Abstract
Early studies have focused on the synthesis of palladium nanoparticles within the periplasmic layer or on the outer membrane of Desulfovibrio desulfuricans and on the S-layer protein of Bacillus sphaericus. However, it has remained unclear whether the synthesis of palladium nanoparticles also takes place in the bacterial cell cytoplasm. This study reports the use of high-resolution scanning transmission electron microscopy with a high-angle annular dark field detector and energy dispersive X-ray spectrometry attachment to investigate the intracellular synthesis of palladium nanoparticles (Pd NPs). We show the intracellular synthesis of Pd NPs within cells of two anaerobic strains of D. desulfuricans and an aerobic strain of B. benzeovorans using hydrogen and formate as electron donors. The Pd nanoparticles were small and largely monodispersed, between 0.2 and 8 nm, occasionally from 9 to 12 nm with occasional larger nanoparticles. With D. desulfuricans NCIMB 8307 (but not D. desulfuricans NCIMB 8326) and with B. benzeovorans NCIMB 12555, the NPs were larger when made at the expense of formate, co-localizing with phosphate in the latter, and were crystalline, but were amorphous when made with H2, with no phosphorus association. The intracellular Pd nanoparticles were mainly icosahedrons with surfaces comprising {111} facets and about 5 % distortion when compared with that of bulk palladium. The particles were more concentrated in the cell cytoplasm than the cell wall, outer membrane, or periplasm. We provide new evidence for synthesis of palladium nanoparticles within the cytoplasm of bacteria, which were confirmed to maintain cellular integrity during this synthesis.
Electronic supplementary material
The online version of this article (doi:10.1007/s11051-015-3067-5) contains supplementary material, which is available to authorized users.
Keywords: Bacillus benzeovorans, Desulfovibrio desulfuricans, Icosahedron, Intracellular, Palladium nanoparticles, Synthesis, Biomedicine
Introduction
There is an increasing demand for precious metals in the areas of electronics and catalysis. Since the introduction of catalytic converters in the USA in 1975 and in Europe in 1986 (Wiseman and Zereini 2009), platinum group metals have been widely used to control emission of harmful pollutants like carbon monoxide, nitrogen oxides and unburnt hydrocarbons from automobiles. The ability of palladium to catalyse a wide range of reactions in chemical synthesis makes it the most sought after precious metal in a range of industrial applications (Holt et al. 1999). As a consequence, the high demand for palladium has led to a parallel increase in price (Cowley 2013) resulting in increasing focus on the clean and sustainable recovery of palladium from waste sources (Cui and Zhang 2008; Macaskie et al. 2011; Lee and Pandey, 2012) with the potential for bioconversion of wastes into new catalytic materials (Mabbet et al. 2006; Murray et al. 2015).
Microbial recovery of palladium is considered more promising than the conventional methods of bulk recovery (Narayanan and Sakthivel 2010; Thakkar et al. 2010). Recovery traditionally involves the dissolution of metals from spent catalysts using acids at high temperatures and then application of strong chemical reductants, which are toxic (De corte et al. 2013) and generate undesirable waste products. Bacterial cells act as reducing agents using enzymes, avoiding the use of toxic reagents, and also controlling particle growth via ‘bio-patterning’ (Mikheenko et al. 2008) while minimizing artefacts from surfactants and capping agents (Narayanan and Sakthivel 2010).
Several studies have focused on the reductive precipitation of soluble precious metal ions to insoluble metals using various pure cultures of dissimilatory sulphate-reducers (de Vargas et al. 2005; Wood et al. 2010; Bennett et al. 2013), E. coli (Deplanche et al. 2010, 2012; Foulkes et al. 2011), other facultative anaerobes (Deplanche et al. 2014) and also aerobic bacteria such as Bacillus sphaericus (Selenska-Pobell et al. 1999; Fahmy et al. 2006; Creamer et al. 2007). Studies on the use of bacteria in the synthesis of metallic nanoparticles have centred mainly on metal reducing bacteria such as Geobacter, Shewanella (De Windt et al. 2005; Law et al. 2008; Heugebaert et al. 2012; Yates et al. 2013) and also sulphate-reducers of the genus Desulfovibrio (Riddin et al. 2009; Bennett et al. 2010).
The precipitation of palladium by the anaerobic sulphate-reducing bacterium D. desulfuricans has been well demonstrated via several studies; e.g. a primary route for nanoparticle synthesis is via hydrogenases (Mikheenko et al. 2008) but other components of the ‘nanofactory’ are not known. Indeed, E. coli stripped genetically of its hydrogenases (Deplanche et al. 2010), or E. coli (Foulkes et al. 2011) and Serratia spp. (Deplanche et al. 2014) grown aerobically (where hydrogenases are not expressed) reduced Pd (II) to Pd (0), while the aerobic Bacillus sphaericus also reduced Pd (II) to Pd (0) nanoparticles very similar to those of D. desulfuricans, with similar catalytic activity in hydrogenation of itaconic acid (Creamer et al. 2007). Bacillus spp. have long been known to accumulate heavy metals selectively (Selenska-Pobell et al. 1999). Their ability to reduce metals has been studied by various authors (Boone et al. 1995; Cheng and Li 2009) but the only known route towards the synthesis of nanosized Pd deposits was via metal deposition on an S-layer protein (Pollman et al. 2005; Fahmy et al. 2006) which, of several strains examined, was only associated with B. sphaericus (Selenska-Pobell et al. 1999). Cells of this strain bind to cationic metals strongly via carboxylic groups (e.g. glutamic acid) and also to phosphate groups as a result of the extensive phosphorylation of its S-layer protein (Pollman et al. 2005; Merroun et al. 2007). However, Pd nanoparticles were also observed to be localized below the S-layer, and Pd deposition was attributed to other factors (Deplanche et al. 2014). Fahmy et al. (2006) showed that carboxylates of aspartate and glutamate residues are coordination sites for Pd (II) in Bacillus spp. S-layer protein. However, the nature and localization of the additional putative “Pd reductase” beneath the S-layer are not known.
The reduction of palladium at the cell surface by various bacterial species has been well documented. However, the formation of intracellular palladium nanoparticles (Pd NPs) has never been previously examined and this forms the focus of this study. Examination of EDX data (Foulkes et al. 2011) showed intracellular Pd located on the inner surface of the plasma membrane of E. coli but details on the nature of the intracellular particles were not discussed further. It has been shown, however, that intracellular metallic nanoparticles could be synthesized by various microorganisms. Beveridge and Murray (1980) reported precipitation of gold nanoparticles within bacterial cells from Au3+ which was confirmed in later work (Southman and Beveridge 1996). Using cells of lactic acid bacteria, Nair and Pradeep (2002) synthesized intracellular nanoparticles of gold, silver and also alloys. Other examples include intracellular gold nanoparticles made via a novel alkalotolerant actinomycete, Rhodococcus species (Ahmad et al. 2003) and Shewanella alga (Konishi et al. 2006). More recent studies continue to report the synthesis of intracellular non-precious metal nanoparticles (Peng et al. 2010; Navarrete et al. 2011; Polti et al. 2011; Ravindranath et al. 2011; Sousa et al. 2013) but the characterization of intracellular Pd NPs has never been performed. Confirmation of the formation of intracellular Pd NPs not associated with the cell membrane will prompt further fundamental questions into the underlying mechanisms of intracellular ‘trafficking’ of non-essential precious metals to key locations within bacteria; identification of the underlying biochemical mechanisms would, in turn, provide tools for the application of synthetic biology towards enhancing nanoparticle production.
This current study examines intracellular Pd (0) deposition by two strains of D. desulfuricans and a strain of B. benzeovorans using hydrogen and formate as electron donors. D. desulfuricans NCIMB 8307 was noted to have a higher biosorption capacity (de Vargas et al. 2005) than other strains of the same genus which is key to making catalytically very active bio-Pd. However, D. desulfuricans NCIMB 8307 has never been compared with a closer relative within the same species. D. desulfuricans NCIMB 8326 was chosen since this strain (unlike NCIMB 8307) lacks fatty acid ethanolamide (Goldsworthy 2011), a candidate species for the initial amide-coordination (de Vargas et al. 2005) of incoming Pd (II). A better understanding of the mechanism of patterning of Pd NPs in D. desulfuricans, and exploiting subtle strain-dependent differences, could provide useful tools in potential molecular design to improve bio-Pd catalyst. This would provide insight as to what improvements could be expected or aimed for, and on how to potentially improve catalyst selectivity in a range of reactions and products. The literature in this area of research continues to expand but there is relatively little focus on the mechanisms of Pd (0) deposition and patterning.
