Abstract
A decline in mitochondrial quality and activity has been associated with normal aging and correlated with the development of a wide range of age-related diseases. Here, we review the evidence that a decline in mitochondria function contributes to aging. In particular, we discuss how mitochondria contribute to specific aspects of the aging process including cellular senescence, chronic inflammation and the age-dependent decline in stem cell activity. Signaling pathways regulating the mitochondrial unfolded protein response and mitophagy are also reviewed with particular emphasis placed on how these pathways might in turn regulate longevity. Taken together, these observations suggest that mitochondria influence or regulate a number of key aspects of aging, and suggest that strategies directed at improving mitochondrial quality and function might have far-reaching beneficial effects.
Introduction
Scientists have long struggled to explain the evolutionary basis of aging. In particular, how can there be a genetic program of aging, if aging manifests itself long after the reproductive period has passed, and therefore after all the forces of natural selection have long-since abated? Potential explanations have come from Medawar’s mutation accumulation hypothesis, Kirkwood’s disposable soma theory and the concept of antagonistic pleiotropy (Hughes and Reynolds, 2005). The latter hypothesis revolves around the notion that genes regulating aging in the old organism actually have a different, antagonistic function when the animal is young (Williams, 1957). In this scenario, those genes positively regulating growth and fertility in the young animal might serve to accelerate senescence and aging in the older animal. While from an evolutionary viewpoint, this concept has been exclusively applied to our genetic inheritance, the notion of antagonistic pleiotropy actually provides a useful framework to understand the role of mitochondria in aging. Perhaps no structure is so intimately and simultaneously connected to both the energy of youth and the decline of the old. The revelation of these complex and antagonistic functions of mitochondria has slowly transformed how we view this subcellular organelle. Mitochondria can no longer be viewed as simple bioenergetics factories, but rather as platforms for intracellular signaling, regulators of innate immunity and modulators of stem cell activity. In turn, each of these properties provides clues as to how mitochondria might regulate aging and age-related diseases. Here, we review how mitochondria participate in aging and how these insights may usher in a new era of mitochondrial-targeted therapies to potentially slow or reverse the aging process.
Mitochondrial function during aging
It has been long appreciated that aging in model organisms is accompanied by a decline in mitochondrial function and that this decline might in turn contribute to the observed age-dependent decline in organ function (Rockstein and Brandt, 1963). Similarly, a decline in mitochondrial function in humans has also been observed, and again, this decrement may pre-dispose to certain age-related diseases (Petersen et al., 2003). It is also known that mitochondrial mutations increase in frequency with age in both animal models and in humans (Cortopassi and Arnheim, 1990; Piko et al., 1988), although the levels and kind of mutations appear to differ between tissues and even within tissues (Soong et al., 1992). While some have speculated that the increased levels of mitochondrial mutations contribute to aging and age-related diseases (Linnane et al., 1989), others have questioned whether these mutations ever reach a significant enough level to contribute to the aging process (Khrapko and Vijg, 2009). Indeed, since mitochondrial DNA exists in hundreds to thousands of copies per cell, the detection of mutant mitochondrial DNA does not in itself imply dysfunction, as it is generally believed that mutational load must exceed a threshold value (perhaps exceeding 60% of all mitochondria within a given tissue) for there to be a significant phenotype (Rossignol et al., 2003). Perhaps the strongest evidence for a potential causative role for mitochondrial DNA (mtDNA) mutations in mammalian aging comes from analyzing the ‘mitochondrial mutator mice’ which are knockin mice containing a mutated (D257A), proofreading-deficient form of the mitochondrial DNA polymerase POLGγ. This nuclear-encoded gene is the sole mitochondrial DNA polymerase and the mutation at amino acid position 257 results in an enzyme that retains normal polymerase function but has impaired proofreading activity. Mice containing one or two copies of this proofreading-deficient POLG accumulate a significant level of mitochondrial mutations and homozygous knockin mice exhibit an accelerated aging phenotype (Kujoth et al., 2005; Trifunovic et al., 2004). Nonetheless, while this model clearly links mitochondrial mutations to aging, it should be noted that the type and magnitude of mitochondrial mutations do not appear to faithfully replicate what is seen during normal aging (Williams et al., 2010). Thus, while the levels of mitochondrial mutations increase with age, it remains unclear whether this increase plays a fundamental role in the aging process.
Irrespective of mitochondrial DNA, in humans, the link between mitochondrial function and aging has been perhaps best studied by analyzing skeletal muscle. While all studies are not in complete concordance, the majority of reports have found that aging is generally accompanied by a decline in activity of mitochondrial enzymes (e.g. citrate synthase), a decrease in respiratory capacity per mitochondria (e.g. substrate-dependent oxygen consumption), an increase in ROS production and a reduced phosphocreatine (PCr) recovery time (an in vivo measurement of mitochondrial respiratory capacity). Nonetheless, the literature is also filled with many counter examples that may reflect differences in how the specific assays were performed, or differences in the muscle fiber type studied (Hepple, 2014). Most studies have also noted that aging is accompanied by an accelerated rate of muscle loss, both in terms of mass and activity (e.g. strength). Although muscle strength over a lifetime declines at an average rate of roughly 1 % per year, for patients in their 70’s, that rate of decline can increase 2-4 fold (Goodpaster et al., 2006). At present, perhaps the best intervention to counteract this age-dependent decline in muscle function, termed sarcopenia, is physical exercise. Indeed, accumulating evidence from epidemiological studies and randomized clinical trials illustrates that regular physical activity and endurance exercise benefits a range of human age-related pathologies including sarcopenia, as well as the age-dependent decline in cardiac and cognitive function (Chakravarty et al., 2008; Kosmadakis et al., 2010; Rowe et al., 2014; Stessman et al., 2009; Willis et al., 2012). Interestingly, endurance exercise also conferred phenotypic protection and prevented the premature mortality observed in the mitochondrial mutator mice mentioned above (Safdar et al., 2011). The therapeutic effects of endurance exercise are accompanied by a number of physiological adaptations, however, one of the most beneficial effects appears to be stimulation of mitochondrial biogenesis in a wide variety of tissues including the brain (Arany et al., 2005; Egan and Zierath, 2013; Rowe et al., 2014; Steiner et al., 2011; Wu et al., 2002). Mitochondrial biogenesis is largely coordinated by the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator-1 α (PGC-1α), (Handschin and Spiegelman, 2008; Ruas et al., 2012). PGC-1α, in turn, regulates the activity of several transcription factors involved in creating new mitochondria including the nuclear respiratory factors (NRF1 and NRF2) and mitochondrial transcription factor A (TFAM) (Austin and St-Pierre, 2012). Increasing PGC-1α levels in mouse skeletal muscle is sufficient to forestall the development of age-dependent sarcopenia, again emphasizing the potential importance of this pathway for aging biology (Wenz et al., 2009). The development of sarcopenia is not, however, solely an issue of impaired mitochondrial biogenesis (Argiles et al., 2015). Recently, it was also shown that phosphorylation and hence activity of ATP citrate lyase (ACL) a key regulator of acetyl-CoA levels, was markedly reduced in sarcopenic muscle (Das et al., 2015). In this study, ACL phosphorylation was stimulated by IGF-1, a growth factor known to increase muscle mass (Egerman and Glass, 2014), and also known to decline in the serum of aging men and woman (O'Connor et al., 1998). Increasing ACL levels in mice resulted in improved mitochondrial function suggesting that this might be a complementary approach to combat the deleterious effects of skeletal muscle aging (Das et al., 2015).
