Abstract
When sun plants, such as Arabidopsis thaliana, are under canopy shade, elongation of stems/petioles will be induced as one of the most prominent responses. Plant hormones mediate the elongation growth. However, how environmental and hormonal signals are translated into cell expansion activity that leads to the elongation growth remains elusive. Through forward genetic study, we identified shade avoidance2 (sav2) mutant, which contains a P287L mutation in β‐TUBULIN 4. Cortical microtubules (cMTs) play a key role in anisotropic cell growth. Hypocotyls of sav2 are wild type‐like in white light, but are short and highly swollen in shade and dark. We showed that shade not only induces cMT rearrangement, but also affects cMT stability and dynamics of plus ends. Even though auxin and brassinosteroids are required for shade‐induced hypocotyl elongation, they had little effect on shade‐induced rearrangement of cMTs. Blocking auxin transport suppressed dark phenotypes of sav2, while overexpressing EB1b‐GFP, a microtubule plus‐end binding protein, rescued sav2 in both shade and dark, suggesting that tub4P287L represents a unique type of tubulin mutation that does not affect cMT function in supporting cell elongation, but may affect the ability of cMTs to respond properly to growth promoting stimuli.
Keywords: Arabidopsis, microtubules, plant hormones, shade
Edited by: Chris J. Staiger, Purdue University, USA
INTRODUCTION
Postembryonic growth of plants is highly plastic, which allows a plant to shape its structure according to environmental cues such as light and temperature. Hypocotyl of Arabidopsis is an embryonic stem that consists of about 20 cells. Elongation of hypocotyl is mostly due to cell expansion, not cell division (Gendreau et al. 1997; Saibo et al. [Link]). Growth of hypocotyl is influenced by both environmental signals such as light, temperature, gravity, and various internal factors such as phytohormones (De Grauwe et al. 2005; Vandenbussche et al. 2005). Thus, the simple architecture and complex regulatory network of hypocotyl make it a perfect system to study interactions between environmental stimuli and plant development.
Plant hormones are key regulators of hypocotyl growth. Auxin, gibberellins (GAs) and brassinosteroids (BRs) are all reported regulators of hypocotyl growth (De Grauwe et al. 2005; Vandenbussche et al. 2005). Environmental changes in light, gravity or temperature regulate hypocotyl growth through altered hormone metabolism and/or signaling. As a shade intolerant plant, hypocotyls of light‐grown Arabidopsis seedlings elongate in response to vegetative shade (Gray et al. 1998; Steindler et al. 1999; Franklin and Whitelam 2005; Vandenbussche et al. 2005). Phytohormones including auxin, GAs and BRs are required for this process (Djakovic‐Petrovic et al. 2007; Kozuka et al. 2010). Biosynthesis of auxin increases in young leaves and cotyledons of shade‐treated seedlings, which is then transported to petioles and hypocotyls, promoting cell elongation (Tao et al. 2008). Mutants that are defective in auxin biosynthesis (such as sav3/taa1), auxin transport (such as pin3) compromised shade‐induced hypocotyl elongation (Keuskamp et al. 2010; Effendi et al. 2013). In Arabidopsis, shade also promotes the expression of genes involved in GA biosynthesis and signaling. DELLA proteins are key negative regulators of GA signaling, and they are also growth repressors. Shade reduces the abundance of DELLA proteins in petioles and hypocotyls, and shade‐induced hypocotyl elongation is absent in GA‐insensitive gai mutant (Djakovic‐Petrovic et al. 2007).
Microtubules are components of cytoskeleton that are polymers assembled from α and β tubulin. Repeating αβ heterodimers align longitudinally to form protofilaments. Through lateral interactions, protofilaments constitute a sheet that rolls up and closes to form a tube structure. The Arabidopsis genome contains six α‐tubulin genes and nine β‐tubulin genes. Although null mutants of tubulin genes exhibit no detectable defects, a large number of semi‐dominant or dominant mutants with missense mutations or small deletions were identified and they displayed multiple defects such as anisotropic cell growth and flower development defects (Thitamadee et al. 2002; Ishida et al. 2007; Hashimoto 2013). It was demonstrated for some of the tubulin mutants that they can incorporate into microtubules along with other wild type tubulin isoforms and may subsequently interfere with microtubule assembly, stability or dynamics, which may account for their semi‐dominant/dominant effect on microtubules (Ishida et al. 2007). Analysis of these tubulin mutants revealed critical residues involved in intradimer or interdimer interaction and residues required for lateral contact between protofilaments (Ishida et al. 2007; Hashimoto 2013). Thus, by analyzing these tubulin mutants, we gained more insights into the organization and function of microtubules in plants.
Cortical microtubules (cMTs) are interphase microtubules that lie just beneath plasma membrane. Transversely aligned cMTs are often associated with rapid cell elongation. They are believed to guide the movement of cellulose synthase complexes, which then produce parallel arrays of the shape controlling cellulose microfibers and generate a mechanically anisotropic cell wall that favors cell elongation and prevents radial expansion (Crowell et al. 2011). Longitudinal or oblique alignment of cMTs is found in cells with slow elongation growth (Chan et al. 2011; Lloyd 2011). Hormones such as auxin, GA and BR, were shown to promote the transverse alignment of cMTs (Blancaflor and Hasenstein 1995; Wenzel et al. 2000; Catterou et al. [Link], [Link]; Wiesler et al. 2002; De Grauwe et al. 2005; Komorisono et al. 2005). Structures and dynamics of microtubules are constructed and regulated by a variety of microtubule‐associated proteins (MAPs) such as end binding protein 1 (EB1), which preferentially binds plus‐end of microtubules and promote tube formation or stabilize tube structure (Mimori‐Kiyosue et al. 2005; Komaki et al. 2010). MAPs, such as MAP65, MDP25, MDP40 and WDL3, were reported to regulate hypocotyl elongation and their activities were subjected to regulations by stimuli such as intracellular calcium level, hormones and light (Li et al. 2011; Wang et al. 2012; Liu et al. 2013). Recently, Locascio and colleagues showed that nuclear‐localized GA repressor protein DELLA modulates microtubule organization by regulating tubulin folding (Locascio et al. 2013). Together, these data indicate that environmental signals may regulate cMTs through phytohormones and MAPs.
Three EB1 genes: ATEB1a (At3g47690), ATEB1b (At5g62500) and ATEB1c (At5g67270) were identified in Arabidopsis (Chan et al. 2003; Bisgrove et al. 2008; Komaki et al. 2010). ATEB1a and 1b were demonstrated to track the growing microtubule plus ends, while ATEB1c localized to the interphase nucleus (Chan et al. 2003; Dixit et al. 2006). As animal EB1s, all three ATEB1s were also able to promote microtubule polymerization in vitro (Komaki et al. 2010). But surprisingly, single, double or triple mutants of ateb1s displayed very mild phenotypes (Bisgrove et al. 2008; Komaki et al. 2010). ATEB1a and EB1b are not required for proper microtubule organization in vivo, whereas ATEB1c is required for spindle proper positioning of spindle poles and chromosome segregation (Komaki et al. 2010).
Here we report the identification and characterization of shade avoidance 2 (sav2) mutant, which carries a P287L mutation in β‐TUBULIN 4 (TUB4) protein. Hypocotyl phenotypes of sav2 are modified by light, indicating that hypocotyl growth regulated by light requires proper functions of microtubules. We then investigated the regulation of cMTs by shade signals and how tub4P287L mutation in sav2 affects cMTs. We showed that shade treatment induces cMT reorganization, affects PPM sensitivity and alters polymerization rate of microtubule plus ends. Plant hormones including auxin, BR and GAs are involved in some but not all of these shade‐induced alterations of cMTs. tub4P287L mutation results in altered sensitivity to microtubule drugs, and it abolished shade‐induced cMT rearrangement. However, this mutation does not abolish the role of cMTs in supporting hypocotyl elongation growth, and it may affect responses of cMTs to external stimuli.
RESULTS
sav2 mutant is defective in shade/dark induced hypocotyl elongation
sav2 mutant was identified through a forward genetic screen designed to discover mutants whose hypocotyls do not elongate in response to simulated shade. Under continuous white light (Wc), hypocotyls of 5‐d‐old sav2 seedlings were morphologically similar to that of the Col‐0 wild type. However, under simulated shade (shade), they were short and swollen instead of being thin and elongated (Figure 1). Similar phenotypes were observed in dark grown sav2 (Figure 1B, D). Using scanning electromicroscopy (SEM), we examined the epidermal cell profile of the hypocotyls. As shown in Figure 1E, under Wc, the epidermal cells of wild type and sav2 hypocotyls were of similar size, and were packed in a well‐organized manner. In the shade or dark, the epidermal cells of wild type hypocotyls elongated extensively, while those of the sav2 hypocotyls showed little elongation. They expanded horizontally and bulged out. Besides the hypocotyl phenotype, we also observed a shade‐dependent petiole twisting of sav2. Under both Wc and simulated shade, the first set of true leaves of wild type seedlings emerged in a direction perpendicular to the pair of cotyledons (Figure 1F). sav2 mutant behaved similarly to the wild type in Wc. However, under simulated shade, petioles of sav2 true leaves displayed a counter‐clockwise rotation (viewing from the top). Taken together, these results indicated that phenotypes of sav2 were modified by shade light or dark conditions triggering hypocotyl elongation.
