Wang D, Lu Y, Wang Y, Schwartz AG, et al. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J Natl Cancer Inst. 2008; 100:1326–1330. 10.1093/jnci/djn268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loukola A, Hallfors J, Korhonen T, Kaprio J. Genetics and smoking. Curr Addict Rep. 2014;1(1):75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallego X, Molas S, Amador-Arjona A, Marks MJ, Robles N, Murtra P, et al. Overexpression of the CHRNA5/A3/B4 genomic cluster in mice increases the sensitivity to nicotine and modifies its reinforcing effects. Amino Acids. 2012;43(2):897–909. 10.1007/s00726-011-1149-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(DOCX)
(SAV)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.