Mitochondria as Regulators of Stem Cell Function
While aging is accompanied by a general decline in mitochondrial function in all tissues, the effects of mitochondrial dysfunction might be particularly important within certain specialized cell types. Since a decline in adult stem cell function is thought to contribute to various aspects of aging (Lopez-Otin et al., 2013), the role of mitochondrial dysfunction in stem cell biology has become a subject of increasing interest. In the case of hematopoietic stem cells (HSCs), perhaps the best studied stem cell population, mitochondria are thought to play a relatively minor role in the resting bioenergetics profile of these cells (Suda et al., 2011). Quiescent HSCs are generally thought to instead rely on glycolytic metabolism as the major source of their ATP, presumably in keeping with the low oxygen environment of the HSC niche, and as a mechanism to minimize the long term deleterious effects of mitochondrial ROS production (Suda et al., 2011). Indeed, there are a number of links that suggest a rise in ROS might be harmful for stem cell function (Ito et al., 2004; Liu et al., 2009; Tothova et al., 2007), although there are also increasing examples in which ROS appear to play a positive and necessary signaling role in stem cell biology (Bigarella et al., 2014).
One clear connection between mitochondria and stem cell function has come from the analysis of the previously described mtDNA mutator mice (Kujoth et al., 2005; Trifunovic et al., 2004). Several reports have analyzed the stem cell function of the POLG knockin mice and found a range of defects. These include the development of a severe and often fatal anemia in the mice, as well as abnormalities in B cells (Chen et al., 2009). A similar impairment was observed in neural stem cell populations derived from POLG knockin mice (Ahlqvist et al., 2012). Several features of these analyses deserve mentioning. First, the stem cell defects could at least be partially ameliorated by the administration of the antioxidant N-acetylcysteine (Ahlqvist et al., 2012). Indeed, follow up studies have demonstrated that POLG knockin cells also have markedly impaired capacity to be reprogrammed into pluripotent stem cells, a defect again related to an increase in mitochondrial ROS production (Hamalainen et al., 2015). The second point to emphasize is that the observed stem cell defects appear to arise because of cell autonomous mitochondrial defects. This mutator mouse model affects a multitude of cell types, including the stem cell, their progeny, as well as the niche. Nonetheless, transplantation of POLG knockin HSCs into a normal host recapitulates the observed defect (Chen et al., 2009) and other mouse models that have large scale mitochondrial deletions only within post-mitotic tissues do not exhibit any stem cell defects (Ahlqvist et al., 2012). Thus, even though stem cells do not seem to rely on oxidative phosphorylation for their energetics, mitochondria are clearly required for long term function of these cells and their progenitors in a cell autonomous capacity. Finally, as mentioned previously, it is important to note that these stem cell defects do not appear to accurately recapitulate aging (Norddahl et al., 2011). Indeed, from a histological viewpoint, the anemia observed in these animals looks less like the anemia of aging, and more like the pre-leukemic abnormality known as myelodysplastic syndrome (Chen et al., 2009). It should also be noted, that the level of mitochondrial mutation seen in these models is also dramatically higher than seen during the normal aging process which may account for why the observed stem cell defects do not faithfully recapitulate what is seen during normal aging.
Another mechanism by which mitochondria might contribute to stem cell maintenance is through regulation of specific metabolites. Increasingly, there is evidence that metabolic intermediates play an important role in regulating the transcriptional and epigenetic state of cells. It is no presumed accident that chromatin modifications are largely dependent on the same carbon intermediates (e.g. methyl, acetyl, etc.) that are generated during normal mitochondrial metabolism. For example, one recent study demonstrated that in mouse embryonic stem (ES) cells, the intracellular ratio of α-ketoglutarate (αKG) to succinate was important in maintaining pluripotency (Carey et al., 2015). Both of these metabolites are generated as a result of tricarboxylic acid (TCA) metabolism in the mitochondrial matrix. In turn, it was shown that levels of αKG modulated distinct chromatin modifications. This modulation was mediated, at least in part, by the activity of αKG-dependent demethylases including Jumonji C (JmjC)-domain-containing enzymes and the ten-eleven translocation (Tet)-dependent DNA demethylases (Kaelin, 2011). Another important set of metabolites that connect stem cells to the mitochondria is the NAD+/NADH ratio. Levels of NAD+ appears to decline in tissues as they age (Mouchiroud et al., 2013; Yoshino et al., 2011). Analysis of neural stem cells (NSCs) has shown that reducing NAD+ levels recapitulates at least some of the phenotypes of stem cell aging, while NAD+ supplementation can restore function to old NSCs (Stein and Imai, 2014). These effects appear to be mediated, in part, by the sirtuin family of NAD-dependent enzymes. This connection has also been observed in HSC biology. SIRT3 is one of seven mammalian sirtuin family members and is found within the mitochondria where it regulates the mitochondrial acetylome in an NAD-dependent fashion (Lombard et al., 2007). Interestingly, SIRT3 is highly enriched in HSCs, although its expression declines with age. Augmenting SIRT3 levels in old HSCs results in improved regenerative capacity in these aging stem cells (Brown et al., 2013). Similar results have been recently observed with overexpression of SIRT7 (Mohrin et al., 2015).