Figure 1.
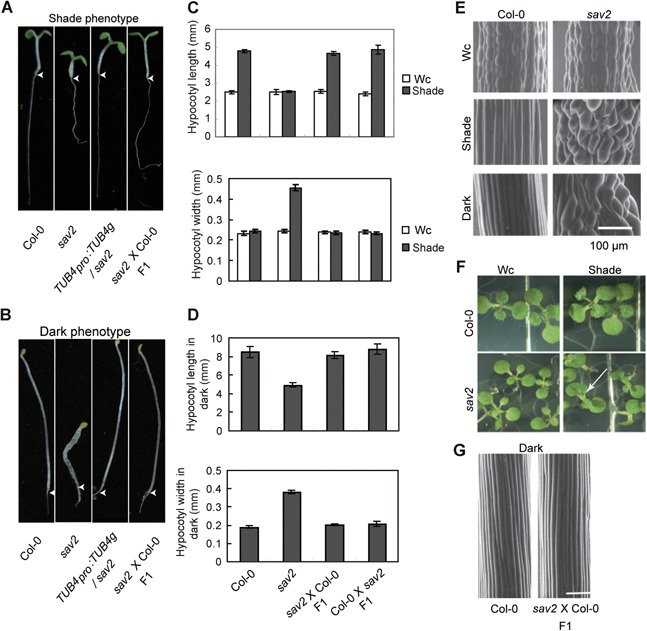
Phenotypic characterization of sav2 mutant (A, B) Hypocotyls of sav2 are short and swollen in shade and dark. sav2 behaved as a recessive mutant and was complemented by genomic fragment of TUB4. Representative seedlings of Col‐0, sav2, TUB4pro::TUB4g/sav2 and sav2(♀)X Col‐0 (♂) F1 heterozygous seedlings (g: genomic) grown in simulated shade (A) and dark (B, 3‐d old) are shown. Arrow head indicates the junction between hypocotyl and root. (C‐D) Quantitative measurements of hypocotyl length and width of Col‐0, sav2, sav2 (♀) X Col‐0 (♂) and Col‐0 (♀) X sav2 (♂) F1 heterozygous seedlings grown in continuous white light (Wc), shade (C) and dark (D). Error bars represent standard error of mean (n ≥ 10). (E) Scanning electron microscopy (SEM) images showing hypocotyl epidermal cells of Col‐0 and sav2 grown in Wc, shade and dark. Scale bars represent 100 µm. (F) sav2 mutants exhibit twisting petioles in shade, but not in Wc. Arrowhead indicates a pair of true leaves with twisted petioles. (G) SEM images showing hypocotyl epidermal cells of Col‐0 and F1 heterozygous seedlings of sav2 crossed to Col‐0 in dark. sav2 behaves as a recessive mutant. Scale bars = 100 μm.
sav2 contains a point mutation in TUB4 gene
We crossed sav2 with Col‐0 and observed that hypocotyls of the F1 seedlings were wild type‐like in Wc, shade or dark (Figure 1A–D, G), suggesting that mutation in sav2 may be recessive. Then, we crossed sav2 with Ler‐erecta ecotype to generate a mapping population. Through map‐based cloning, we narrowed down the mutation to a region between 17.8 Mb‐17.9 Mb on chromosome 5 that contains 17 genes. We then identified a C‐to‐T mutation in a β‐tubulin gene, TUB4 (At5g44340, tubulin beta chain 4) through direct sequencing. This mutation results in a P287L conversion (Figure S1A). Complementation test was performed using full length genomic DNA of TUB4 with its own promoter and 3′UTR. As shown in Figures 1A, B and S1B, TUB4 genomic DNA fully complemented sav2 and rescued its hypocotyl phenotype in shade and dark, indicating that the mutant phenotypes of sav2 indeed resulted from the P287L mutation in TUB4. Mutant with the exact same mutation in TUB4 was previously recovered by Ishida and colleagues through screening of mutants with root skewing phenotypes upon PPM (propyzamide, a MT disrupting drug) treatment (Ishida et al. 2007). In Ishida's study, the mutant displayed strong root phenotypes constitutively. However, in our study, the mutant had short and swollen hypocotyl only in shade/dark, but not in the regular growth conditions. We thus further characterized this mutant.
According to the published structure of the αβ tubulin dimmer, P287 locates in Helix 9 (Figure S1A) (Nogales et al. 1998). Helix 9 not only marks the border of M‐loop, which is a key structure involved in lateral contact between microtubule protofilaments, but also directly participates in the lateral interaction (Lowe et al. 2001; Ravelli et al. 2004). As the inter‐protofilament interaction is believed to define the stability and mechanical properties of microtubules (Sui and Downing 2010), we examined whether cMT stability was altered in sav2. We tested the sensitivity of sav2 to Taxol (a MT stabilizing drug) and PPM in both Wc and shade. Seedlings were grown on 1/2MS supplemented with PPM/Taxol. They were first grown in Wc for 5 d and then transferred to Wc or shade for 3 d. Taxol strongly inhibited the growth of wild type seedlings, while sav2 seedlings were much larger than the wild types under both Wc and shade on medium supplemented with Taxol, indicating they were more resistant to Taxol (Figure 2A, B). In the presence of PPM, sav2 and Col‐0 wild type seedlings were of similar size. However, hypocotyls of sav2 were more sensitive to PPM‐induced hypocotyl expansion, suggesting that sav2 was more sensitive to PPM than wild type (Figure 2A, B). To visualize cMTs directly, we crossed 35Spro:GFP‐TUA6 (Ueda K 1999) into sav2 and tested PPM sensitivity by examining PPM‐induced microtubule depolymerization. As shown in Figure 2C, 5‐d‐old light grown 35Spro:GFP‐TUA6 or 35Spro:GFP‐TUA6 /sav2 seedlings were soaked in 30 µM of PPM. After 30 min of PPM treatment, cMT microfibrils in Col‐0 only showed slight dissociation, while the majority of the microfibrils in sav2 mutants lost their fibrous structure, indicating that the tub4 P287L mutation resulted in increased PPM sensitivity and may destabilize cMTs.
Figure 2.
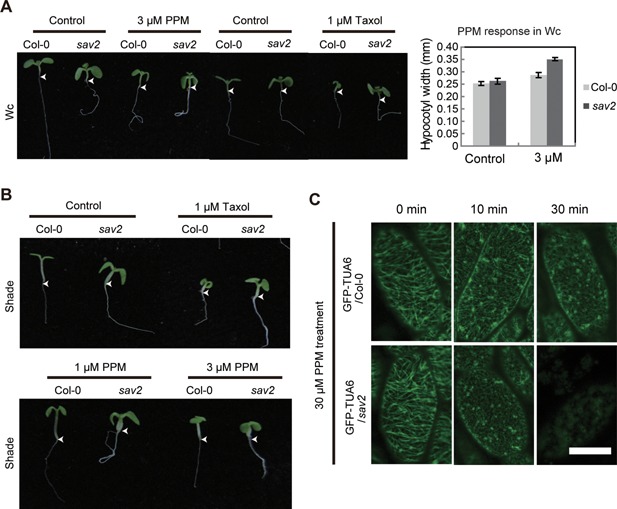
sav2 exhibits altered sensitivity to microtubule drugs (A, B) Phenotypes of Col‐0 and sav2 seedlings growing on 1/2MS medium supplemented with PPM or Taxol under Wc (A) or shade (B) are shown. Quantitative measurement of hypocotyl width of seedlings grown on control and PPM under Wc is shown on the right panel of (A). Error bars represent standard error of mean (n ≥ 10). Arrowhead indicates the junction between hypocotyl and root. (C) PPM induced cMT dissociation is enhanced in sav2 mutant. 5‐d‐old light grown seedlings (35S::GFP‐TUA6 in Col‐0 and sav2 background) were soaked in 30 µM of PPM for indicated amount of time. Patterns of GFP‐TUA6 are shown. Scale bars = 20 μm.
Shade induced cMT rearrangement is abolished in sav2
Identification of sav2 as a shade avoidance mutant suggests that cMTs are required for shade‐induced hypocotyl elongation. We thus examined how shade influences cMTs. We first examined cMT organization in hypocotyl epidermal cells under both Wc and shade using 35Spro:GFP‐TUA6 in wild type and in sav2 background. cMTs of cells in the middle region of the hypocotyls were examined as they respond rapidly to changes in light (Sambade et al. 2012). As shown in Figure 3A, the majority of the cMT arrays exhibited random or oblique patterns in both wild type and sav2 seedlings grown in Wc. After 12‐h of shade treatment, cMT patterns in wild type cells became mostly parallel (with respect to the longitudinal elongation axis), while cMTs in most of the sav2 cells showed a swirling, cotton ball‐like pattern (Figure 3A), indicating that tub4P287L affects shade‐induced cMT rearrangement.