This study compares two methods for the synthesis of intracellular bio-Pd nanoparticles in Desulfovibrio and Bacillus. The first method involves the reduction of Pd (II) in the presence of hydrogen as the electron donor at room temperature, while in the second method, the temperature was set at 30 °C in order to increase the reaction rate using formate (Shi et al. 2013). A marked difference in the patterning or formation of the bio-Pd nanoparticles was sought as the first indication of the potential for nanoparticle “steerage” according to the condition of biomanufacture.
Materials and methods
Bacterial strains, culture conditions and harvest
The following bacteria were used in this study: two strains of anaerobic Desulfovibrio desulfuricans (NCIMB 8307 and NCIMB 8326) and one strain of aerobic Bacillus benzeovorans NCIMB 12555. The D. desulfuricans were grown anaerobically under oxygen-free nitrogen in Postgate’s medium C (pH 7.5 ± 0.2) at 30 °C (inoculated from a 24 h pre-culture, 10 %v/v) in sealed anaerobic bottles (Deplanche et al. 2010), while B. benzeovorans was grown aerobically (rotary shaker; 180 rpm, 30 °C) in nutrient medium (pH 7.3 ± 0.2) of the following composition: 1.0 g beef extract (Sigma-Aldrich), 2.0 g yeast extract (Sigma-Aldrich), 5.0 g peptone (Sigma-Aldrich) and 15.0 g NaCl per litre of distilled water. Cells were harvested (Beckman Coulter Avati J-25 Centrifuge, U.S.A) by centrifugation (9094×g, 15 min at 4 °C) at mid-exponential phase (OD600 0.5–0.7 for D. desulfuricans; OD600 0.7–1.0 for B. benzeovorans), washed three times in air with 20 mM MOPS (4-morpholinepropanesulfonic acid)-NaOH buffer, pH 7.0 and then concentrated in a small amount of the same buffer to usually 20–30 mg/ml as determined by reference to a pre-determined OD600 to dry weight conversion as previously described (Deplanche et al. 2010). The concentrated cells of known OD600 were degassed and stored under oxygen-free nitrogen (OFN) at 4 °C until use, usually within 24 h.
Preparation of palladium-challenged cells
A known volume of a concentrated resting cell suspension was transferred into an appropriate volume of degassed (30 min) 2 mM Pd (II) salt solution (Na2PdCl4, pH 2 ± 0.1 adjusted with 0.01 M HNO3) to make a 20 % loading (1:5) on biomass of D. desulfuricans and B. benzeovorans. Complete removal of Pd onto cells was determined by assay (below). The cells and palladium mixture were allowed to stand in a water bath (30 min, 30 °C) for biosorption of the Pd (II) onto the cell surfaces of each strain with occasional shaking. Hydrogen and formate were used separately as exogenously applied electron donors for Pd (II) reduction to Pd (0). The first method involves reduction under hydrogen at room temperature by bubbling hydrogen through the suspension (15 min) and then leaving in the head-space for reduction to reach completion (15 min). In the second method, sodium formate was added to a final concentration of 20 mM from a 1 M stock with shaking at 30 °C (overnight). The products were harvested by centrifugation (9094×g, 4 °C, 15 min) prior to examination by electron microscopy.
Pd (II) assay using tin (II) chloride
Prior to harvesting the palladized cells, the complete removal of Pd (II) from the solution was confirmed by the tin (II) chloride assay method as described previously (Deplanche et al. 2010) spectrophotometrically (Ultraspec III, Pharmacia Biotech, Little Chalfront, Buckinghamshire, UK). Stock tin (II) chloride was prepared by dissolving 29.9 g SnCl2 powder into 500 ml HCl and diluted fresh (aq) with the samples for use as required.
High-resolution scanning transmission electron microscopy (STEM) with HAADF (high-angle annular dark field) detector and EDX analysis
Pd-loaded cells of D. desulfuricans and B. benzeovorans prepared using both hydrogen and formate as electron donors were harvested as above, washed twice with distilled water and fixed with 2.5 % (w/v) glutaraldehyde fixative in 0.1 M cacodylate buffer (pH 7.0) at 4 °C and stained with 1 % aq osmium tetraoxide. For scanning and transmission electron microscopy (STEM) and energy dispersive X-ray spectroscopy (EDX), the Pd-loaded cells were dehydrated using an ethanol series and washed twice in propylene oxide (Deplanche et al. 2010). The cells were embedded in epoxy resin and cut into sections (100–150 nm thick) and mounted on carbon-coated copper TEM grids. Electron opaque deposits were examined by EDX with peaks sought corresponding to X-ray emission energies of Pd (Yong et al. 2002). STEM and EDX were done using a FEI image Cs-corrector configuration Titan™ G2 60-300 STEM microscope equipped with HAADF detector, accelerating voltage of 300 kV.
Image processing, lattice spacing and particle size analysis
The images obtained with HAADF–STEM were imported into image processing software, “ImageJ” and analysed (Abramoff et al. 2004; Schneider et al. 2012). The bandpass filter function was applied in order to distinguish the Pd nanoparticles on cells of the three bacteria from background signals and artefacts by adjustment. To avoid uneven illumination of the images, image thresholding was done, the total number of the Pd nanoparticles on bacterial cells was counted and the particles size diameter was calculated. The particle size distribution was then estimated (“Origin C”). From these values, the dispersity of Pd nanoparticles was calculated using a Gaussian function. The lattice spacings were also determined using “ImageJ” through profiling of high-resolution HAADF–STEM images and compared against lattice spacing of bulk palladium from the database generated using “Powder cell 2.4″ software.
Results
Examination of Pd nanoparticles by electron microscopy and EDX
High-resolution HAADF–STEM (high-angle annular dark field–scanning transmission electron microscopy) with EDX was used to confirm the identity of cellular nanoparticles as Pd within cells of B. benzeovorans and D. desulfuricans (Fig. 1a–l). Sections of samples produced with hydrogen as electron donor (Fig. 1a, b, e, f, i, and j) show bacteria with intracellular nanosized particles identified as Pd (below) and occasional larger inclusions. Cells unchallenged with Pd (II) showed no accumulated nanoparticles at their surfaces or intracellularly (S1). Few Pd nanoparticles were visible at the surface of B. benzeovorans NCIMB 12555 (Fig. 1a, b), while D. desulfuricans NCIMB 8307 (Fig. 1e, f) supported surface-bound Pd (0) but none was apparent on D. desulfuricans NCIMB 8326 (Fig. 1i, j). In all three strains, there were more numerous Pd nanoparticles located within the intracellular matrices than in the outer layers. At high magnification, HAADF–STEM images revealed many intracellular Pd nanoparticles in B. benzeovorans NCIMB 12555 (Fig. 1b) and D. desulfuricans NCIMB 8307 (Fig. 1f) but fewer in D. desulfuricans NCIMB 8326 (Fig. 1j).
Fig. 1.
HAADF-STEM micrographs of intracellular Pd nanoparticles synthesized using 20 % by mass Pd loading (1:5) on bacterial cells from 2 mM Na2PdCl4 solution, in 0.01 M HNO3 using hydrogen (left) and formate (right) as electron donors: ordinary TEM images of Pd nanoparticle deposits on bacteria (a, c, e, g, i, k); HAADF–STEM of Pd nanoparticles in cells coupled with high-resolution HAADF–STEM of a section of intracellular Pd nanoparticles (b, d, f, h, j, l) in B. benzeovorans NCIMB 12555 (top), D. desulfuricans NCIMB 8307 (middle) and D. desulfuricans NCIMB 8326 (bottom)
A similar trend was observed when formate was used as the electron donor, (Fig. 1c, d, g, h, k, and l) with some extracellular Pd nanoparticles present in each case. However, under hydrogen or formate as electron donors, only D. desulfuricans NCIMB 8307 had a major deposition of surface-bound nanoparticles (Fig. 1e–h). While mostly invisible in TEM micrographs (Fig. 1a, e, i, c, g, k), intracellular nanoparticles were visible in all three strains at high resolution using electron backscattering (Fig. 1b, f, j, d, h, l).