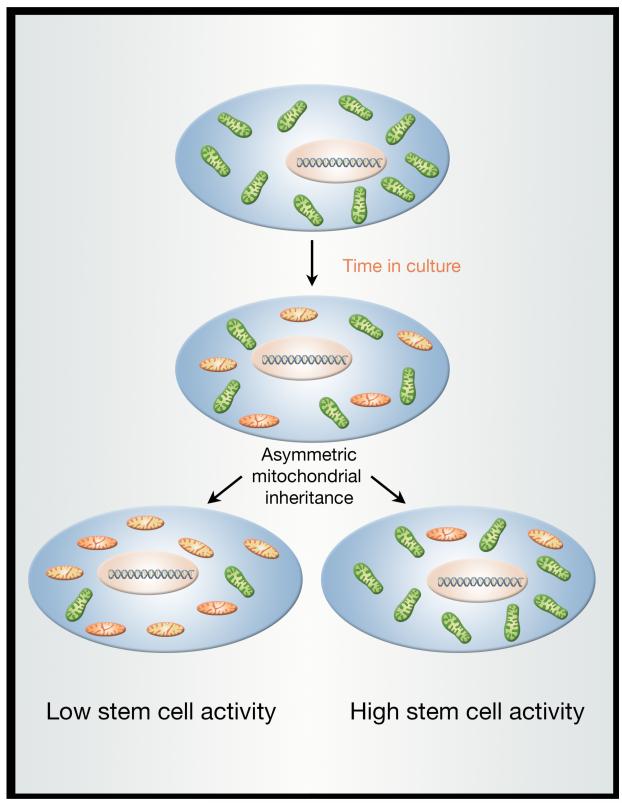
The unique properties of stem cells suggest that these cells might have mechanisms to ensure that these critical cells do not accumulate old and dysfunctional mitochondria. Preliminary evidence suggests that in the brain, areas enriched for NSCs appear to have augmented rates of mitophagy (Sun et al., 2015). Another potential mechanism appears to be a unique capacity of adult stem cells to exclude older mitochondria. Indeed, a recent report studying the stem-like cells within immortalized human mammary epithelial cell cultures noted that there was an uneven distribution of mitochondria after cell division (Katajisto et al., 2015). This asymmetry was not a difference in mitochondrial number between the two daughter cells but rather, a difference in the segregation of young and old mitochondria (Figure 1). Stem-like cells getting young mitochondria maintained their stem cell properties much more robustly then those cells receiving older mitochondria. This unequal distribution of mitochondria based on the age of the mitochondria was only seen within the stem-like cells in the culture, not the more differentiated mammary epithelial cells. In addition, this property was only seen with mitochondrial segregation and not with other organelles such as lysosomes or ribosomes or with cellular components such as chromatin. It is currently unclear whether this property is present in vivo and if so, whether it is present in all, or just some, types of stem cells. It should however be noted that in yeast, where there is asymmetric division between the mother cell and the bud, there is also evidence of a corresponding asymmetric inheritance of both mitochondria (McFaline-Figueroa et al., 2011) and misfolded proteins (Clay et al., 2014).
Figure 1.
Stem cells exhibit asymmetric mitochondrial inheritance. Analysis of stem-like cells within immortalized, transformed epithelial cultures revealed that young mitochondria (shown in green) and old mitochondria (depicted in orange) are not symmetrically distributed after the stem-like cell divides. Moreover, the daughter cell inheriting the younger mitochondria, also exhibits higher stem-like activity. The molecular basis for this asymmetric mitochondrial distribution is not clear, nor is it known whether similar mechanisms exist in vivo.
Mitochondria and Cellular Senescence
As noted in the discussion of stem cell biology, mitochondria can regulate cellular aging through the modulation of the metabolic profile of the cell. Cellular senescence is accompanied by profound changes in the metabolome and although different triggers of senescence all have a similar morphological appearance, the metabolic profile of oncogene-induced senescence and replicative senescence appear distinct (Quijano et al., 2012). There is increasing evidence that these metabolic changes are casually related to the senescent state. For instance, p53 plays an important role in senescence and evidence suggests that it can also repress expression of mitochondrial malic enzyme (ME2) which converts the TCA metabolite malate to pyruvate via oxidative decarboxylation (Jiang et al., 2013). Moreover, knockdown of ME2 results in the induction of senescence, while forced expression allows cells to escape from senescence. This argues that the ability of p53 to mediate senescence may partially be through its ability to modulate TCA metabolism. Interestingly, previous observations have established that overexpression of malate dehydrogenase also results in lifespan extension in yeast (Easlon et al., 2008). The link between mitochondrial metabolism and senescence is also observed in oncogene-induced senescence (OIS). Analysis of cells undergoing OIS precipitated by expression of the BRAF oncogene demonstrated an increase in pyruvate oxidation that contributed to the generation of increased mitochondrial ROS and entry into the senescent state (Kaplon et al., 2013). This increase in pyruvate utilization was due to alteration in the phosphorylation state, and hence activity, of the mitochondrial pyruvate dehydrogenase (PDH) complex. Again, gain and loss of function studies suggest these metabolic changes appear to be necessary for BRAF-induced senescence. Interestingly, the PDH complex is also regulated by the mitochondrial sirtuins, particularly SIRT3 and SIRT4 (Fan et al., 2014; Mathias et al., 2014). Similarly, from an organismal context, a recent large-scale screen of yeast single-gene deletion mutants uncovered a number of enzymes involved in the TCA cycle as potent regulators of lifespan (McCormick et al., 2015). Together, these argue that mitochondrial-induced metabolic changes might be necessary, and in some cases sufficient, to trigger cellular senescence and potentially to regulate overall longevity.