Figure 3.
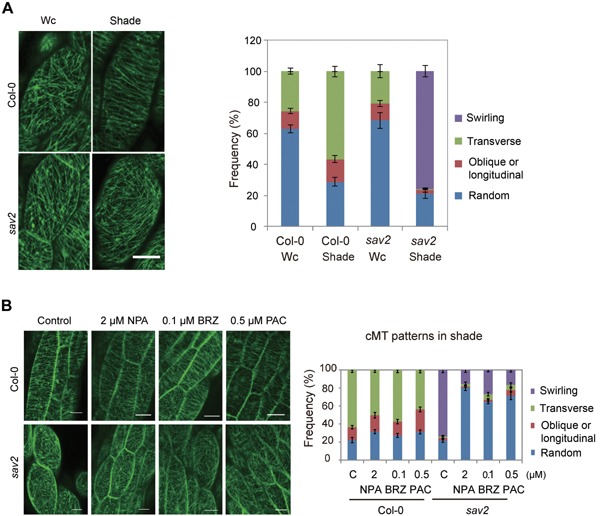
Shade induces cMT rearrangement (A) cMT patterns in Col‐0 and sav2 under Wc and shade. Quantitative data are shown on the right. Error bars represent standard error of mean (n ≥ 20). (B) Effects of hormone inhibitors: NPA, BRZ and PAC on shade‐induced cMT rearrangement in wild type and sav2. Quantitative data are shown on the right. Error bars represent standard error of mean (n ≥ 24 for Col‐0 samples, n ≥ 10 for sav2 samples). Scale bars = 20 µm.
Phytohormones including auxin, BRs and GAs were regulators of shade avoidance responses. We first examined if these hormones were required for shade‐induced cMT rearrangement using NPA (Auxin transporter inhibitor), BRZ (BR biosynthesis inhibitor brassinazol) and PAC (GA biosynthesis inhibitor Paclobutrazol). Interestingly, despite the strong inhibitory effect of NPA, BRZ and PAC on shade‐induced hypocotyl elongation in wild type seedlings (Figure S2A), we observed only slight reduction in the percentage of cells exhibiting transversely aligned microtubule fibrils (Figure 3B), suggesting that first, shade induced formation of transversely aligned cMTs can be uncoupled from hypocotyl elongation and it is not sufficient to promote hypocotyl elongation in shade. Second, transversely aligned cMTs may be induced through pathways other than these hormone signaling pathways. Alternatively, hormones may affect only the dynamics of cMT rearrangement, but are not the ultimate driving force for cMT rearrangement.
Although these hormone inhibitors exhibited limited effects on shade‐induced cMT rearrangement in wild type, all of them inhibited the formation of the swirling cMT in shade‐grown sav2 (Figure 3B) and increased percentage of cells with randomly organized cMTs, indicating that formation of the swirling cMTs in sav2 is hormone‐dependent. We speculated that it is the shade‐induced swirling cMTs that leads to hypocotyl swelling in sav2. To test this, we grew seedlings in the shade in the presence of various concentrations of NPA, PAC and BRZ and examined their hypocotyl phenotypes. As shown in Figure S2A, these chemicals all inhibited hypocotyl swelling of sav2. To test if these growth hormones are sufficient to induce hypocotyl expansion in sav2, we grew seedlings on 1/2 MS medium supplemented with picloram (an auxin analogue), epi‐brassinolide (BL) or GA3 under Wc. We found that all of these hormones induced hypocotyl elongation in wild type seedlings and promoted hypocotyl expansion in sav2 (Figure S2B), suggesting that these hormones are all required and sufficient for inducing hypocotyl expansion in sav2. To further examine the involvement of auxin and BR in shade‐induced hypocotyl expansion of sav2, we crossed sav2 to sav3‐1(taa1) and sav1‐1(dwf4) mutants, which contain mutations in key enzymes involved in the IAA and BR biosynthesis, respectively (Tao et al. 2008). As shown in Figure S2C, the double mutants had short, but significantly reduced hypocotyl width compared to the sav2, indicating that the shade‐induced hypocotyl swelling of sav2 does require these two hormones.
Shade‐grown seedlings are more sensitive to drugs disturbing cMT stability/dynamics
Dynamics of microtubules are critical for their functions. Both PPM and Taxol are microtubule drugs that interfere with microtubule stability/dynamics and they also inhibit hypocotyl elongation in the shade (Figure 2B). We thus tested if shade alters cMT stability/dynamics. First, we examined if PPM/Taxol sensitivity of wild type seedlings is affected by shade. Seedlings were grown on 1/2 MS medium supplemented with various concentrations of PPM/Taxol in Wc for 5 d, they were then either kept in Wc or transferred to shade and grown for 3 more days. Hypocotyl width was then measured. As shown in Figures 4A and S3A, increasing concentrations of PPM promotes radial expansion of wild type hypocotyls. Such an effect was enhanced in seedlings grown in the shade compared to those grown in Wc. Similar results were obtained in Taxol treated seedlings (Figure S3B). These data suggest that shade‐grown seedlings are more sensitive to perturbation of microtubule stability/dynamics.
Figure 4.
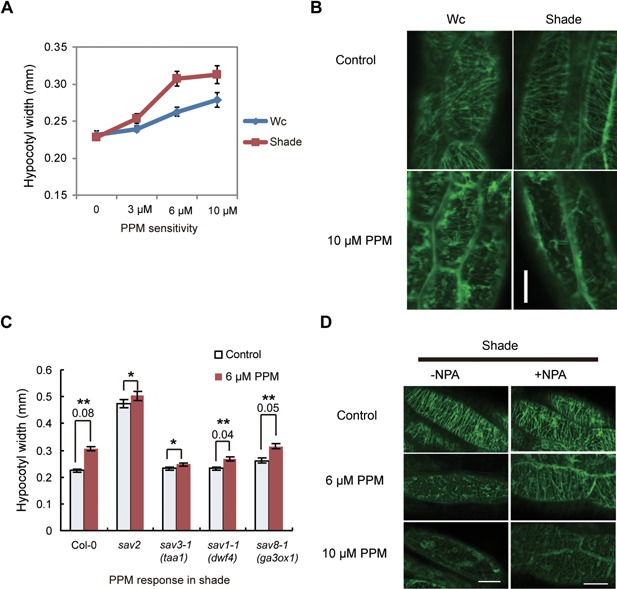
Shade‐grown seedlings are hypersensitive to perturbation of cMT dynamics (A) Hypocotyl width of wild type seedlings grown on 1/2MS supplemented with various concentrations of PPM under Wc and shade. Error bars represent standard error of mean (n = 20). (B) Responses of cMTs (visualized using GFP‐TUA6) to PPM in hypocotyl cells of seedlings (35S::GFP‐TUA6) grown in Wc and shade. 5‐d‐old light grown seedlings were treated with Wc or shade for 24 h, they were then soaked in 10 μM of PPM. The figures show representative pattern of GFP‐TUA6 in these seedlings in response to PPM treatment. (C) Responses of Col‐0, taa1, dwf4 and ga3ox1 to PPM in shade. Seedlings were grown on 1/2MS supplemented with various concentrations of PPM under shade. Hypocotyl width was measured. Error bars represent standard error of mean (n = 30). *Student's t‐test results indicate that there is no significant difference between control and PPM‐treated samples with P > 0.1. **t‐test results show there is significant difference between control and PPM‐treated samples with P < 0.001. The number below ** is the absolute difference in hypocotyl width (mm) between PPM treated and control samples. (D) Responses of cMTs (visualized using GFP‐TUA6) to PPM in hypocotyl cells in the presence of NPA. 35S::GFP‐TUA6 seedlings grown on 1/2MS medium supplemented with/without 2 μM of NPA for 5 d. They were then treated with Wc or shade for 24 h prior to PPM treatment. Scale bars=20 μm.
High doses of PPM induces de‐polymerization of microtubules in a short time, which allows us to directly visualize the sensitivity of microtubules to PPM. We used 5‐d‐old light‐grown, 35S::GFP‐TUA6 seedlings and treated them with Wc or shade for 24 h to allow changes in microtubules dynamics to be induced. Seedlings were then treated with 10 µM of PPM for 1 h and patterns of microtubules were visualized using confocal microscopy. As shown in Figure 4B, PPM‐promoted dissociation of cMT microfibrils occurred much faster in seedlings grown in the shade than those grown in Wc, confirming that shade enhanced PPM sensitivity.