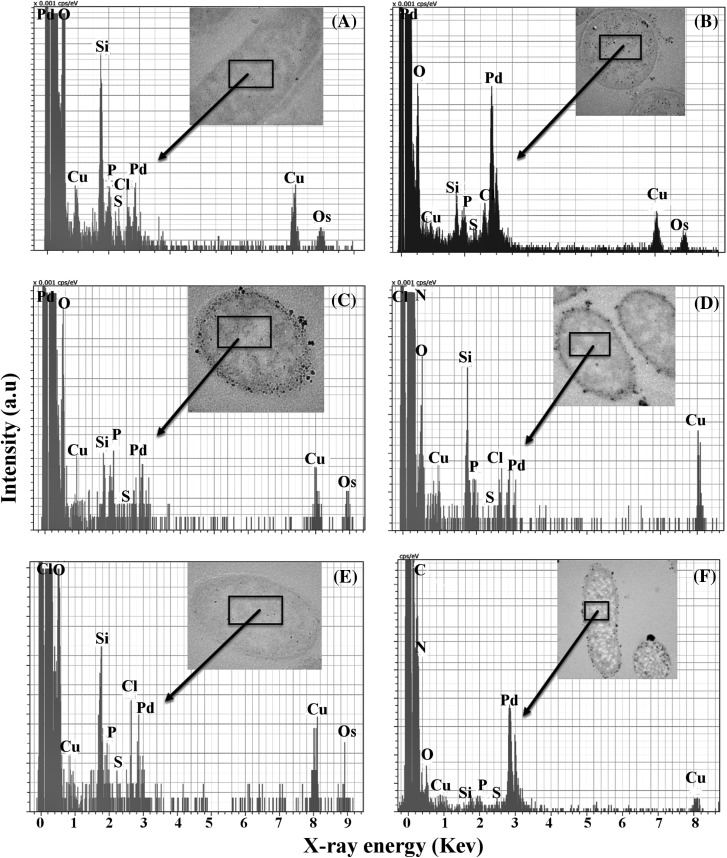
It was confirmed that irrespective of the electron donors used, Pd-NPs were present in the cytoplasm of cells by using STEM–EDX mapping of palladized bacteria. This allowed the identification of Pd clusters with phosphorus (P) and sulphur (S), as the major inorganic elements (Fig. 2a–f). Elemental mapping (Fig. 2; S2) shows that only cells that had significant detectable Pd also showed S and P detected above the background. The co-localization of Pd and S in larger deposits within D. desulfuricans (Fig. 3a) was assumed to be PdS as a result of biogenic H2S production but was not investigated further. No clear co-localization of S otherwise, nor of P, was seen in D. desulfuricans (Fig. 3a) but B. benzeovorans evidenced co-localization of Pd and P (Fig. 3b, circled). The presence of S is seen as a normal constituent of bacterial proteins and P within deoxyribonucleic acids, ribonucleic acids, phospholipids, techoic acids and other compounds (Southman and Beveridge 1996) but it is not clear why these elements are more evident in cells that showed internal Pd deposition. Southman and Beveridge (1996) showed the co-appearance of intracellular gold crystals with S and P in B. subtilis, in which it was suggested that the signals from S and P could have been contributed due to organic film from the bacteria and coatings on the Au crystals used. It is highly plausible to suggest that the complexation of macromolecules with Pd depletes cellular reserves of P and S as it coordinates to these groups and as a result would stimulate the cells to produce more in order to set a cellular balance. It could also mean a detoxification mechanism which reduces the mobility, and in turn, the toxicity of Pd nanoparticles by neutralizing the negative charges of Pd nanoparticles by P and S containing molecules in the cell. The incorporation of palladium nanoparticles within organic materials may help to stabilize them and control their size e.g. proteins within the network of calcium carbonates in Myxococcus xanthus have been shown to stabilize the structure of these biogenic Ca carbonates (Rodriguez-Navarro et al. 2007). We conclude that, while surface-bound Pd NPs have been reported extensively using many microorganisms (Creamer et al. 2007; Foulkes et al. 2011; Bennett et al. 2013; De Corte et al. 2013; Deplanche et al. 2014), we now show evidence for intracellular localization of palladium and hence an inference of the ability of Pd (II) to be translocated via cellular transport and deposition mechanisms which await further study. Other studies, to be reported in a subsequent publication (see S3), used flow cytometry to confirm continued cell integrity and metabolism during the process of Pd deposition.
Fig. 2.
HAADF–STEM–EDX analysis of intracellular Pd nanoparticles (selected areas shown) with phosphorus (P) and sulphur (S) in B. benzeovorans NCIMB 12555 (a, b top), D. desulfuricans NCIMB 8307 (c, d middle) and D. desulfuricans NCIMB 8326 (e, f bottom) respectively, with hydrogen (left) and formate (right) as electron donors. The Cu is from the TEM grid, while the silicon results from the oil in the diffusion pump of the column of the TEM system
Fig. 3.
EDX Elemental mapping showing Pd, P and S in D. desulfuricans NCIMB 8307 (a) and B. benzeovorans (b). Only B. benzeovorans shows co-localization of Pd with P (white circle) but not S within the cell cytoplasm
Dispersity and distribution of intracellular Pd nanoparticles
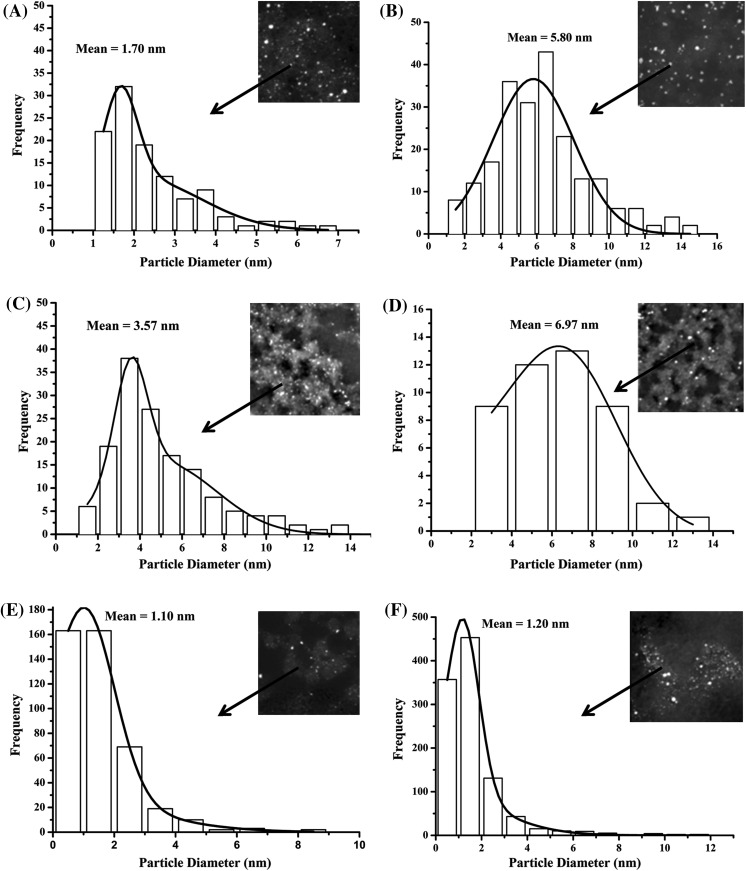
High-resolution images allowed the calculation of particle size distribution using “ImageJ” software (Abramoff et al. 2004; Schneider et al. 2012). The mean particle sizes of the intracellular Pd nanoparticles were calculated as shown in Fig. 4a–f and Table 1. When hydrogen was used as the electron donor, the ranking of the mean particle size followed the order D. desulfuricans NCIMB 8307 > B.benzeovorans NCIMB 12555 > D. desulfuricans NCIMB 8326 (Fig. 4a, c, e). A similar ranking of the mean particle diameter of the Pd NPs produced was observed using formate (Fig. 4b, d, f; Table 1). The particle size distributions were similar in the case of B. benzeovorans and D. desulfuricans NCIMB 8307 (Fig. 4a–d), being a normal distribution (cells in Fig. 1b, d) from formate but skewed towards small NPs when Pd (II) was reduced under H2 (cells in Fig. 1a, c). Very little difference was apparent using the two electron donors with D. desulfuricans NCIMB 8326 (Fig. 4e, f) skewed in each case, although larger NPs were produced at the expense of formate by the other two strains (Table 1).
Fig. 4.
Particle size distribution of intracellular Pd nanoparticles made using hydrogen (left) and formate (right) as electron donors with B. benzeovorans NCIMB 12555 (a, b top), D. desulfuricans NCIMB 8307 (c, d middle) and D. desulfuricans NCIMB 8326 (e, f bottom), respectively, and >100 particles were counted in each case
Table 1.