There is also a strong link between mitochondrial metabolism, ROS generation and the senescent state. Almost four decades ago, it was noted that the lifespan of human cells in culture could be significantly extended by culturing the cells in a low oxygen environment (Packer and Fuehr, 1977). A similar effect was also observed in mouse cells (Parrinello et al., 2003). Similarly, OIS triggered by Ras expression results in an increase in ROS levels and OIS can be prevented by growing these cells in a low oxygen state or supplementing the media with an antioxidant (Lee et al., 1999). Similar relationships have been observed between other regulators of senescence and ROS including the p53 target and cell cycle regulator p21 which also appears to regulate senescence in a redox-dependent fashion (Macip et al., 2002; Passos et al., 2010). All of these observations fit well with the long standing notions of the free radical theory of aging that postulated a causal role for ROS in the aging process (Harman, 1956). Nonetheless, there are a number of observations that suggest that the cellular effects of ROS with regards to inducing senescence do not unequivocally transfer to organismal aging. For instance, while in some animal models, scavenging mitochondrial oxidants appears to extend lifespan (Schriner et al., 2005), in other cases, a consistent relationship between ROS levels and lifespan was seemingly absent (Sanz et al., 2010; Yang et al., 2007). Moreover, in some cases, a rise in ROS appears to actually increase, rather than reduce, overall lifespan (Yang and Hekimi, 2010; Zarse et al., 2012).
The Mitochondrial Unfolded Protein Response and longevity
Genetic screens in C. elegans have found that disruption of mitochondrial function often leads to an increase in overall lifespan (Dillin et al., 2002; Lee et al., 2003). In many ways, these observations seemed counterintuitive, especially given the wealth of data, as previously discussed, that suggests a decline in mitochondrial function occurs with aging. Nonetheless, there are a growing number of experimental observations that suggest in a wide range of organisms, a modest decline or impairment in mitochondrial function leads to lifespan extension (Liu et al., 2005; Owusu-Ansah et al., 2013; Yee et al., 2014). Insight into this seeming paradox, perhaps another example of antagonistic pleiotropy, first came from examining worms that were long lived due to knockdown of a nuclear-encoded cytochrome C oxidase subunit (cco-1). Following cco-1 knockdown, the impairment of electron transport in these animals appeared to trigger activation of the mitochondrial unfolded protein response (Durieux et al., 2011). The UPRmt is a stress response pathway initially characterized in mammalian cells in which there was either a depletion of the mitochondrial genome or accumulation of misfolded proteins within the mitochondria (Martinus et al., 1996; Zhao et al., 2002). In either case, it was noted that this mitochondrial perturbation triggered a nuclear transcriptional response that included the increased expression of mitochondrial chaperone proteins. While initially described in mammalian cells (Zhao et al., 2002), the biochemistry and genetics of this pathway have been predominantly studied in C. elegans. It is now clear that the UPRmt regulates a large set of genes that not only involve protein folding but also involve changes in ROS defenses, metabolism, regulation of iron sulfur cluster assembly and, as will discussed below, modulation of the innate immune response (Nargund et al., 2015; Schulz and Haynes, 2015). In general terms, all of these changes allow for a restoration of mitochondrial function while at the same time re-wiring the cell to temporarily survive as best as possible without the benefit of full mitochondrial capacity. Nonetheless, the existence of this broad transcriptional response demonstrates that a means of communication and coordination exists between the mitochondria and the nucleus.
It is now known that in worms, the UPRmt is regulated, in part, by a unique transcription factor termed Activating Transcription Factor associated with Stress-1. ATFS-1 was identified initially in a screen for factors that mediate the UPRmt in C. elegans (Haynes et al., 2010). It was subsequently demonstrated that ATFS-1 has both a nuclear localization targeting sequence, as well as a mitochondrial targeting sequence (Nargund et al., 2012). While a mitochondrial localization predominates under basal conditions, mitochondrial stress results in reduced importation of ATFS-1, leading to nuclear accumulation and the transcriptional response delineated above. In addition to ATFS-1, in worms, the UPRmt appears to require a number of other factors including the homeobox transcription factor DVE-1, the ubiquitin-like protein UBL-5, the mitochondrial protease ClpP and the inner mitochondrial membrane transporter HAF-1 (Jensen and Jasper, 2014).
The link between the UPRmt and lifespan was made initially in the setting of attempting to explain why mutant worms, such as those with knockdown of cco-1, live longer (Durieux et al., 2011). These results demonstrated that this mutant, as well as other long lived mitochondrial mutants all appeared to require activation of the UPRmt for their lifespan extension. In contrast, other long lived mutants, such as those involved in insulin/IGF signaling appeared to extend lifespan independent of UPRmt activation (Durieux et al., 2011). Remarkably, when cco-1 was knocked down in one tissue (e.g. neurons), it appeared to activate induction of the UPRmt in other, distal tissues (e.g. intestine). This suggested the existence of a circulating factor that signals, and perhaps coordinates metabolism between tissues. The authors called this factor a mitokine (Durieux et al., 2011), although to date, its molecular makeup remains undefined. Whether or not such factors exist in higher organisms is unclear but there is clearly a growing interest in circulating factors that regulate aging, as evidence by the renewed interest in parabiosis studies (Conboy et al., 2013).This notion of mitochondrial dysfunction in one tissue acting as a signal for other tissues has also been observed in Drosophila. In a recent example, muscle-specific impairment of a component of Complex I resulted in an increase in overall lifespan of the fly (Owusu-Ansah et al., 2013). This mitochondrial stress resulted in the activation for at least two separate pathways that appeared to contribute to the observed longevity effects. In the muscle itself, disruption of Complex I resulted in the induction of the UPRmt through what appeared to involve a redox-sensitive pathway. Indeed, overexpression of hydrogen peroxide scavenging enzymes such as catalase or glutathione peroxidase suppressed the induction of the UPRmt and also abrogated the increased longevity seen with Complex I inhibition (Owusu-Ansah et al., 2013). These negative effects of redox scavengers are reminiscent of similar observations in humans, where for instance the beneficial effects of exercise appear to be abrogated by antioxidant supplementation (Ristow et al., 2009). In addition to the induction of the UPRmt in muscle, the authors also observed a systemic effect on insulin signaling mediated by changes in the level of a particular circulating IGF binding partners (Owusu-Ansah et al., 2013). Again, these results argue that mitochondrial dysfunction in one tissue can signal through secreted factors in the circulation to alter the function of distal tissues. This inter-organ communication appears to be ultimately required for the observed increase in lifespan.