The involvement of hormones in shade‐induced PPM/Taxol hypersensitivity was also examined. We first compared the PPM sensitivity of sav3‐1 (taa1), sav1‐1 (dwf4) and sav8‐1 (ga3ox1) to the wild type in shade by measuring hypocotyl width. As shown in Figure 4C, sav3‐1 was insensitive to PPM induced hypocotyl swelling, while both sav1‐1 and sav8‐1 had reduced, but detectable level of PPM sensitivity. Similar results were obtained for Taxol sensitivity assay (Figure S3C). Since all three mutants were short in shade and their hypocotyls are of similar length (Figure S3D), we propose that auxin may be a key component required for shade‐induced hypersensitivity to perturbation of cMT stability/dynamics.
To verify the role of auxin, we also examined if PPM sensitivity is affected by NPA. We grew 35S::GFP‐TUA6 seedlings on medium supplemented with or without NPA in Wc for 5 d, then treated seedlings with shade for 24 h followed by 1 h of PPM treatment. As shown in Figure 4D, 6 µM of PPM treatment induced cMT to disassemble in control seedlings, while in NPA‐treated seedlings, cMT fibrils were still visible. Such a difference is also clear in 10 µM of PPM treated samples. In addition, when 35S::GFP‐TUA6 seedlings were grown on medium supplemented with Picloram under Wc, they exhibited enhanced PPM sensitivity compared to the control seedlings (Figure S3E). These results are consistent with our previous results and indicate that auxin is involved in shade‐induced PPM hypersensitivity.
Dynamics of microtubule plus ends is reduced in shade
Dynamics of microtubule plus ends can be tracked using plus end‐binding protein, EB1 (Chan et al. 2003; Vaughan 2005). We used 35Spro::EB1b‐GFP transgenic line to study how shade influences the dynamics of cMT plus ends. With short exposure, EB1b appeared as a dot with a cometary tail (Figure 5A left panel). As the mobility of EB1 directly reflects microtubule polymerization rate at the plus end, we measured the velocity of EB1b over a fixed time period using confocal microscopy. We scanned eight images with 3.5‐sintervals. For the ease of measurement, we stacked images 1,3,5,7 using Image J software (http://imagej.net/Welcome). Examples of obtained images are shown in Figure 5A left panel. A line connecting all four dots was drawn, which represents the path of a microtubule plus end in 24.5 s. By measuring the length of these lines, we calculated the velocity of EB1b‐GFP. The results were shown in Figure 5A right panel. Under continuous white light, average velocity of EB1b proteins was 9.7 ± 0.10 µm/min. Upon shade treatment, EB1b velocity decreased gradually. By 24‐h of treatment, average velocity of EB1b was 7.2 ± 0.11 µm/min, which was significantly lower than that in the Wc. This result suggests that the polymerization rate of microtubule plus ends is reduced in shade. As overexpressing EB1b‐GFP may influence dynamics of cMTs, we further examined dynamics of cMTs using ATEB1bpro::ATEB1b‐GFP transgenic line (Dixit et al. 2006). As shown in Figure S4A, after 24 h of shade treatment, a significant reduction in EB1b velocity was also observed, which confirmed that shade treatment did reduce the polymerization rate of cMT plus ends. To test if shade‐altered EB1b velocity is hormone regulated, we measured velocity of EB1b in shade‐grown 35Spro::EB1b‐GFP seedlings treated with NPA, BRZ or PAC. As shown in Figure S4B, none of these inhibitors increased the EB1b velocity in shade, suggesting that reduced plus end polymerization in shade does not require these hormones.
Figure 5.
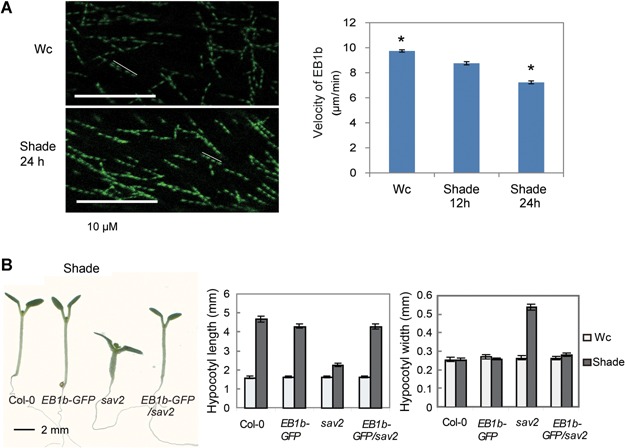
Regulation of EB1 by shade (A) Stacked confocal images of EB1b‐GFP, showing the velocity of EB1b is reduced in shade. White lines represent the movement of one EB1b‐GFP labeled cMT plus end within 24.5 s. Quantitative results are shown in the right panel. Error bars represent standard error of mean (n ≥ 65) *t‐test result showing these samples are significantly different with P < 0.001. (B) Overexpression of EB1b‐GFP in sav2 rescued hypocotyl phenotypes of sav2 in shade. Representative seedlings were shown on the left panel. Quantitative measurements of hypocotyl length and width are shown in the middle and right panel, respectively. Error bars represent standard error of mean (n = 20).
As EB1 can affect stability/dynamics of microtubules, we further investigated if EB1is regulated by shade. To investigate if the protein stability of EB1b is regulated by shade, we used 35Spro::EB1b‐GFP transgenic line. No significant difference in EB1b protein level was detected in shade treated samples (Figure S4C), suggesting that shade may not affect steady state EB1b protein level.
To examine if the dynamics of EB1b is affected in sav2, we crossed 35Spro::EB1b‐GFP into sav2. Genotype of sav2 was confirmed using dCAPS marker (Figure S4E). 35Spro::EB1b‐GFP/sav2 seedlings exhibited no significant difference in Wc compared to its two parental lines (Figure S4F). Interestingly, overexpression of EB1b rescued sav2 in the shade and dark (Figures 5B, S4D), suggesting that overexpression of EB1b counteracted the effect of sav2 mutation. As phenotypes of sav2 were rescued almost to the wild type level, we did not observe any difference in EB1b dynamics between 35Spro::EB1b‐GFP and 35Spro::EB1b‐GFP /sav2 and thus cannot further investigate the effect of sav2 mutation on EB1b dynamics.
tub4P287L mutation affects responses of cMTs to growth‐promoting stimuli
As overexpressing EB1b rescued sav2, it indicates that the tub4P287L mutation does not affect the role of cMTs in supporting hypocotyl elongation. sav2 mutants are short and swollen in the dark as well. As elongation of hypocotyls in dark was not affected by NPA (Jensen et al. 1998), we examined how NPA would affect sav2 in the dark. Interestingly, NPA not only inhibited the hypocotyl swelling, but also promoted the hypocotyl elongation of sav2 in a concentration dependent manner (Figure 6A). As in shade, cMTs in many hypocotyl cells of dark grown sav2 showed a swirling pattern, which could also be rescued by NPA (Figure S5A, B). Further analysis revealed that although high concentrations of BRZ/PAC inhibited hypocotyl elongation of wild type seedlings in the dark, when used at low concentration, both BRZ and PAC partially rescued sav2 and promoted hypocotyl elongation (Figure 6B). They both rescued the swirling cMT pattern in sav2 as well (Figure S5A, B). The above results also indicated that tub4P287L mutation does not abolish functions of cMTs in elongation growth of hypocotyl cells. We propose that tub4P287L may represent a unique type of mutation that affects the responses of cMTs to growth promoting stimuli, producing swirling cMT, which subsequently causes hypocotyl expansion and inhibits hypocotyl elongation.
Figure 6.
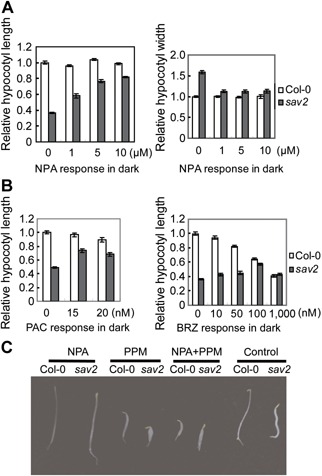
Dark phenotypes of sav2 (A) Hypocotyl length and width of sav2 in the dark in the presence of various concentrations of NPA, showing that NPA can rescue sav2 in the dark. (B) Relative hypocotyl length of sav2 in the dark in the presence of low concentrations of PAC or BRZ, showing both PAC and BRZ can partially rescue sav2 in dark. (C) Hypocotyl phenotypes of dark‐grown Col‐0 and sav2 in the presence of NPA (2 µM), PPM (2 µM) or both.
PPM or Taxol treated dark grown wild type seedlings were short (Figures 6C, S5C), indicating proper function of cMTs is required for hypocotyl elongation in the dark. However, NPA did not rescue PPM or Taxol induced short hypocotyl of wild type seedlings (Figures 6C, S5C), which indicated that the rescuing effect is specific to the tub4P287L mutation.