Particle size distribution of intracellular Pd nanoparticles
| Cell type | Reducing agents | Dispersity | ||||
|---|---|---|---|---|---|---|
| Mean particle size (nm) | Polydispersity index | Monodispersity (%) | ||||
| Hydrogen | Formate | Hydrogen | Formate | Hydrogen | Formate | |
| B. benzeovorans | 1.70 ± 0.41 | 5.80 ± 2.24 | 0.24 | 0.39 | 76 | 61 |
| NCIMB 12555 | ||||||
| D. desulfuricans | 3.57 ± 0.77 | 6.97 ± 2.46 | 0.22 | 0.35 | 78 | 65 |
| NCIMB 8307 | ||||||
| D. desulfuricans | 1.10 ± 0.95 | 1.20 ± 0.69 | 0.86 | 0.58 | 14 | 42 |
| NCIMB 8326 | ||||||
Mean particle size distribution of intracellular Pd nanoparticles produced by three strains of bacteria using hydrogen and formate as electron donors. The polydispersity index was calculated from the particle size distribution using a Gaussian function, and the monodispersity values were converted into percentage values. In all, >100 particles were counted in each case using “ImageJ” software. 20 % by mass Pd loading (1:5) on cells of each bacterium was used from 2 mM Na2PdCl4 solution, in 0.01 M HNO3. Data are mean ± standard deviation of the means
In order to determine the degree of dispersity of the Pd nanoparticles, the polydispersity indices were calculated from the size distribution of the nanoparticles, while the monodispersity was presented in percentages (Table 1). D. desulfuricans NCIMB 8326 produced the most heterogeneous Pd nanoparticles with a polydispersity index of 0.86, resulting in 14 % monodispersity (Table 1) under hydrogen as electron donor. In contrast, the Pd NPs of D. desulfuricans NCIMB 8307 and B. benzeovorans NCIMB 12555 were less polydispersed with similar polydispersity indices of 0.22 and 0.24, respectively, meaning that 78 and 76 % (Table 1) of the Pd nanoparticles were monodispersed. Overall, the degree of homogeneity of intracellular Pd nanoparticles produced by the three strains of bacteria followed the order D. desulfuricans NCIMB 8307 > B. benzeovorans NCIMB 12555 > D. desulfuricans NCIMB 8326.
The polydispersity indices of the intracellular Pd nanoparticles reduced with formate were higher than the indices of Pd NPs reduced by using hydrogen except for D. desulfuricans NCIMB 8326 (Table 1). Under formate-mediated reduction, the polydispersity indices calculated for B. benzeovorans NCIMB 12555, D. desulfuricans NCIMB 8307 and D. desulfuricans NCIMB 8326 Pd NPs were 0.39, 0.35 and 0.58, respectively, corresponding to a degree of homogeneity (monodispersity) of the Pd nanoparticles of 61, 65 and 42 %, respectively (Table 1), revealing that D. desulfuricans NCIMB 8326 had the least homogeneously distributed intracellular Pd nanoparticles. According to Nidhin et al. (2008), a polydispersity index of >0.7 is associated with samples with very broad size distribution and only the NPs of D. desulfuricans NCIMB 8326 exceeded this value under hydrogen. Using formate as an electron donor, Shi et al. (2013) chemically synthesized Au–Pd alloy nanocrystals and obtained very large (~20 nm) and irregularly shaped nanoparticles when compared with stronger reducing agents like NaBH4 and ascorbic acid. This was attributed to the differences in reaction kinetics (Shi et al. 2013). Formate is a weaker reducing agent than hydrogen taking more time for bioreduction to reach completion using bacterially mediated reduction (I.P Mikheenko and L.E. Macaskie, unpublished). Hence, the reduction of Pd (II) by formate was done at 30 °C with constant agitation to increase the reaction rate, while hydrogen reduction was accomplished at room temperature by bubbling it through the cell suspension. The rates of uptake and reduction of Pd (II) in each case were not measured.
Crystallinity and lattice spacing of intracellular Pd nanoparticles
High-resolution TEM images revealed that, while the Pd nanoparticles produced by B. benzeovorans (Fig. 5a) were amorphous under hydrogen as electron donor, those produced by this strain from formate and in both cases by D. desulfuricans NCIMB 8307 (Fig. 5c) and D. desulfuricans NCIMB 8326 (Fig. 5e) were crystalline. The lattice spacings of representative crystalline particles were 0.250 nm, 0.243 nm and 0.258 nm for D. desulfuricans NCIMB 8307 (Fig. 5c), which is consistent with the {111} facet of Pd, exposing mainly the {111} facets to the surface (Fig. 5c) with some particles appearing to be icosahedral in shape (Bennett et al. 2013) comprising mainly single crystal structures for B.benzeovorans NCIMB 12555 (Fig. 5b) and multiply twinned structures for D. desulfuricans NCIMB 8307 (Fig. 5c, d) and D. desulfuricans NCIMB 8326 (Fig. 5e, f) which may have formed as a result of particle growth via coalescence (Lim et al. 2010). The values of the Pd planes showed a distortion of about 5 % when compared with the values generated for bulk palladium from the database using “Powder cell 2.4” software. About 10 % distortion of the {111} facet for extracellular Pd nanoparticles from D. desulfuricans NCIMB 8307 (Bennett et al. 2013) and a 12.5 % expansion of the {200} facet of Pd nanoparticles synthesized chemically on amorphous carbon (José-Yacamán et al. 2001) were reported. Shapes are characterized according to the type of facets exposed to the surface; octahedrons and icosahedrons have their surfaces covered mainly by {111} facets (Zhang et al. 2013). Only D. desulfuricans NCIMB 8326 reduced under hydrogen showed a mix-facet arrangement of Pd nanoparticles with lattice spacings of 0.208 and 0.248 nm (Fig. 5e), corresponding to the {200} and {111} facets, respectively (Fig. 5e). This Pd nanostructure is similar to a cubo-octahedron or its truncation (José-Yacamán et al. 2001). The reason for the production of amorphous Pd nanoparticles by B. benzeovorans under hydrogen (Fig. 5a) remains unclear and is the focus of an ongoing study which also aims to compare the catalytic activities of the different Pd-types. Reduction via formate, however, showed that the resulting intracellular Pd NPs were crystalline and very similar (Fig. 5b, d, f) for all three strains. The lattice spacing for these nanocrystals was analysed for B. benzeovorans NCIMB 2555 (0.250 nm), D. desulfuricans NCIMB 8307 (0.250 nm) and D.desulfuricans NCIMB 8326 (0.241 and 0.250 nm) corresponding to the {111} facets exposed to the surface as shown by representative TEM images (Fig. 5b, d, f) with similar distortions as the Pd reduced by hydrogen (Fig. 5a, c, e).
Fig. 5.
High-resolution TEM images of intracellular Pd crystals with insets revealing lattice spacing in crystals made using hydrogen (left) and formate (right) as reducing agents; B. benzeovorans NCIMB 12555 (top), D. desulfuricans NCIMB 8307 (middle) and D. desulfuricans NCIMB 8326 (bottom), respectively
Discussion
This study focuses on the synthesis and characterization of intracellular palladium nanoparticles in aerobic and anaerobic bacteria made via hydrogen and formate as electron donors, whereas all previous studies have been focused on surface Pd NPs. Hydrogen and formate are metabolized differently with the possibility of affecting the synthesis and patterning of Pd nanoparticles in bacterial cells. Chemical reduction of palladium (II) under hydrogen or formate using killed bacterial cells was significantly slower (Mikheenko et al. 2008). A substantial number of surface-bound Pd nanoparticles were shown on the surface of D. desulfuricans NCIMB 8307 but not apparent in the cells of NCIMB 8326 using H2 as electron donor which is supported by previous reports of Pd binding on cell surfaces and within the surface periplasmic layer (Bennett et al. 2010). For use as catalysts, the palladized bacteria are routinely washed in acetone which destabilizes and removes lipid membrane structures, to allow access to intracellular as well as surface-localized NPs, but during NP synthesis, continued membrane integrity (and hence the permeability barrier) was confirmed using flow cytometry (see S3). Hydrogenases are located predominantly periplasmically in D. desulfuricans (Hatchikian et al. 1990; Casalot et al. 1998) and have been implicated to play important roles in Pd (II) reduction (Mikheenko et al. 2008). In the case where hydrogen was used as electron donor, it is possible that hydrogenases play a dominant role (alongside other enzymes and electron-generating biomolecules) in determining the nature of the final Pd NPs in the two strains of D. desulfuricans (Fig. 5c, e) but the absence of surface-bound Pd in D. desulfuricans NCIMB 8326 suggests that alternative mechanism(s) predominate in this strain. In B. benzeovorans (grown aerobically where hydrogenases are not present), only amorphous Pd nanoparticles were visible (Fig. 5a) suggesting a possible involvement of other mechanisms. In previous studies, Mikheenkho et al. (2008) observed that Desulfovibrio fructosovorans deficient in its periplasmic hydrogenases relocated the bio-Pd (0) deposits to the cytoplasmic membrane, which was the site of the remaining hydrogenases. This was further supported by similar work involving hydrogenase-deficient E. coli mutants (Deplanche et al. 2010), where alternative supplementary mechanisms were implied.