Another apparent way in which the UPRmt appears to be activated is when there is a stoichiometric imbalance between mitochondrial and nuclear proteins (Houtkooper et al., 2013). This imbalance can be achieved experimentally in worms by knocking down a mitochondrial ribosomal gene (Mrps5), resulting in the selective translational impairment of mitochondrial transcripts. It was observed that such a knockdown or treatment with certain antibiotics that differentially effect mitochondrial and nuclear proteins, triggers induction of the UPRmt and an increase in lifespan (Houtkooper et al., 2013). These authors also observed a strong correlation between expression of Mrps5 and murine lifespan. This suggests, as does other evidence (Wu et al., 2014), that elements of the UPRmt appear to very well conserved, even though the mammalian equivalent of ATFS-1 remains elusive. Additional evidence comes from analysis of the long-lived Surf1 knockout mice (Dell'agnello et al., 2007). Surf1 is a cytochrome c oxidase assembly factor, and Surf1−/− mice live 20% longer than controls, a phenotype that appears to be linked to the activation of a mitochondrial stress response pathway (Pulliam et al., 2014). Other potential relevant observations include a recent re-analysis of CLK-1, a monooxygenase that catalyzes the hydroxylation of 5-demethoxyubiquinone, an important step in the synthesis of ubiquinone. Clk-1 null worms live longer (Felkai et al., 1999), as do mice who have lost one allele of the mammalian ortholog COQ7 (Liu et al., 2005). Evidence suggests that the lifespan extension observed in clk-1 null worms appears to involve activation of the UPRmt (Nargund et al., 2012). While it was assumed that CLK-1 was exclusively mitochondrial, recent evidence suggests that CLK-1, as well as it mammalian ortholog COQ7, can also be found in the nucleus (Monaghan et al., 2015). In the nucleus, CLK-1 appears to help lower ROS levels and suppress the UPRmt. Much like ATFS-1, CLK-1 and the mammalian COQ7 can exist in both the nucleus and mitochondria and thereby appear uniquely suited to modulate the UPRmt. The list of such factors is likely to grow. It will be of interest to see whether other mitochondrial mutants that are also long-lived, including those with alterations in iron-sulfur proteins in Complex III (Hughes and Hekimi, 2011) or the outer mitochondrial membrane (Wu et al., 2012), also activate the UPRmt or related pathways. Finally, two important caveats are worth noting. First, while a number of observations support a role for UPRmt activation in modulating the aging process, it is important to note that this pathway is incompletely characterized at present, and evidence suggests that UPRmt activation may not by itself be sufficient to extend lifespan (Bennett et al., 2014). Secondly, and perhaps relatedly, as recent observations in yeast suggest (Wang and Chen, 2015; Wrobel et al., 2015), the cellular response to mitochondrial perturbation is undoubtedly more complex than the currently conceived model of the UPRmt.
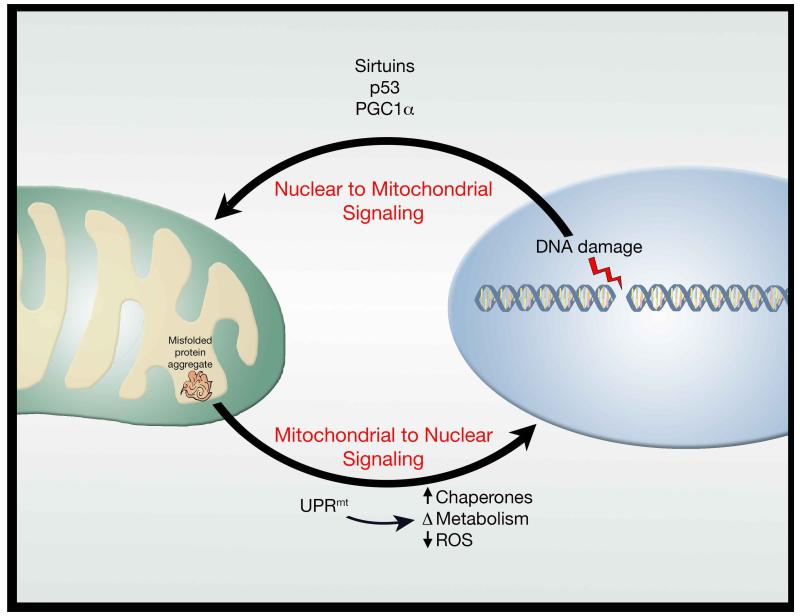
Finally, while as noted above, there is considerable interest in mitochondrial to nucleus signaling, there is also an important role for nuclear to mitochondrial signaling. For instance, telomere dysfunction results in impaired mitochondrial biogenesis through a pathway involving both p53 and PGC-1α (Sahin et al., 2011). Conversely, cells lacking intact nuclear excision DNA repair (xeroderma pigmentosum group A (XPA)) also exhibit mitochondrial dysfunction due to impaired mitophagy, mediated by a decline in NAD+ levels and sirtuin activity (Fang et al., 2014). Interestingly, XPA, and the related Cockayne syndrome, are DNA repair disorders that phenotypically manifest as accelerated aging conditions. Nonetheless, there is increasing evidence that these primary nuclear DNA repair disorders have profound metabolic consequences (Scheibye-Knudsen et al., 2014). Thus, between the nucleus and the mitochondria, signaling occurs in both directions and further dissection of these pathways will likely yield important clues about organismal aging (Figure 2). Furthermore, signaling between the mitochondria and other organelles (e.g. lysosomes) is also emerging as a potential critical determinant of lifespan (Hughes and Gottschling, 2012).
Figure 2.
Bidirectional signaling between the nucleus and mitochondria. Communication exists between the nucleus and the mitochondria with evidence that nuclear stresses, such as DNA damage, trigger a mitochondrial response. Similarly, mitochondrial stresses, such as protein aggregates, stimulate a retrograde response to the nucleus. Both directions of this signaling paradigm appear intimately linked to longevity.