DISCUSSION
In our study, sav2 was identified as a recessive mutant defective in shade induced hypocotyl elongation. It contains a P287L mutation in the TUB4 protein. Mutant with the same mutation in TUB4 was previously recovered by Ishida and colleagues (Ishida et al. 2007) as a semi‐dominant mutant. Consistent with Ishida's result, we also found that when grown on vertical plates with 1.5% agar, roots of sav2 slanted to the right (viewing from the top), while roots of Col‐0 skewed slightly to the left (Figure S6A). In addition, roots of sav2 were short and highly swollen (Figure S6A–C). All of these sav2 root phenotypes were visible in seedlings grown in Wc. Roots of tub4P287L heterozygous seedlings also skewed to the right on hard agar, but to a lesser degree (Figure S6A). Moreover, root width of heterozygous seedlings was in‐between Col‐0 and sav2 (Figure S6D). Interestingly, root length of the heterozygous seedlings was wild type‐like (Figure S6A, C). Thus, depending on the phenotypes examined, tub4P287L mutant behaved as a recessive or semi‐dominant mutant. Ishida and colleagues showed that YFP tagged tub4P287L could incorporate into microtubule polymers and overexpressing tub4P287L in wild type recapitulated the mutant phenotype. Thus, tub4P287L is likely to be a gain‐of‐function mutant, which explains why a point mutation in one of the nine β‐Tubulins would produce such strong phenotypes. We propose that the phenotype severity of tub4P287L mutant may be influenced by other factors such as hormones and activities of microtubule‐associated proteins such as EB1. These factors may be responsive to environmental signals and vary in concentrations or activities in different tissues, which would then explain why tub4P287L behaves differently when different phenotypes are examined.
Shade induced hypocotyl elongation involves co‐operative actions of multiple hormones including auxin, BRs and GAs. All of these hormones were previously reported to be able to promote the formation of transversely aligned cMTs (Blancaflor and Hasenstein 1995; Wenzel et al. 2000; Catterou et al. [Link], [Link]; Wiesler et al. 2002; De Grauwe et al. 2005; Komorisono et al. 2005). Although inhibitors of these hormone pathways all rescued the swirling cMT pattern and swollen hypocotyls of sav2 in the shade, they had limited effect on shade‐induced cMT rearrangement in the wild type (Figures 3, S2). This suggests that shade‐induced cMT reorganization requires other regulatory factors. Alternatively, hormones may only modify the dynamics of cMT rearrangement, but are not the driving forces for it. Furthermore, we discovered that shade grown seedlings exhibited higher sensitivity to PPM and reduced EB1b velocity, which indicated that regulation of cMTs by shade occurs at multiple levels. EB1 was reported to stabilize the seam between the first and the 13th protofilaments and promote microtubule formation or stabilize tube structure. Further analysis on EB1s and ateb1a,b,c triple mutant would be needed to test if shade‐induced PPM hypersensitivity correlates with the shade‐induced reduction in EB1 velocity.
P287 locates in Helix 9 and is potentially involved in lateral contacts between microtubule protofilaments. sav2 is hypersensitive to PPM and hyposensitive to Taxol, indicating the mutant protein may reduce cMT stability/dynamics (Figure 2). Hypocotyls of sav2 mutants were short and swollen in shade, but were wild type‐like in Wc (Figure 1). To investigate if the light‐regulated hypocotyl phenotypes of sav2 resulted from shade‐induced changes in the TUB4 expression level or not, we examined the mRNA and the protein levels of TUB4. As shown in Figure S7A, in response to shade, no significant change in TUB4 mRNA level was detected in wild type or sav2 seedlings. To detect changes in TUB4 at the protein level, we used transgenic plants overexpressing YFP‐TUB4 and did not detect significant changes at protein level either (Figure S7B). These results indicated that the light‐regulated hypocotyl phenotypes of sav2 were not due to changes at TUB4 level. We thus hypothesize that shade‐induced phenotypes of sav2 may be due to altered responses of cMTs containing tub4P287L to shade‐derived signals, such as hormones.
Despite the fact that hypocotyls of sav2 were short in both shade and dark, we were surprised to find that NPA rescues only the swollen hypocotyls of sav2 in the shade, but completely rescued sav2 in the dark (Figures 6, S2). In addition, overexpressing EB1b‐GFP in sav2 rescued its phenotypes in both dark and shade (Figures 5B, S4D), These results revealed that first, the P287L mutation in TUB4 does not affect the role of cMTs in supporting hypocotyl elongation; and second, phenotypes of sav2 may result from altered responses of cMTs to growth promoting stimuli; alternatively, the mutation may affect downstream signaling mediated by EB1 or auxin.
It was previously reported that auxin transport was required for hypocotyl elongation in light‐grown but not in dark grown Arabidopsis (Jensen et al. 1998). With 2 µM of NPA, we observed a strong reduction in hypocotyl length of shade‐treated seedlings, but observed not much change in dark‐grown seedlings treated with up to 5 µM of NPA treatment (Figures 6, S2), which was consistent with the previous report. The fact that NPA rescued sav2 in dark indicating that tub4P287L does not abolish the function of cMTs in supporting hypocotyl growth and the short hypocotyls may result from altered responses of cMTs to auxin in the dark. Incorporation of tub4P287L may promote the formation of cotton‐ball shaped cMT array in the presence of auxin, which causes hypocotyl expansion and subsequently blocks hypocotyl elongation. NPA treatment inhibited the formation of cotton‐ball shaped cMT array, released its inhibition on hypocotyl elongation. As NPA itself did not inhibit hypocotyl elongation in the dark, it thus could rescue sav2 in the dark. In the shade, although NPA treatment also prevented the formation of cotton‐ball like cMTs as it does in the dark, it may also block the function of other elongation promoting factors regulated by auxin since NPA is required for hypocotyl elongation of wild type seedlings in the shade. Consequently, NPA only rescued hypocotyl swelling, but not hypocotyl elongation of sav2 in shade.
Overexpressing EB1b‐GFP, on the other hand, rescued sav2 in both shade and dark. One simple explanation could be that P287L mutation may reduce the binding affinity of EB1b to cMTs, as the mutated residue locates at Helix 9, which is likely to be required for interaction between protofilaments and interacts with EB1. Overexpressing EB1b‐GFP may rescue sav2 by compensating for the reduced affinity of tub4P287L to EB1b. As EB1s function by both regulating microtubule stability/dynamics and interacting with other MAPs or cargo proteins, reduced interaction between EB1b and tub4P287L may affect both cMT stability/dynamics and downstream signaling (Sandblad et al. 2006). Further study is required to unravel the exact impact of tub4P287L on EB1.
In summary, P287 of TUB4 is a residue critical for cMTs to mount proper responses to growth promoting stimuli. This residue could either be directly involved in interactions with various MAPs, such as EB1 or it is critical for shade‐induced changes in cMT.
MATERIALS AND METHODS
Plant materials and growth conditions
35S::GFP‐TUA6 (Ueda K 1999), 35S::EB1b‐GFP (overexpressing Arabidopsis EB1b, a gift from Dr. Tongling Mao), ATEB1bpro::EB1b‐GFP (Dixit et al. 2006) sav3‐1, sav1‐1 (Tao et al. 2008), sav8‐1 (unpublished ga3ox1 mutant isolated in the lab, mutant phenotypes can be complemented with GA3OX1 genomic DNA) were used and crossed with sav2. Seeds were sown on 1/2 MS media supplemented with 0.8% agar, and then imbibed at 4°C for 2−4 d. All seedlings were grown at 22°C. For shade treatment, seedlings were first grown in Wc (66 µmol m−2 s−1) for 5 d, they were then transferred to simulated shade (Red: 13 µmol m−2 s−1, Blue: 3.5 µmol m−2 s−1, Far red: 21 µmol m−2 s−1) and grew for 3 d. Hypocotyls of dark grown seedlings were measured after seedlings were grown in the dark for 3.0−3.5 d. Quantitative measurements of hypocotyls were performed on scanned images (100% original size for hypocotyl length measurement and 400% enlarged images for hypocotyl width measurement) of seedlings using scion image software (http://www.scioncorp.com). In all figures, error bars represent standard error of mean.
For hormone and chemical treatments, Picloram and GA3 (BBI, Markham, Canada) were dissolved in ethanol, epi‐BL (Sigma, St. Louis, MO, USA), BRZ, NPA and Paclobutrazol (BBI) were dissolved in dimethylsulfoxide (DMSO), Ethephon and AVG (Sigma) were dissolved in water. Taxol (Sangon, Shanghai, China) and PPM (Sigma) were dissolved in DMSO. For hypocotyl measurements and cMT pattern assays, seeds were sown on 1/2MS medium supplemented with hormones/chemicals at indicated concentrations and they were then grown under conditions described above. For PPM/Taxol sensitivity assay, seedlings were grown on 1/2MS supplemented with PPM/Taxol for 5 d and they were then treated with shade for 2 d before hypocotyl width measurement. For PPM‐induced cMT dissociation assay, light‐grown, 5‐d old seedlings were first treated with shade for 24 h, seedlings were then soaked in PPM for 1 h or the indicated amount of time, cMT patterns were subsequently visualized using confocal microscopy.
sav2 genotyping
A dCAPS marker was designed (http://helix.wustl.edu/dcaps/dcaps.html) to genotype sav2. Primer sequences were as follows: sav2‐dCAPS‐F 5′‐ACCTTAGGAAACTCGCTGTGAA‐3′, sav2‐dCAPS‐R 5′‐ACATCTGCTGGGTCAGTCCA‐3′. A 135 bp polymerase chain reaction (PCR) product was amplified followed by ScrFI (Thermo, Waltham, MA, USA) digestion. Resulting fragments were separated in 3% agarose gel. A 115 and a 20 bp fragment can be generated using Col‐0 wild type genomic DNA and an undigested 135bp will be obtained using sav2 genomic DNA.