While there were differences in terms of the shapes of the intracellular Pd NPs under hydrogen, the shapes of the Pd NPs using formate as an electron donor appeared to be mainly crystalline icosahedrons. However, this major, thermodynamically stable, nanostructure (formed due to its lowest surface energies and slower growth rate: Xiong et al. 2007; Zhang et al. 2013) differs between the bacterial types. Most of the final Pd nanocrystals made within Gram-positive B. benzeovorans were single nanocrystals while those within the two Gram-negative strains of D. desulfuricans could be seen as multiply twinned nanoparticles (Ascencion et al. 1998). It is possible that an association with cellular phosphate structures (Fig. 3b) reduced the mobility of the NPs within the cells of the former and stabilized them against agglomeration, whereas no apparent associations were seen in the Gram-negative cells. Chemically synthesized nanoparticles of Pd on carbon have been shown with four basic shapes: FCC, cubo-octahedral, icosahedral and twinned shapes (José-Yacamán et al. 2001). Shapes, together with the atoms at corners and edges, define the facets and surface structure of nanoparticles e.g. icosahedrons are formed only by {111} planes (Xiong et al. 2007). Several factors like the metal salt precursors, capping agents, stabilizers and reducing agents can influence the final shapes of nanoparticles. For example, the chemical reduction of palladium (II) acetylacetonate (Pd (acac)2) in the presence of tetraethylene glycol (TTEG) resulted in the formation of tetrahedral Pd NPs, while octahedrons with {111} facets (Wang et al. 2013) were formed with Na2PdCl4, and a Pd cubo-octahedron was synthesized in the presence of ethylene glycol (EG) and polyvinylpyrrolidone (PVP) (Zhang et al. 2013). Icosahedral Pd nanoparticles were formed chemically by reducing H2PdCl4 in the presence of TTEG (Chen et al. 2009) which was suggested to be due to the higher viscosity and milder reducing property of TTEG than EG. That could indicate a reason for the formation of mainly icosahedral crystalline intracellular Pd nanostructures by the three bacteria in the presence of formate, a milder reducing agent than hydrogen, within heavily hydrated gel-type cellular matrices. However, it is equally possible that, intracellularly, formate is oxidized enzymatically with another biomolecule effecting the final electron transfer onto Pd (II).
Palladium nanoparticles made via using formate as electron donor were shown to be similar (crystalline) for each bacterial strain. Formate, like hydrogen, must be broken down to release the electrons needed for the reduction of Pd (II). The first deposited Pd (0) can split formate chemically by weakening the C–H bond due to interaction of a delocalized electron pair with the surface of Pd (0) (Rhodin and Ertl 1979). This mechanism releases CO2 and H2. H2 is then split on the surface of Pd (0) to yield single electrons for the subsequent reduction of Pd (II) to Pd (0) (Livingstone, 1973); hence, enzymatic involvement is not obligatory.
Biochemically, in anaerobic bacteria, formate bio-oxidation provides electrons for pathways that utilize final electron acceptor molecules other than dioxygen (Mota et al. 2011), while NAD+ is an intermediate electron/proton acceptor in aerobes using oxygen as the terminal electron acceptor. However, it was shown that an anaerobic growth condition lowers the production of NAD+ in Bacillus cereus (de Sarrau et al. 2012) at low temperature suggesting a possible use of other alternative electron carriers under these conditions. Enzymatic formate oxidation is made possible by formate dehydrogenases (FDH) (Costa et al. 1997; Ding et al. 2011). An early study (Weimer 1984) reported that formate, a product of glucose fermentation in Bacillus macerans, was gradually depleted with a resultant detection of CO2 and H2; the addition of exogenous formate led to the formation of CO2 and H2. Pyruvate formate lyase and formate dehydrogenase (FDH) activities were then detected in cell extracts which confirmed the enzymatic oxidation of formate in this strain. In anaerobes, formate hydrogen-lyase (FHL) (Axley et al. 1990) is responsible for splitting formate to make hydrogen. The metal-containing FDH e.g. the molybdenum-containing FDH in E.coli (Jormakka et al. 2002) and tungsten-containing FDH in D. gigas (Raaijmakers et al. 2002) are typical of anaerobic bacteria, while the NAD+-dependent FDH found in e.g. Bacillus spp. F1 (Ding et al. 2011) and Rhodobacter capsulatus (Hartmann and Leimkuhler 2013) are located in the cytoplasm and differ in composition and in the mechanism of formate metabolism (Hartmann and Leimkuhler 2013). Regardless of the specific enzymatic mechanism, the process of formate oxidation provides the source of electrons needed for the reduction of Pd (II) into nanoparticles by the bacteria.
Intracellular Pd nanoparticles were visible in each strain using both electron donors. The intracellular nanoparticles, generally too small to be readily visible in TEM micrographs, were shown to be present via electron backscattering and confirmed as Pd via EDX analysis. The behaviour of B. benzeovorans and D. desulfuricans NCIMB 8307 was similar in terms of the dispersity of their intracellular Pd NPs when compared with D. desulfuricans NCIMB 8326. Deplanche et al. (2014) highlighted a possible ‘Pd (II)-trafficking’ across the cytoplasmic membrane developed from a report (Foulkes et al. 2011) whereby Pd (II) reduction by anaerobic E. coli was attributed in part to the Pd (II) travelling across the cytoplasmic membrane by an unknown translocation mechanism. However, this was not characterized, nor the mechanism by which further transport into and across the cytoplasm was facilitated. Ni (II) is a key component of metalloenzymes like hydrogenases and ureases (Mulrooney and Hausinger 2003; Liermann et al. 2007) and both enzymes are commonly found in the bacterial cytoplasm. Since Pd (II) shares some similar chemistry with Ni (II), it was suggested that Pd (II) could be transported through Ni (II) ‘trafficking’ systems (Deplanche et al. 2014) as a possible route to its unproductive delivery into intracellular enzymes, with deposition as cytoplasmic inclusions as a possible method for ‘packaging’ physiologically redundant but potentially cytotoxic metals. Like Pt, Pd is well known to bind to DNA (Shoukry and Mohamed 2012). In this context, although the occasional larger Pd-inclusions visible in D. desulfuricans (Fig. 3a) resemble cell nuclei, this is unlikely to be the explanation as the DNA of bacteria is distributed around the cell and not within nuclear bodies and, furthermore, DNA binding would account for the co-occurrence of Pd with P in B. benzeovorans but not S. Molecular “destinations” for Pd (II) in lieu of Ni (II) would provide a possible rationale for the presence of intracellular Pd nanoparticles as many metalloproteins can select the wrong metals when presented with a mixture of other metals because protein metal-coordination sites can be quite receptive to inorganic or non-essential partners (Waldron and Robinson 2009). For example, Cd (II) can replace Zn (II) in carbonic anhydrase in Thalassiosira weissflogii (a diatom), a possible evolution of enzyme capable of working with Cd in prokaryotes in Cd-polluted environments (Xu et al. 2008). Also, the high affinity of Hg and Cd to thiols makes them liable to outcompete Zn for protein-binding sites that contain cysteine which can result in the inactivation of Zn-requiring proteins (Frausto da Silva and Williams 2001) just as Ag and Au (non-essential metals) can inactivate Cu in proteins (Gupta et al. 1999). However, as far as we are aware, Pd (II) cannot substitute biochemically for Ni (II), and hence in this study, the cell would be left with a burden of Pd (II). Since Pd (II) (redox potential of +0.99 V) (Hayes, 1993) is reduced more easily than Ni (II) (−0.27 V), it is possible that in order to maintain the homeostasis of the intracellular environment, the cell attempts to expel Pd (II) as a consequence of a stress response via Ni exporters such as the ABC transporter or P-type ATPases (Silver and Phung 1996; Hendricks and Mobley 1997) but instead fortuitously reduces Pd (II) and deposits Pd (0) within the intracellular matrix as a by product of a failed attempt to incorporate Pd (II) instead of Ni (II) into enzymes or proteins. It is still unknown how Pd (II) is trafficked within the cell. The literature on Ni-trafficking is extensive and the reader is referred to other published works (see Maroney 1999; Eitinger and Mandrand-Berthelot 2000; Waldron and Robinson 2009) as the mechanistic aspects of Pd-metabolism are beyond the scope of this paper.