Mitophagy in aging
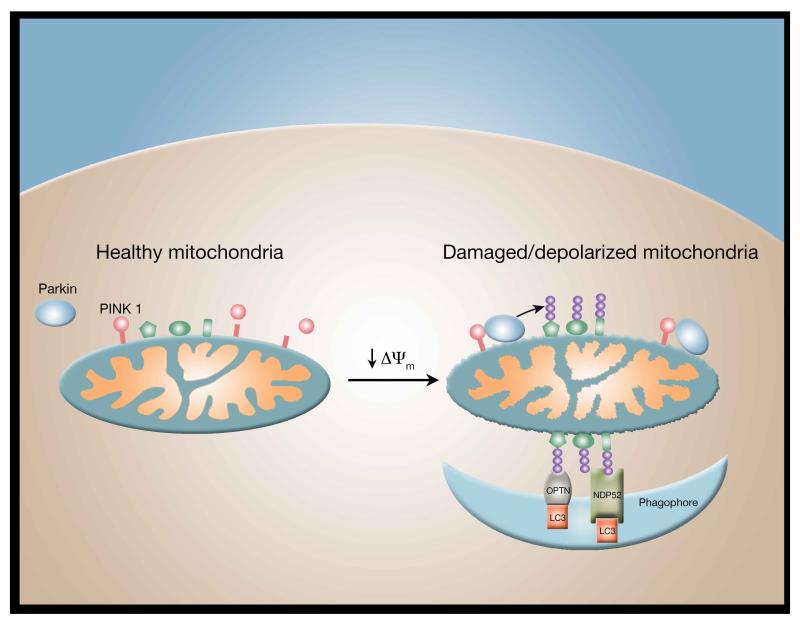
If misfolded proteins stemming from mitochondrial DNA mutations or proteotoxic stress accumulate to a level that exceeds the capacity of the UPRmt, autophagy of mitochondria (mitophagy), or piecemeal autophagy of mitochondrial subdomains, appears to mitigate mitochondrial impairment. The biochemical steps of one mitophagy pathway have been mapped out in some detail (Pickrell and Youle, 2015). In higher eukaryotes, including man and insects, a mitochondrial kinase PINK1 senses damage and signals this to the cytosolic E3 ligase Parkin. Mitochondrial damage caused by misfolded mitochondrial matrix proteins or other stresses that lead to inner mitochondrial membrane depolarization inhibits protein import through the inner mitochondrial membrane. By avoiding mitochondrial import, PINK1 circumvents proteolytic degradation and accumulates on the impaired mitochondria with the kinase domain facing the cytosol (Figure 3). There, it phosphorylates ubiquitin attached to mitochondrial outer membrane proteins. These phospho-ubiquitin chains bind to Parkin recruiting it from the cytosol to the mitochondria and activating its latent E3 ubiquitin ligase activity. Parkin further ubiquitinates mitochondrial outer membrane proteins to recruit receptors such as optineurin and NDP52 that signal autophagosome assembly proximal to individual damaged mitochondria (Itakura et al., 2012; Lazarou et al., 2015). In humans, loss of function mutations in either PINK1 or Parkin lead to early onset Parkinson’s disease, normally a disease associated with aging, suggesting that insufficient mitophagy may directly lead to the loss of dopaminergic neurons that causes the motor phenotype. Interestingly, the effects of Parkin can be reversed by a family of mitochondrial deubiquitinating enzymes (DUBs) including Ub-specific protease 8 (USP8), USP15 and USP30 (Durcan and Fon, 2015). The best evidence to date comes from analyzing USP30 which appears to antagonize Parkin function as evidenced by the fact that genetic inhibition of USP30 rescues Parkin-deficient flies (Bingol et al., 2014). Pharmacological manipulation of mitochondrial ubiquitination would therefore appear to be an attractive therapeutic avenue.
Figure 3.
Parkin-dependent mitophagy. In healthy mitochondria, the PINK1 kinase is constitutively degraded. A fall in mitochondrial membrane potential (Δψm) stabilizes PINK1 facilitating the recruitment of cytosolic Parkin to the mitochondrial outer membrane. Activation of Parkin results in the ubiquitinization (purple balls) of multiple outer mitochondrial membrane proteins (shown in green). Once ubiquinated, these proteins are recognized by specific mitophagy receptors such as optineurin (OPTN) and NDP52, which along with LC3, directs the phagophore to surround the damaged mitochondria allowing for its ultimate delivery to the lysosome for degradation via mitophagy.
In contrast to man, loss of Parkin and PINK1 in mice does not lead to a neuronal phenotype. To assess endogenous Parkin function in mice under mitochondrial stress, POLG mutator mice were crossed into a Parkin null background. Although neither POLG mutator mice nor Parkin null mice display dopaminergic neuron loss, mutator mice in a Parkin null background lost ~40% of substantia nigral dopaminergic neurons by 1 year of age (Pickrell et al., 2015). The Mutator/Parkin null mice also displayed a substantial motor phenotype that was rescued by L-dopa treatment. Thus, endogenous Parkin preserves dopaminergic neurons from death stemming from mitochondrial DNA mutation accumulation. However, Parkin did not rescue the mutator mice from the accumulation of mtDNA mutations suggesting that Parkin compensates for mitochondrial mutation accumulation, perhaps by clearing damaged proteins by autophagy. This model of Parkin removal of damaged proteins is consistent with the finding that ΔOTC, a misfolded matrix protein (deletion mutant of ornithine transcarbamylase) that induces the UPRmt (Zhao et al., 2002), also induces PINK1 accumulation and Parkin translocation to mitochondria without depolarizing mitochondria (Jin and Youle, 2013). Parkin expression diminishes misfolded ΔOTC accumulation suggesting that Parkin may function downstream of mitochondrial DNA mutation accumulation to clear proteotoxic stress during aging. Additional evidence that misfolded, mutated or oxidized proteins can be selectively removed from mitochondria come from studies in Drosophila showing that Parkin functions via the autophagy machinery to eliminate select respiratory chain complex proteins (Vincow et al., 2013). How selective removal of mitochondrial proteins via autophagy occurs is not clear but may involve asymmetric mitochondrial fission (Twig et al., 2008) or mitochondrial derived vesicles (Sugiura et al., 2014). Consistent with the suggestion that mitophagy protects animals from loss of mitochondrial function during aging, mitophagy rates decrease in the dentate gyrus with age and upon human huntingtin overexpression (Sun et al., 2015).