Microscopy
For cMT pattern analysis using 35S::GFP‐TUA6 line, cMTs of cells in the middle region of 4.5‐d‐old seedling hypocotyls were examined and imaged using confocal laser scanning microscope (Zeiss LSM 780, Oberkochen, Germany, 40× objective). The excitation wavelength was 488 nm, and for GFP emission a 505−530 nm band path filter was used. Patterns of cMTs were classified into four groups: transverse (perpendicular to the growth axis), oblique or longitudinal, random and swirling. For each treatment, at least 16 hypocotyls were examined and imaged. For PPM sensitivity analysis using confocal microscopy, two hypocotyl cells from at least eight seedlings were imaged and analyzed.
For EB1 moving rate measurement, 5‐d‐old 35S::EB1b‐GFP or ATEB1bpro::ATEB1b‐GFP transgenic seedlings were used. We scanned eight images (Zeiss LSM 780, 63× oil immersion objective) with 3.5 s interval, and images 1, 3, 5 and 7 were stacked using Image J (http://rsb.info.nih.gov/ij/). A freehand line was drawn to connect the four dots and the distance between the first and the 7th dot was measured, which represented moving distance of EB1b‐GFP within 24.5 s. Moving rates were then calculated. In each experiment, 8−18 seedlings were observed. The whole experiment was repeated more than three times.
Scanning electron microscopy images were obtained using a XL30 ESEM scanning electron microscope (Philips, Amsterdam, Netherlands). Fresh seedlings grown under Wc or shade as described above were picked up from medium, and immediately washed by MES (pH5.6) buffer. Whole seedlings were scanned at 4°C, under 4.9 mBar, and middle regions of hypocotyls were imaged.
Quantitative RT‐PCR
Total RNAs were extracted following Roche TriPure RNA isolation protocol (www.roche-applied-science.com). One microgram of total RNAs were reverse transcribed using the First Strand cDNA Synthesis Kit (Thermo). Quantitative real‐time PCR was performed using SYBR green method with a Stratagene M×3000P real‐time system cycler (Agilent, Santa Clara, CA, USA). A 40‐cycle, three‐step amplification protocol (10 s at 95°C, 20s at 60°C and 25 s at 72°C) was used for all measurement. REF3 (At1g13320) was used as reference gene (Czechowski et al. 2005). At least three replicates were included in each experiment. Primers used were as below: REF3‐f 5′‐ GGAGCCAACTAGGACGGATCTGG‐3′, REF3‐r 5′‐ GTAGATCAATCCCAATAACCTGGTTCACTT‐3′. TUB4‐f 5′‐TAAGATGTTGTCAATGGCTCCCT‐3′, TUB4‐r 5′‐AATACACGCAAAAGTTTAACAAATCC‐3′.
Western
Total proteins were extracted following tripure protein isolation protocol (www.roche-applied-science.com), and separated on 10% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) gel. EB1b‐GFP, YFP‐TUB4 and YFP‐mTUB4 proteins were probed with anti‐GFP antibody (www.Beyotime.com).
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web‐site:
Figure S1. sav2 can be complemented by TUB4 (A) P287 locates in Helix 9. Protein sequence alignment of Arabidopsis TUB4 and β tubulin (sequence obtained from PDB: 1TUB) Black line marks region of helix 9 and the arrowhead indicates P287 residue. (B) Quantitative measurements of hypocotyl length and width of seedlings grown in shade and dark, showing phenotypes of sav2 in shade and dark are complemented by genomic fragment of TUB4 gene. Col‐0, sav2 and TUB4pro::TUB4g/sav2 (g: genomic sequence of TUB4 with its own promoter and 3'UTR) were measured. Error bars represent standard error of mean (n ≥ 8).
Figure S2. Shade‐induced hypocotyl swelling of sav2 requires various hormones (A) Quantitative measurements of hypocotyl width and length of Col‐0 and sav2 seedlings grown on 1/2MS medium supplemented with various concentrations of NPA, BRZ, PAC or AVG in shade. (B) Quantitative measurements of hypocotyl width and length of Col‐0 and sav2 seedlings grown on 1/2MS medium supplemented with various concentrations of Picloram, BL or GA3 in Wc. (C) Hypocotyl width and length of sav2 sav1‐1 and sav2 sav3‐1 double mutants in Wc and shade, showing mutation in TAA1 (sav3‐1) or DWF4 (sav1‐1) suppresses hypocotyl swelling of sav2 in shade. Error bars represent standard error of mean (n ≥ 10).
Figure S3. cMTs in shade‐grown seedlings are hypersensitive to perturbations of cMT dynamics (A) Representative wild type seedlings grown on 1/2 MS medium supplemented with/without PPM under Wc or shade. (B) Hypocotyl width of wild type seedlings grown on 1/2m MS medium supplemented with various concentrations of Taxol in Wc or shade. (C) Hypocotyl width of Col‐0, sav2, sav3‐1, sav1‐1 and sav8‐1 grown in shade with/without Taxol treatment. (D) Hypocotyl length of sav3‐1, sav1‐1, sav8‐1 in Wc and shade. (E) Responses of cMTs (visualized using GFP‐TUA6) to PPM in hypocotyl cells of seedlings grown in Wc on 1/2MS medium supplemented with/without 1 µM of picloram. Seedlings were soaked in PPM for 1 h. Scale bars = 20 µm. Error bars represent standard error of mean (n = 20–30).
Figure S4. Shade‐induced changes in EB1b and overexpressing EB1b‐GFP suppresses sav2 (A) Velocity of EB1b is reduced in response to shade. 5‐d‐old ATEB1bpro::ATEB1b‐GFP transgenic seedlings were treated with Wc or shade for 24 h. Moving rate of more than 220 EB1b‐labelled plus ends from 16 seedlings were measured. Error bars represent standard error of mean. Student's t‐test analysis revealed that there is significant difference between EB1 Wc‐ and shade‐treated samples with P < 0.002. (B) Velocity of EB1b in NPA, BRZ or PAC‐treated, shade‐grown (24 h) 35S::EB1b‐GFP seedlings. Error bars represent standard error of mean (n = 30). (C) Protein levels of EB1b‐GFP in 35S::EB1b‐GFP seedlings grown in Wc and shade, showing protein stability of EB1b‐GFP is not altered by shade. (CBB: commassie brilliant blue staining). (D) Representative seedlings of Col‐0, 35S::EB1b‐GFP, sav2 and 35S::EB1b‐GFP/sav2 in dark. Quantitative measurement of hypocotyl width and length of these seedlings are shown in the middle and right panel, respectively. Error bars represent standard error of mean (n = 20–30). (E) Genotyping of 35S::EB1b‐GFP/sav2 using a dCAPS marker. (F) Representative seedlings of Col‐0, 35S::EB1b‐GFP, sav2 and 35S::EB1b‐GFP/sav2 in Wc. (G) Western blot showing ATEB1b‐GFP protein level in ATEB1bpro::ATEB1b‐GFP transgenic plants. 5‐d‐old light grown seedlings were treated with Wc or shade for 24 h, soluble protein was extracted and probed with anti‐GFP antibody. (CBB, commassie brilliant blue staining).
Figure S5. Dark phenotypes of sav2 can be rescued by NPA (A) cMT patterns in dark grown sav2 in response to NPA, BRZ or PAC. Bar, 20 µm. (B) Quantitative measurement of percentage of cells with different patterns of cMTs in dark grown seedlings. 35S::GFP‐TUA6 in wild type or sav2 background were grown on 1/2 MS medium supplemented with or without various hormone inhibitors (NPA, BRZ or PAC) for 3 d. Error bars represent standard error of mean. (n ≥ 277 hypocotyls, from more than 20 seedlings). (C) Responses of dark grown Col‐0 and sav2 to NPA (2 µM), Taxol (0.5 µM) or both.
Figure S6. Root phenotypes of sav2 (A) Root phenotypes of Col‐0, sav2 and F1 heterozygous seedlings of sav2 crossed to Col‐0. Seedlings were grown vertically on 1.5% hard agar under Wc. Pictures were taken from the top. (B) Roots of Col‐0 and sav2 grown in Wc, showing that roots of sav2 are wider than that of the wild type. (C‐D) Quantitative measurement of root length (C) and root width (D) of Col‐0, sav2 and F1 heterozygous seedlings of sav2 crossed to Col‐0. Error bars represent standard error of mean (n = 8).