Conclusions
This study has demonstrated the synthesis of intracellular Pd nanoparticles by B. benzeovorans NCIMB 12555 (Gram-positive, aerobic bacterium), D. desulfuricans NCIMB 8307 and D. desulfuricans NCIMB 8326 (Gram-negative, anaerobic bacteria). The structure of the Pd NPs was analysed to establish the influence of factors such as the nature of the electron donor and bacterial cell type. The fundamentally different D. desulfuricans NCIMB 8307 and B. benzeovorans produced similar homogeneously distributed (monodispersed) intracellular Pd NPs, with those produced via formate oxidation being larger (5–7 nm) than those produced via oxidation of hydrogen (1–4 nm). The intracellular Pd nanoparticles produced by the three bacteria in the presence of formate were mainly crystalline icosahedrons enclosed by {111} facets. In B. benzeovorans, amorphous intracellular Pd NPs were observed under hydrogen. The mechanism of Pd (II) trafficking in the cell is unknown, but this study indicates a possible ‘trafficking’ of palladium ions into the cytoplasm of both aerobic and anaerobic bacteria which may serve as a platform for ‘fine-tuning’ and biomanufacture of novel targeted catalysts. In this respect, the intracellular nanoparticles produced by ethanolamide-deficient D. desulfuricans NCIMB 8326 were smaller than in NCIMB 8307, and the polydispersity index was higher; the comparative catalytic activities of the two types of ‘bio-Pd’ will be reported in subsequent publications.
Electronic supplementary material
Acknowledgments
The authors acknowledge with thanks a Commonwealth scholarship to JBO and to Maria del Mar Abad Ortega (Centro de Instrumentación Científica, University of Granada, Spain) with technical assistance on the STEM/HAADF measurement. They also acknowledge the financial support of EPSRC (grant number EP/J008303/1).
Conflict of interest
The authors report no conflict of interest.
References
- Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Ahmad A, Senapati S, Khan MI, Kumar R, Ramani R, Srinivas V, Sastry M. Intracellular synthesis of gold nanoparticles by a novel alkalotolerant actinomycete, Rhodococcus species. Nanotechnology. 2003;14:824–828. doi: 10.1088/0957-4484/14/7/323. [DOI] [Google Scholar]
- Ascencio JA, Gutierrez-Wing C, Espinosa ME, Marin M, Tehuacanero S, Zorrilla C, Jose-Yacaman M. Structure determination of small particles by HREM imaging: theory and experiment. Surf Sci. 1998;396:349–368. doi: 10.1016/S0039-6028(97)00689-4. [DOI] [Google Scholar]
- Axley MJ, Grahame DA, Stadtman TC. Escherichia coli formate-hydrogen lyase purification and properties of the selenium-dependent formate dehydrogenase component. J Biol Chem. 1990;265:18213–18218. [PubMed] [Google Scholar]
- Bennett JA, Creamer NJ, Deplanche K, Macaskie LE, Shannon IJ, Wood J. Palladium supported on bacterial biomass as a novel heterogeneous catalyst: a comparison of Pd/Al2O3 and bio-Pd in the hydrogenation of 2-pentyne. Chem Eng Sci. 2010;65:282–290. doi: 10.1016/j.ces.2009.06.069. [DOI] [Google Scholar]
- Bennett JA, Mikheenko IP, Deplanche K, Shannon IJ, Wood J, Macaskie LE. Nanoparticles of palladium supported on bacterial biomass: new re-usable heterogeneous catalyst with comparable activity to homogeneous colloidal Pd in the heck reaction. Appl Catal B. 2013;140–141:700–707. doi: 10.1016/j.apcatb.2013.04.022. [DOI] [Google Scholar]
- Beveridge TJ, Murray RG. Sites of metal deposition in the cell wall of Bacillus subtilis. J Bacteriol. 1980;141:876–887. doi: 10.1128/jb.141.2.876-887.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone DR, Liu YT, Zhao ZJ, Balkwill DL, Drake GR, Stevens TO, Aldrich HC. Bacillus-infernus sp-nov, an Fe(III)-reducing and Mn(IV)-reducing anaerobe from the deep terrestrial subsurface. Int J Syst Bacteriol. 1995;45:441–448. doi: 10.1099/00207713-45-3-441. [DOI] [PubMed] [Google Scholar]
- Casalot L, Hatchikian EC, Forget N, de Philip P, Dermoun Z, Bélaich J-P, Rousset M. Molecular study and partial characterization of iron-only hydrogenase in Desulfovibrio fructosovorans. Anaerobe. 1998;4:45–55. doi: 10.1006/anae.1997.0137. [DOI] [PubMed] [Google Scholar]
- Chen Y, He B, Huang T, Liu H. Controlled synthesis of palladium icosahedra nanocrystals by reducing H2PdCl4 with tetraethylene glycol. Colloids Surf A. 2009;348:145–150. doi: 10.1016/j.colsurfa.2009.07.007. [DOI] [Google Scholar]
- Cheng GJ, Li XH. Bioreduction of chromium (VI) by Bacillus sp. isolated from soils of iron mineral area. Eur J Soil Biol. 2009;45:483–487. doi: 10.1016/j.ejsobi.2009.06.009. [DOI] [Google Scholar]
- Costa C, Teixeira M, LeGall J, Moura JJG, Moura I. Formate dehydrogenase from Desulfovibrio desulfuricans ATCC 27774: isolation and spectroscopic characterization of the active sites (heme, iron-sulfur centers and molybdenum) J Biol Inorg Chem. 1997;2:198–208. doi: 10.1007/s007750050125. [DOI] [Google Scholar]
- Cowley A. Platinum 2013 interim review. Hertfordshire: Johnson Matthey PLC; 2013. pp. 2–35. [Google Scholar]
- Creamer NJ, Mikheenko IP, Yong P, Deplanche K, Sanyahumbi D, Wood J, Pollmann K, Merroun M, Selenska-Pobell S, Macaskie LE. Novel supported Pd hydrogenation bionanocatalyst for hybrid homogeneous/heterogeneous catalysis. Catal Today. 2007;128:80–87. doi: 10.1016/j.cattod.2007.04.014. [DOI] [Google Scholar]
- Cui J, Zhang L. Metallurgical recovery of metals from electronic waste: a review. J Hazard Mater. 2008;158:228–256. doi: 10.1016/j.jhazmat.2008.02.001. [DOI] [PubMed] [Google Scholar]
- De Corte S, Bechstein S, Lokanathan AR, Kjems J, Boon N, Meyer RL. Comparison of bacterial cells and amine-functionalized abiotic surfaces as support for Pd nanoparticle synthesis. Colloids Surf B. 2013;102:898–904. doi: 10.1016/j.colsurfb.2012.08.045. [DOI] [PubMed] [Google Scholar]
- de Sarrau B, Clavel T, Clerte C, Carlin F, Ginies C, Nguyen-The C. Influence of anaerobiosis and low temperature on Bacillus cereus growth, metabolism, and membrane properties. Appl Environ Microbiol. 2012;78:1715–1723. doi: 10.1128/AEM.06410-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vargas I, Sanyahumbi D, Ashworth MA, Hardy CM, Macaskie LE (2005) Use of X-ray photoelectron spectroscopy to elucidate the mechanism of palladium and platinum biosorption by Desulfovibrio desulfuricans biomass. In: Harrison STL, Rawlings DE, Petersen J (eds) 16th international biohydrometallurgy symposium. University of Cape Town, pp 605–616
- De Windt W, Aelterman P, Verstraete W. Bioreductive deposition of palladium (0) nanoparticles on Shewanella oneidensis with catalytic activity towards reductive dechlorination of polychlorinated biphenyls. Environ Microbiol. 2005;7:314–325. doi: 10.1111/j.1462-2920.2005.00696.x. [DOI] [PubMed] [Google Scholar]
- Deplanche K, Caldelari I, Mikheenko IP, Sargent F, Macaskie LE. Involvement of hydrogenases in the formation of highly catalytic Pd(0) nanoparticles by bioreduction of Pd(II) using Escherichia coli mutant strains. Microbiol-SGM. 2010;156:2630–2640. doi: 10.1099/mic.0.036681-0. [DOI] [PubMed] [Google Scholar]