Although loss of Parkin expression has not been reported to exacerbate the aging phenotype of wild type or POLG mutator mice, loss of Parkin expression in Drosophila decreases animal lifespan (Greene et al., 2003) and Parkin overexpression extends fly longevity without impairing fertility or food consumption (Rana et al., 2013). Parkin overexpression also reduces the levels of ubiquitin/protein aggregates that normally accumulate in Drosophila muscle with age. Thus, PINK1/Parkin-mediated mitophagy appears to mitigate deleterious consequences of mitochondria DNA mutation accumulation in mammals and foster longevity in flies.
PINK1/Parkin-independent mitophagy pathways have been also identified. One mitophagy process that occurs during mammal development induces the wholesale elimination of mitochondria from red blood cells. Expression levels of Nix, also called BNIP3L, increase dramatically during reticulocyte development and mice lacking Nix retain mitochondria in mature RBCs (Schweers et al., 2007). Nix is localized on the outer mitochondrial membrane and exposes a domain toward the cytosol that binds to LC3 on autophagosomes and that participates to some extent in autophagic engulfment of mitochondria. Whether Nix functions only constitutively to eliminate mitochondria or is regulated post-translationally to mediate mitophagy remains unclear.
Expression of a predicted Nix homologue, PINK1 and Parkin in C. elegans appears to promote longevity (Palikaras et al., 2015). Although loss of Pink1, PDR-1 (a Parkin orthologue) and DCT-1, an outer mitochondrial membrane protein with domain organization similar to that of NIX, does not affect lifespan in wild type worms, it decreases the lifespan of the long-lived daf-2 mutant animals with a disrupted insulin-like signaling cascade, and the feeding-impaired and thus calorie-restricted eat-2 mutant worms. This suggests that mitophagy is required for multiple distinct pathways that extend lifespan. Interestingly, loss of PINK1 or DCT-1 does decrease lifespan in C. elegans lacking a homologue of NRF2 called SKN-1. This indicates that mitochondrial biogenesis, mediated through SKN-1, compensates for a lack of mitophagy in wild-type nematodes. In mammals, the PINK1-Parkin axis requires SIRT1, the NAD-dependent deacetylase previously linked to aging (Giblin et al., 2014), for full activity. Inhibition of SIRT1 decreases activation of PGC-1α leading to defective PINK1- and Parkin-mediated mitophagy (Fang et al., 2014).
Another link between mitochondrial quality control and lifespan has been observed in Podospora anserine, a well-established fungal model of aging. In this model, increased expression of the mitochondrial matrix AAA+ protease LON can substantially extend lifespan without impairing growth, respiration, or fertility (Luce and Osiewacz, 2009). Both the LON protease, and the ATP-dependent Clp protease (CLPP), are essential for protein homeostasis in the mitochondrial matrix, and deficits in their activity is tightly linked to a decline in mitochondrial function and to aging (Quiros et al., 2015). Interestingly, the LON protease is also known to regulate mitochondrial levels of PINK1 (Thomas et al., 2014), as well as being the dominant protease responsible for initially handling misfolded and aggregated proteins in the mitochondrial matrix (Bezawork-Geleta et al., 2015). The latter stimulus is the classic activator of the UPRmt. Thus, while for clarity we have discussed mitophagy and the mitochondrial unfolded protein response as distinct regulatory pathways, the above observation with LON proteases, as well as other evidence (Jin and Youle, 2013), suggests the existence of substantial cross regulation between these various mitochondrial quality control pathways.
Mitochondria and Inflammation
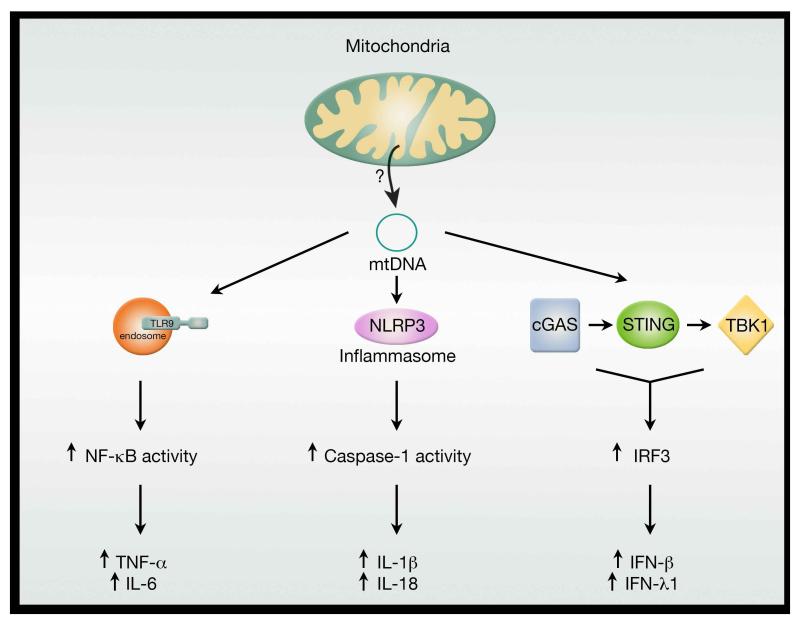
One of the hallmarks of aging is the development of a low-grade, chronic, sterile inflammatory state often deemed ‘inflammaging’. The development of this state, characterized in part by increased circulating inflammatory biomarkers such as IL-6 and C-reactive protein, is a known risk factor for increased morbidity and mortality in the elderly (Franceschi et al., 2000). Increasingly, there is a connection between mitochondrial function and the activation of this enhanced age-dependent immune response. Mechanistically, this connection can perhaps be traced back to the bacterial origins of the present day mitochondria. As opposed to nuclear DNA, mitochondrial DNA (like bacterial DNA) is not methylated. The immune system has adapted to this subtle difference and has evolved strategies to recognize non-methylated DNA, primarily through members of the Toll-like receptors including TLR9. This response presumably allows rapid activation of the immune system in the setting of bacterial infection. Besides releasing non-methylated DNA, damaged mitochondria, like bacteria, can release formyl peptides that can signal through the formyl peptide receptor-1 to trigger an immune response. Both mitochondrial DNA and mitochondrial formylated peptide can be viewed as mitochondrial-derived damage associated molecular patterns (DAMPs) that are known to stimulate the innate immune system. The importance of this mitochondrial-elicited TLR9 response can be seen in a number of important medical inflammatory states including trauma (Zhang et al., 2010) and heart failure (Oka et al., 2012). Mitochondrial DNA can also activate the NLRP3 inflammasome (Nakahira et al., 2011; Shimada et al., 2012). The inflammasome is a large multi-protein complex that controls caspase-1 activation, a step that is required for the subsequent processing and secretion of IL-1β and IL-18. Interestingly, macrophages lacking mitochondrial DNA have severely impaired secretion of IL-1β (Shimada et al., 2012). Moreover, genetic ablation of Nlrp3 resulted in a diminished age-dependent activation of the innate immune system and protected animals from a number of age-related pathologies including bone loss, thymic involution and loss of glycemic control (Youm et al., 2013). It is tempting to speculate that in older tissues, the slow, chronic release of mitochondrial DNA or mitochondrial proteins might contribute to the age-dependent activation of the inflammasome and thereby contribute to the ‘inflammaging’ milieu.