Figure S7. Expression of TUB4 at mRNA and protein levels (A) Quantitative RT‐PCR results showing relative expression level of TUB4 in Col‐0 and sav2 under Wc and shade. Five‐d‐old light‐grown seedlings were treated with Wc or simulated shade for 2 h. Error bars represent standard error of mean (n = 3). (B) Protein levels of TUB4 did not change in response to shade. Five‐day‐old transgenic 35S::YFP‐TUB4 seedlings were treated with white light or simulated shade for 2 h (left panel) or indicated amount of time (right panel). Line 1 and line 2 were different overexpression lines. YFP‐TUB4 was detected by western using anti‐GFP antibody. Commassie blue (CBB) stain of total proteins are shown as loading control.
ACKNOWLEDGEMENTS
We thank Dr. Rong Yu for providing 35S::GFP‐TUA6 seeds; Dr. Takashi Hashimoto for providing tub4P287L, tub4E288K and other tubulin mutants; Dr. Tonglin Mao for critical reading and providing 35S::EB1b‐GFP and ATEB1bpro::ATEB1b‐GFP seeds. Early stages of this work were supported by the National Institutes of Health, 5 RO1GM52413 to J.C. and the Howard Hughes Medical Institute. Later work was funded by the Science and Technology Program of Fujian Province, 2008F3102 to Y.T.; the National Natural Science Foundation of China, 30870210, 90917013 to Y.T; Fundamental Research Funds for the Central Universities 2010121090 to Y.T. This work was also supported by 111 Project B12001.
Yu J, Qiu H, Liu X, Wang M, Gao Y, Chory J, Tao Y ( 2015) Characterization of tub4P287L, a β‐tubulin mutant, revealed new aspects of microtubule regulation in shade. J Integr Plant Biol 57: 757–769. 10.1111/jipb.12363
Available online on Apr. 21, 2015 at www.wileyonlinelibrary.com/journal/jipb
REFERENCES
- Bisgrove SR, Lee YR, Liu B, Peters NT, Kropf DL ( 2008) The microtubule plus‐end binding protein EB1 functions in root responses to touch and gravity signals in Arabidopsis . Plant Cell 20: 396–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancaflor EB, Hasenstein KH ( 1995) Time‐course and auxin sensitivity of cortical microtubule reorientation in maize roots. Protoplasma 185: 72–82 [DOI] [PubMed] [Google Scholar]
- Catterou M, Dubois F, Schaller H, Aubanelle L, Vilcot B, Sangwan‐Norreel BS, Sangwan RS (2001a) Brassinosteroids, microtubules and cell elongation in Arabidopsis thaliana. I. Molecular, cellular and physiological characterization of the Arabidopsis bul1 mutant, defective in the Delta(7)‐sterol‐C5‐desaturation step leading to brassinosteroid biosynthesis. Planta 212: 659–672 [DOI] [PubMed] [Google Scholar]
- Catterou M, Dubois F, Schaller H, Aubanelle L, Vilcot B, Sangwan‐Norreel BS, Sangwan RS (2001b) Brassinosteroids, microtubules and cell elongation in Arabidopsis thaliana. II. Effects of brassinosteroids on microtubules and cell elongation in the bul1 mutant. Planta 212: 673–683 [DOI] [PubMed] [Google Scholar]
- Chan J, Calder GM, Doonan JH, Lloyd CW ( 2003) EB1 reveals mobile microtubule nucleation sites in Arabidopsis . Nat Cell Biol 5: 967–971 [DOI] [PubMed] [Google Scholar]
- Chan J, Eder M, Crowell EF, Hampson J, Calder G, Lloyd C ( 2011) Microtubules and CESA tracks at the inner epidermal wall align independently of those on the outer wall of light‐grown Arabidopsis hypocotyls. J Cell Sci 124: 1088–1094 [DOI] [PubMed] [Google Scholar]
- Crowell EF, Timpano H, Desprez T, Franssen‐Verheijen T, Emons AM, Hofte H, Vernhettes S ( 2011) Differential regulation of cellulose orientation at the inner and outer face of epidermal cells in the Arabidopsis hypocotyl. Plant Cell 23: 2592–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR ( 2005) Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis . Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Grauwe L, Vandenbussche F, Tietz O, Palme K, Van Der Straeten D ( 2005) Auxin, ethylene and brassinosteroids: Tripartite control of growth in the arabidopsis hypocotyl. Plant Cell Physiol 46: 827–836 [DOI] [PubMed] [Google Scholar]
- Dixit R, Chang E, Cyr R ( 2006) Establishment of polarity during organization of the acentrosomal plant cortical microtubule array. Mol Biol Cell 17: 1298–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic‐Petrovic T, de Wit M, Voesenek LA, Pierik R ( 2007) DELLA protein function in growth responses to canopy signals. Plant J 51: 117–126 [DOI] [PubMed] [Google Scholar]
- Effendi Y, Jones AM, Scherer GF ( 2013) AUXIN‐BINDING‐PROTEIN1 (ABP1) in phytochrome‐B‐controlled responses. J Exp Bot 64: 5065–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC ( 2005) Phytochromes and shade‐avoidance responses in plants. Ann Bot‐London 96: 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, Hofte H ( 1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana . Plant Physiol 114: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M ( 1998) High temperature promotes auxin‐mediated hypocotyl elongation in Arabidopsis . Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasezawa S, Ueda K, Kumagai F ( 2000) Time‐sequence observations of microtubule dynamics throughout mitosis in living cell suspensions of stable transgenic Arabidopsis – Direct evidence for the origin of cortical microtubules at M/G(1) interface. Plant Cell Physiol 41: 244–250 [DOI] [PubMed] [Google Scholar]
- Hashimoto T ( 2013) Dissecting the cellular functions of plant microtubules using mutant tubulins. Cytoskeleton (Hoboken) 70: 191–200 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kaneko Y, Iwano M, Hashimoto T ( 2007) Helical microtubule arrays in a collection of twisting tubulin mutants of Arabidopsis thaliana . Proc Natl Acad Sci USA 104: 8544–8549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PJ, Hangarter RP, Estelle M ( 1998) Auxin transport is required for hypocotyl elongation in light‐grown but not dark‐grown Arabidopsis . Plant Physiol 116: 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LA, Peeters AJ, Pierik R ( 2010) Auxin transport through PIN‐FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci USA 107: 22740–22744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki S, Abe T, Coutuer S, Inze D, Russinova E, Hashimoto T ( 2010) Nuclear‐localized subtype of end‐binding 1 protein regulates spindle organization in Arabidopsis . J Cell Sci 123: 451–459 [DOI] [PubMed] [Google Scholar]
- Komorisono M, Ueguchi‐Tanaka M, Aichi I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M, Sazuka T ( 2005) Analysis of the rice mutant dwarf and gladius leaf 1. Aberrant katanin‐mediated microtubule organization causes up‐regulation of gibberellin biosynthetic genes independently of gibberellin signaling. Plant Physiol 138: 1982–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuka T, Kobayashi J, Horiguchi G, Demura T, Sakakibara H, Tsukaya H, Nagatani A ( 2010) Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol 153: 1608–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Wang XL, Qin T, Zhang Y, Liu XM, Sun JB, Zhou Y, Zhu L, Zhang ZD, Yuan M, Mao TL ( 2011) MDP25, a novel calcium regulatory protein, mediates hypocotyl cell elongation by destabilizing cortical microtubules in Arabidopsis . Plant Cell 23: 4411–4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Qin T, Ma Q, Sun J, Liu Z, Yuan M, Mao T ( 2013) Light‐regulated hypocotyl elongation involves proteasome‐dependent degradation of the microtubule regulatory protein WDL3 in Arabidopsis . Plant Cell 25: 1740–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C ( 2011) Dynamic microtubules and the texture of plant cell walls. Int Rev Cel Mol Bio 287: 287–329 [DOI] [PubMed] [Google Scholar]
- Locascio A, Blazquez MA, Alabadi D ( 2013) Dynamic regulation of cortical microtubule organization through prefoldin‐DELLA interaction. Curr Biol 23: 804–809 [DOI] [PubMed] [Google Scholar]
- Lowe J, Li H, Downing KH, Nogales E ( 2001) Refined structure of alpha beta‐tubulin at 3.5 A resolution. J Mol Biol 313: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Mimori‐Kiyosue Y, Grigoriev I, Lansbergen G, Sasaki H, Matsui C, Severin F, Galjart N, Grosveld F, Vorobjev I, Tsukita S, Akhmanova A ( 2005) CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus‐end dynamics at the cell cortex. J Cell Biol 168: 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH ( 1998) Structure of the alpha beta tubulin dimer by electron crystallography. Nature 391: 199–203 [DOI] [PubMed] [Google Scholar]
- Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M ( 2004) Insight into tubulin regulation from a complex with colchicine and a stathmin‐like domain. Nature 428: 198–202 [DOI] [PubMed] [Google Scholar]
- Saibo NJM, Vriezen WH, Beemster GTS, Van der Straeten D (2003) Growth and stomata development of Arabidopsis hypocotyls are controlled by gibberellins and modulated by ethylene and auxins. Plant J 33: 989–1000 [DOI] [PubMed] [Google Scholar]
- Sambade A, Pratap A, Buschmann H, Morris RJ, Lloyd C ( 2012) The influence of light on microtubule dynamics and alignment in the Arabidopsis hypocotyl. Plant Cell 24: 192–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandblad L, Busch KE, Tittmann P, Gross H, Brunner D, Hoenger A ( 2006) The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell 127: 1415–1424 [DOI] [PubMed] [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I ( 1999) Shade avoidance responses are mediated by the ATHB‐2 HD‐Zip protein, a negative regulator of gene expression. Development 126: 4235–4245 [DOI] [PubMed] [Google Scholar]
- Sui H, Downing KH ( 2010) Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure 18: 1022–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong FX, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, Cheng YF, Lim J, Zhao YD, Ballare CL, Sandberg G, Noel JP, Chory J ( 2008) Rapid synthesis of auxin via a new tryptophan‐dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thitamadee S, Tuchihara K, Hashimoto T ( 2002) Microtubule basis for left‐handed helical growth in Arabidopsis . Nature 417: 193–196 [DOI] [PubMed] [Google Scholar]
- Ueda K MT, Hashimoto T ( 1999) Visualization of microtubules in living cells of transgenic Arabidopsis thaliana . Protoplasma 206: 201–206 [Google Scholar]
- Vandenbussche F, Verbelen JP, Van der Straeten D (2005) Of light and length: Regulation of hypocotyl growth in Arabidopsis . Bioessays 27: 275–284 [DOI] [PubMed] [Google Scholar]
- Vaughan KT ( 2005) TIP maker and TIP marker; EB1 as a master controller of microtubule plus ends. J Cell Biol 171: 197–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XL, Zhang J, Yuan M, Ehrhardt DW, Wang ZY, Mao TL ( 2012) Arabidopsis MICROTUBULE DESTABILIZING PROTEIN40 is involved in brassinosteroid regulation of hypocotyl elongation. Plant Cell 24: 4012–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel CL, Williamson RE, Wasteneys GO ( 2000) Gibberellin‐induced changes in growth anisotropy precede gibberellin‐dependent changes in cortical microtubule orientation in developing epidermal cells of barley leaves. Kinematic and cytological studies on a gibberellin‐responsive dwarf mutant, M489. Plant Physiol 124: 813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesler B, Wang QY, Nick P ( 2002) The stability of cortical microtubules depends on their orientation. Plant J 32: 1023–1032 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web‐site:
Figure S1. sav2 can be complemented by TUB4 (A) P287 locates in Helix 9. Protein sequence alignment of Arabidopsis TUB4 and β tubulin (sequence obtained from PDB: 1TUB) Black line marks region of helix 9 and the arrowhead indicates P287 residue. (B) Quantitative measurements of hypocotyl length and width of seedlings grown in shade and dark, showing phenotypes of sav2 in shade and dark are complemented by genomic fragment of TUB4 gene. Col‐0, sav2 and TUB4pro::TUB4g/sav2 (g: genomic sequence of TUB4 with its own promoter and 3'UTR) were measured. Error bars represent standard error of mean (n ≥ 8).
Figure S2. Shade‐induced hypocotyl swelling of sav2 requires various hormones (A) Quantitative measurements of hypocotyl width and length of Col‐0 and sav2 seedlings grown on 1/2MS medium supplemented with various concentrations of NPA, BRZ, PAC or AVG in shade. (B) Quantitative measurements of hypocotyl width and length of Col‐0 and sav2 seedlings grown on 1/2MS medium supplemented with various concentrations of Picloram, BL or GA3 in Wc. (C) Hypocotyl width and length of sav2 sav1‐1 and sav2 sav3‐1 double mutants in Wc and shade, showing mutation in TAA1 (sav3‐1) or DWF4 (sav1‐1) suppresses hypocotyl swelling of sav2 in shade. Error bars represent standard error of mean (n ≥ 10).
Figure S3. cMTs in shade‐grown seedlings are hypersensitive to perturbations of cMT dynamics (A) Representative wild type seedlings grown on 1/2 MS medium supplemented with/without PPM under Wc or shade. (B) Hypocotyl width of wild type seedlings grown on 1/2m MS medium supplemented with various concentrations of Taxol in Wc or shade. (C) Hypocotyl width of Col‐0, sav2, sav3‐1, sav1‐1 and sav8‐1 grown in shade with/without Taxol treatment. (D) Hypocotyl length of sav3‐1, sav1‐1, sav8‐1 in Wc and shade. (E) Responses of cMTs (visualized using GFP‐TUA6) to PPM in hypocotyl cells of seedlings grown in Wc on 1/2MS medium supplemented with/without 1 µM of picloram. Seedlings were soaked in PPM for 1 h. Scale bars = 20 µm. Error bars represent standard error of mean (n = 20–30).
Figure S4. Shade‐induced changes in EB1b and overexpressing EB1b‐GFP suppresses sav2 (A) Velocity of EB1b is reduced in response to shade. 5‐d‐old ATEB1bpro::ATEB1b‐GFP transgenic seedlings were treated with Wc or shade for 24 h. Moving rate of more than 220 EB1b‐labelled plus ends from 16 seedlings were measured. Error bars represent standard error of mean. Student's t‐test analysis revealed that there is significant difference between EB1 Wc‐ and shade‐treated samples with P < 0.002. (B) Velocity of EB1b in NPA, BRZ or PAC‐treated, shade‐grown (24 h) 35S::EB1b‐GFP seedlings. Error bars represent standard error of mean (n = 30). (C) Protein levels of EB1b‐GFP in 35S::EB1b‐GFP seedlings grown in Wc and shade, showing protein stability of EB1b‐GFP is not altered by shade. (CBB: commassie brilliant blue staining). (D) Representative seedlings of Col‐0, 35S::EB1b‐GFP, sav2 and 35S::EB1b‐GFP/sav2 in dark. Quantitative measurement of hypocotyl width and length of these seedlings are shown in the middle and right panel, respectively. Error bars represent standard error of mean (n = 20–30). (E) Genotyping of 35S::EB1b‐GFP/sav2 using a dCAPS marker. (F) Representative seedlings of Col‐0, 35S::EB1b‐GFP, sav2 and 35S::EB1b‐GFP/sav2 in Wc. (G) Western blot showing ATEB1b‐GFP protein level in ATEB1bpro::ATEB1b‐GFP transgenic plants. 5‐d‐old light grown seedlings were treated with Wc or shade for 24 h, soluble protein was extracted and probed with anti‐GFP antibody. (CBB, commassie brilliant blue staining).
Figure S5. Dark phenotypes of sav2 can be rescued by NPA (A) cMT patterns in dark grown sav2 in response to NPA, BRZ or PAC. Bar, 20 µm. (B) Quantitative measurement of percentage of cells with different patterns of cMTs in dark grown seedlings. 35S::GFP‐TUA6 in wild type or sav2 background were grown on 1/2 MS medium supplemented with or without various hormone inhibitors (NPA, BRZ or PAC) for 3 d. Error bars represent standard error of mean. (n ≥ 277 hypocotyls, from more than 20 seedlings). (C) Responses of dark grown Col‐0 and sav2 to NPA (2 µM), Taxol (0.5 µM) or both.
Figure S6. Root phenotypes of sav2 (A) Root phenotypes of Col‐0, sav2 and F1 heterozygous seedlings of sav2 crossed to Col‐0. Seedlings were grown vertically on 1.5% hard agar under Wc. Pictures were taken from the top. (B) Roots of Col‐0 and sav2 grown in Wc, showing that roots of sav2 are wider than that of the wild type. (C‐D) Quantitative measurement of root length (C) and root width (D) of Col‐0, sav2 and F1 heterozygous seedlings of sav2 crossed to Col‐0. Error bars represent standard error of mean (n = 8).
Figure S7. Expression of TUB4 at mRNA and protein levels (A) Quantitative RT‐PCR results showing relative expression level of TUB4 in Col‐0 and sav2 under Wc and shade. Five‐d‐old light‐grown seedlings were treated with Wc or simulated shade for 2 h. Error bars represent standard error of mean (n = 3). (B) Protein levels of TUB4 did not change in response to shade. Five‐day‐old transgenic 35S::YFP‐TUB4 seedlings were treated with white light or simulated shade for 2 h (left panel) or indicated amount of time (right panel). Line 1 and line 2 were different overexpression lines. YFP‐TUB4 was detected by western using anti‐GFP antibody. Commassie blue (CBB) stain of total proteins are shown as loading control.