- Deplanche K, Merroun ML, Casadesus M, Tran DT, Mikheenko IP, Bennett JA, Zhu J, Jones IP, Attard GA, Wood J, Selenska-Pobell S, Macaskie LE. Microbial synthesis of core/shell gold/palladium nanoparticles for applications in green chemistry. J R Soc Interface. 2012;9:1705–1712. doi: 10.1098/rsif.2012.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplanche K, Bennett JA, Mikheenko IP, Omajali J, Wells AS, Meadows RE, Wood J, Macaskie LE. Catalytic activity of biomass-supported Pd nanoparticles: influence of the biological component in catalytic efficacy and potential application in ‘green’ synthesis of fine chemicals and pharmaceuticals. Appl Catal B. 2014;147:651–665. doi: 10.1016/j.apcatb.2013.09.045. [DOI] [Google Scholar]
- Ding HT, Liu DF, Li ZL, Du YQ, Xu XH, Zhao YH. Characterization of a thermally stable and organic solvent-adaptative NAD+-dependent formate dehydrogenase from Bacillus sp. F1. J Appl Microbiol. 2011;111:1075–1085. doi: 10.1111/j.1365-2672.2011.05124.x. [DOI] [PubMed] [Google Scholar]
- Eitinger T, Mandrand-Berthelot MA. Nickel transport systems in microorganisms. Arch Microbiol. 2000;173:1–9. doi: 10.1007/s002030050001. [DOI] [PubMed] [Google Scholar]
- Fahmy K, Merroun M, Pollmann K, Raff J, Savchuk O, Hennig C, Selenska-Pobell S. Secondary structure and Pd-(II) coordination in S-layer proteins from Bacillus sphaericus studied by infrared and X-ray absorption spectroscopy. Biophys J. 2006;91:996–1007. doi: 10.1529/biophysj.105.079137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes JM, Malone KJ, Coker VS, Turner NJ, Lloyd JR. Engineering a biometallic whole cell catalyst for enantioselective deracemization reactions. ACS Catal. 2011;11:1589–1594. doi: 10.1021/cs200400t. [DOI] [Google Scholar]
- Frausto da Silva JJR, Williams RJP. Biological chemistry of the elements: the inorganic chemistry of life. Oxford: Oxford University Press; 2001. [Google Scholar]
- Goldsworthy MJH (2011) Hydrocarbon biosynthesis in Mycobacterium spp NCIMB 10403 and Desulfovibrio desulfuricans. PhD Thesis. University of Exeter
- Gupta A, Matsui K, Lo JF, Silver S. Molecular basis for resistance to silver cations in Salmonella. Nat Med. 1999;5:183–188. doi: 10.1038/5545. [DOI] [PubMed] [Google Scholar]
- Hartmann T, Leimkuhler S. The oxygen-tolerant and NAD (+)-dependent formate dehydrogenase from Rhodobacter capsulatus is able to catalyze the reduction of CO2 to formate. FEBS J. 2013;280:6083–6096. doi: 10.1111/febs.12528. [DOI] [PubMed] [Google Scholar]
- Hatchikian EC, Traore A, Fernandez VM, Cammack R. Characterization of the nickel-iron periplasmic hydrogenase from Desulfovibrio fructosovorans. Eur J Biochem. 1990;187:635–643. doi: 10.1111/j.1432-1033.1990.tb15347.x. [DOI] [PubMed] [Google Scholar]
- Hayes PC. Process principles in minerals and materials production. 2. Brisbane: Hayes Publishing Co; 1993. Standard reduction potentials; pp. 660–665. [Google Scholar]
- Hendricks JK, Mobley HLT. Helicobacter pylori ABC transporter: effect of allelic exchange mutagenesis on urease activity. J Bacteriol. 1997;179:5892–5902. doi: 10.1128/jb.179.18.5892-5902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heugebaert TSA, De Corte S, Sabbe T, Hennebel T, Verstraete W, Boon N, Stevens CV. Biodeposited Pd/Au bimetallic nanoparticles as novel Suzuki catalysts. Tetrahedron Lett. 2012;53:1410–1412. doi: 10.1016/j.tetlet.2012.01.030. [DOI] [Google Scholar]
- Holt EM, Kelly GJ, King F. Catalyst design and manufacture. In: King MB, Winterbotton JM, editors. Reactor design for chemical engineers. Cheltenham: Stanley Thornes Ltd; 1999. pp. 276–300. [Google Scholar]
- Jormakka M, Tornroth S, Byrne B, Iwata S. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science. 2002;295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- José-Yacamán M, Marín-Almazo M, Ascencio JA. High resolution TEM studies on palladium nanoparticles. J Mol Catal A. 2001;173:61–74. doi: 10.1016/S1381-1169(01)00145-5. [DOI] [Google Scholar]
- Konishi Y, Tsukiyama T, Ohno K, Saitoh N, Nomura T, Nagamine S. Intracellular recovery of gold by microbial reduction of AuCl4− ions using the anaerobic bacterium Shewanella alga. Hydrometallurgy. 2006;81:24–29. doi: 10.1016/j.hydromet.2005.09.006. [DOI] [Google Scholar]
- Law N, Ansari S, Livens FR, Renshaw JC, Lloyd JR. Formation of nanoscale elemental silver particles via enzymatic reduction by Geobacter sulfurreducens. Appl Environ Microbiol. 2008;74:7090–7093. doi: 10.1128/AEM.01069-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-C, Pandey BD. Bio-processing of solid wastes and secondary resources for metal extraction: a review. Waste Manag. 2012;32:3–18. doi: 10.1016/j.wasman.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Liermann LJ, Hausrath EM, Anbar AD, Brantley SL. Assimilatory and dissimilatory processes of microorganisms affecting metals in the environment. J Anal At Spectrom. 2007;22:867–877. doi: 10.1039/b705383e. [DOI] [Google Scholar]
- Lim B, Kobayashi H, Camargo PC, Allard L, Liu J, Xia Y. New insights into the growth mechanism and surface structure of palladium nanocrystals. Nano Res. 2010;3:180–188. doi: 10.1007/s12274-010-1021-5. [DOI] [Google Scholar]
- Livingstone SE. Palladium: the element. In: Bailer JC, Nyholm R, Trotman-Dickenson AF, editors. Comprehensive Inorganic Chemistry. Oxford: Pergamon Press; 1973. pp. 1274–1276. [Google Scholar]
- Mabbett AN, Sanyahumbi D, Yong P, Macaskie LE. Biorecovered precious metals from industrial wastes: single step conversion of a mixed metal liquid waste to a bioinorganic catalyst with environmental applications. Environ Sci Technol. 2006;40:1015–1021. doi: 10.1021/es0509836. [DOI] [PubMed] [Google Scholar]
- Maroney MJ. Structure/function relationships in nickel metallobiochemistry. Curr Opin Chem Biol. 1999;3:188–199. doi: 10.1016/S1367-5931(99)80032-5. [DOI] [PubMed] [Google Scholar]
- Merroun M, Rossberg A, Hennig C, Scheinost AC, Selenska-Pobell S. Spectroscopic characterization of gold nanoparticles formed by cells and S-layer protein of Bacillus sphaericus JG-A12. Mater Sci Eng C. 2007;27:188–192. doi: 10.1016/j.msec.2006.05.001. [DOI] [Google Scholar]
- Mikheenko IP, Rousset M, Dementin S, Macaskie LE. Bioaccumulation of palladium by Desulfovibrio fructosovorans wild-type and hydrogenase-deficient strains. Appl Environ Microbiol. 2008;74:6144–6146. doi: 10.1128/AEM.02538-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota CS, Rivas MG, Brondino CD, Moura I, Moura JJG, Gonzalez PJ, Cerqueira N. The mechanism of formate oxidation by metal-dependent formate dehydrogenases. J Biol Inorg Chem. 2011;16:1255–1268. doi: 10.1007/s00775-011-0813-8. [DOI] [PubMed] [Google Scholar]
- Mulrooney SB, Hausinger RP. Nickel uptake and utilization by microorganisms. FEMS Microbiol Rev. 2003;27:239–261. doi: 10.1016/S0168-6445(03)00042-1. [DOI] [PubMed] [Google Scholar]
- Murray AJ, Taylor S, Zhu J, Wood J, Macaskie LE (2015) A novel biorefinery: biorecovery of precious metals from spent automotive catalyst leachates into new catalysts effective in metal reduction and in the hydrogenation of 2-pentyne. Min Eng (in press)
- Nair B, Pradeep T. Coalescence of nanoclusters and formation of submicron crystallites assisted by Lactobacillus strains. Cryst Growth Des. 2002;2:293–298. doi: 10.1021/cg0255164. [DOI] [Google Scholar]
- Narayanan KB, Sakthivel N. Biological synthesis of metal nanoparticles by microbes. Adv Colloid Interface Sci. 2010;156:1–13. doi: 10.1016/j.cis.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Navarrete JU, Borrok DM, Viveros M, Ellzey JT. Copper isotope fractionation during surface adsorption and intracellular incorporation by bacteria. Geochim Cosmochim Acta. 2011;75:784–799. doi: 10.1016/j.gca.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nidhin M, Indumathy R, Sreeram KJ, Nair BU. Synthesis of iron oxide nanoparticles of narrow size distribution on polysaccharide templates. Bull Mater Sci. 2008;31:93–96. doi: 10.1007/s12034-008-0016-2. [DOI] [Google Scholar]
- Macaskie LE, Mikheenko IP, Yong P, Deplanche K, Murray AJ, Paterson-Beedle M, Coker VS, Pearce CI, Cutting R, Pattrick RAD, et al. et al. Today’s wastes, tomorrow’s materials for environmental protection. In: Moo-Young B, et al.et al., editors. Comprehensive biotechnology. New York: Pergamon; 2011. pp. 719–725. [Google Scholar]
- Peng X, Zhou H, Li J, Li J, Chen S, Yao H, Wu Z. Intracellular and extracellular mineralization of a microbial community in the Edmond Deep-Sea Vent field environment. Sedim Geol. 2010;229:193–206. doi: 10.1016/j.sedgeo.2010.06.003. [DOI] [Google Scholar]
- Pollmann K, Raff J, Schnorpfeil M, Radeva G, Selenska-Pobell S. Novel surface layer protein genes in Bacillus sphaericus associated with unusual insertion elements. Microbiol-SGM. 2005;151:2961–2973. doi: 10.1099/mic.0.28201-0. [DOI] [PubMed] [Google Scholar]
- Polti MA, Amoroso MJ, Abate CM. Intracellular chromium accumulation by Streptomyces sp. MC1. Water Air Soil Pollut. 2011;214:49–57. doi: 10.1007/s11270-010-0401-5. [DOI] [Google Scholar]
- Raaijmakers H, Macieira S, Dias JM, Teixeira S, Bursakov S, Huber R, Moura JJG, Moura I, Romao MJ. Gene sequence and the 1.8 Å crystal structure of the tungsten-containing formate dehydrogenase from Desulfovibrio gigas. Structure. 2002;10:1261–1272. doi: 10.1016/S0969-2126(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Ravindranath SP, Henne KL, Thompson DK, Irudayaraj J. Surface-enhanced Raman imaging of intracellular bioreduction of chromate in Shewanella oneidensis. PLoS ONE. 2011 doi: 10.1371/journal.pone.0016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodin TN, Ertl G. The nature of the surface chemical bond. Oxford: North-Holland; 1979. pp. 104–108. [Google Scholar]
- Riddin TL, Govender Y, Gericke M, Whiteley CG. Two different hydrogenase enzymes from sulphate-reducing bacteria are responsible for the bioreductive mechanism of platinum into nanoparticles. Enzyme Microbial Technol. 2009;45:267–273. doi: 10.1016/j.enzmictec.2009.06.006. [DOI] [Google Scholar]
- Rodriguez-Navarro C, Jimenez-Lopez C, Rodriguez-Navarro A, Gonzalez-Muñoz MT, Rodriguez-Gallego M. Bacterially mediated mineralization of vaterite. Geochim Cosmochim Acta. 2007;71:1197–1213. doi: 10.1016/j.gca.2006.11.031. [DOI] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH image to imageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenska-Pobell S, Panak P, Miteva V, Boudakov I, Bernhard G, Nitsche H. Selective accumulation of heavy metals by three indigenous Bacillus strains, B-cereus, B-megaterium and B-sphaericus, from drain waters of a uranium waste pile. FEMS Microbiol Ecol. 1999;29:59–67. doi: 10.1111/j.1574-6941.1999.tb00598.x. [DOI] [Google Scholar]
- Shi LH, Wang AQ, Zhang T, Zhang BS, Su DS, Li HQ, Song YJ. One-step synthesis of Au–Pd alloy nanodendrites and their catalytic activity. J Phys Chem C. 2013;117:12526–12536. doi: 10.1021/jp4013202. [DOI] [Google Scholar]
- Shoukry AA, Mohamed MS. DNA binding, spectroscopic and antimicrobial studies of palladium(II) complexes containing 2,2′-bipyridine and 1-phenylpiperazine. Spectrochim Acta A. 2012;96:586–593. doi: 10.1016/j.saa.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Silver S, Phung LT. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- Sousa T, Chung AP, Pereira A, Piedade AP, Morais PV. Aerobic uranium immobilization by Rhodanobacter A2-61 through formation of intracellular uranium–phosphate complexes. Metallomics. 2013;5:390–397. doi: 10.1039/c3mt00052d. [DOI] [PubMed] [Google Scholar]
- Southam G, Beveridge TJ. The occurrence of sulfur and phosphorus within bacterially derived crystalline and pseudocrystalline octahedral gold formed in vitro. Geochim Cosmochim Acta. 1996;60:4369–4376. doi: 10.1016/S0016-7037(96)00235-9. [DOI] [Google Scholar]
- Thakkar KN, Mhatre SS, Parikh RY. Biological synthesis of metallic nanoparticles. Nanomedicine. 2010;6:257–262. doi: 10.1016/j.nano.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Waldron KJ, Robinson NJ. How do bacterial cells ensure that metalloproteins get the correct metal? Nat Rev Microbiol. 2009;7:25–35. doi: 10.1038/nrmicro2057. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xie SF, Liu JY, Park J, Huang CZ, Xia YN. Shape-controlled synthesis of palladium nanocrystals: a mechanistic understanding of the evolution from octahedrons to tetrahedrons. Nano Lett. 2013;13:2276–2281. doi: 10.1021/nl400893p. [DOI] [PubMed] [Google Scholar]
- Weimer PJ. Control of product formation during glucose fermentation by Bacillus macerans. Microbiology. 1984;130:103–111. doi: 10.1099/00221287-130-1-103. [DOI] [Google Scholar]
- Wiseman CLS, Zereini F. Airborne particulate matter, platinum group elements and human health: a review of recent evidence. Sci Total Environ. 2009;407:2493–2500. doi: 10.1016/j.scitotenv.2008.12.057. [DOI] [PubMed] [Google Scholar]
- Wood J, Bodenes L, Bennett J, Deplanche K, Macaskie LE. Hydrogenation of 2-butyne-1,4-diol using novel bio-palladium catalysts. Ind Eng Chem Res. 2010;49:980–988. doi: 10.1021/ie900663k. [DOI] [Google Scholar]
- Xiong Y, McLellan JM, Yin Y, Xia Y. Synthesis of palladium icosahedra with twinned structure by blocking oxidative etching with citric acid or citrate ions. Angew Chem. 2007;119:804–808. doi: 10.1002/ange.200604032. [DOI] [PubMed] [Google Scholar]
- Xu Y, Feng L, Jeffrey PD, Shi YG, Morel FMM. Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms. Nature. 2008;452:56–61. doi: 10.1038/nature06636. [DOI] [PubMed] [Google Scholar]
- Yates MD, Cusick RD, Logan BE. Extracellular palladium nanoparticle production using Geobacter sulfurreducens. ACS Sustain Chem Eng. 2013;1:1165–1171. doi: 10.1021/sc4000785. [DOI] [Google Scholar]
- Yong P, Rowson NA, Farr JPG, Harris IR, Macaskie LE. Bioaccumulation of palladium by Desulfovibrio desulfuricans. J Chem Technol Biotechnol. 2002;77:593–601. doi: 10.1002/jctb.606. [DOI] [PubMed] [Google Scholar]
- Zhang H, Jin MS, Xiong YJ, Lim B, Xia YN. Shape-controlled synthesis of Pd nanocrystals and their catalytic applications. Acc Chem Res. 2013;46:1783–1794. doi: 10.1021/ar300209w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.