The sensing of free, intracellular mitochondrial DNA is not confined to TLR9 or the inflammasome, as recently, a third pathway involving the adaptor protein STING has been described (Figure 4). In this pathway, the cytosolic sensor cGAS recognizes mitochondrial DNA and through the adaptor protein STING and the kinase TKB1 activates the transcription factor IRF3 to induce production of type 1 interferons (IFN) and IFN-stimulated gene products. A previous link between IFN signaling and mitochondria has been made when it was noted that an important component of retinoic-acid-inducible protein I (RIG-1)-like receptor (RLR) signaling was associated with mitochondria. In particular, the RLR adaptor protein MAVS (mitochondrial antiviral-signaling protein) was found to form a scaffold for signaling on the outer mitochondrial membrane surface (Seth et al., 2005). The role of mitochondrial DNA in activating the STING pathway first came to light in more recent studies involving Bax/Bak mediated apoptosis (Rongvaux et al., 2014; White et al., 2014). In these studies, caspase activation during apoptotic cell death was shown to suppress IFN production by preventing the ability of mitochondrial DNA to activate the cGAS-STING pathway. This caspase-mediated suppression ensures that apoptosis is immunologically silent. These results have been recently extended in a study characterizing the effect of haploinsufficiency of TFAM. Among other things, TFAM regulates mitochondrial nucleoid structure, abundance and segregation. Cells expressing only one allele of TFAM (Tfam+/−) were shown to have approximately 50% less mitochondrial DNA but no resting bioenergetics deficit (West et al., 2015). Interestingly, Tfam+/− mouse embryonic fibroblasts exhibited constitutive activation of the cGAS-STING-IRF3 pathway (West et al., 2015). Moreover, in wild type cells, herpes virus infection appears to trigger mitochondrial stress including reducing TFAM levels, and this mitochondrial stress appears to be required to mount the full antiviral response (West et al., 2015). Again, these results argue for a central role of mitochondria, and particularly released mitochondrial DNA, in regulating the innate immune response. The precise nature however as to how mitochondrial DNA is released under these various conditions has not been well characterized.
Figure 4.
Mitochondria as regulators of the innate immune response. Release of mitochondrial DNA appears to trigger at least three distinct pathways linked to inflammation. The precise mechanism by which free mitochondrial DNA enters the cytosol to engage with various intracellular DNA sensors is currently unclear. Nonetheless, age-dependent breakdown of the mitochondrial membrane might allow escape of mtDNA and thereby help fuel the chronic, sterile inflammation associated with aging.
While the above discussion has focused on seemingly permanent damage to mitochondria resulting in presumably rupture of the inner mitochondrial membrane and the subsequent release of mtDNA, other more reversible form of mitochondrial dysfunction can also trigger an immune response. Those investigators interested in probing mitochondrial function have classically used chemicals such as antimycin and cyanide to block electron transport. The natural sources of these inhibitors are bacteria which use these small molecules to disable their host, allowing for more productive infections. Interestingly, when C. elegans are directly challenged with these mitochondrial inhibitors this transient mitochondrial dysfunction appears to be interpreted as a pathogen attack and is sufficient to activate an innate immune response (Liu et al., 2014). In a related set of observation, infection of worms with the bacteria Pseudomonas aeruginosa was also shown to result in mitochondrial dysfunction and UPRmt activation (Pellegrino et al., 2014). The latter response was shown to be critical for the worm to mount an effective immune response. Both observations suggest that in C. elegans, mitochondrial dysfunction triggers activation of the innate immune response.
Conclusion
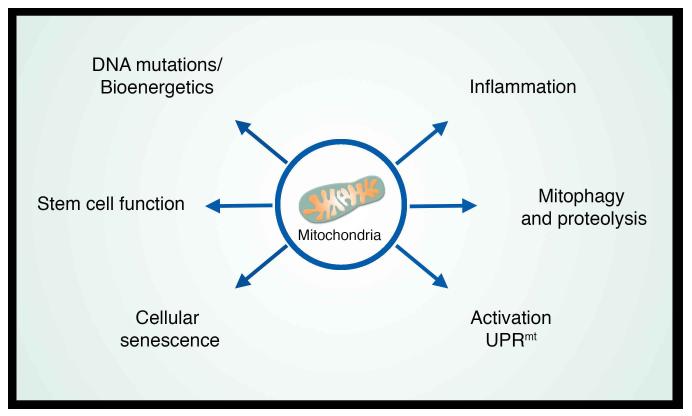
Taken together, these observations suggest that mitochondria can be intimately linked to a wide range of processes associated with aging including senescence, inflammation, as well as the more generalized age-dependent decline in tissue and organ function (Figure 5). What specific perturbations of mitochondrial function are most relevant for the aging process requires additional clarification. As we have discussed, early studies in skeletal muscle concentrated on the accumulation of DNA mutations and the concomitant decline in electron transport function. Recent evidence has implicated triggering of the UPRmt and quality control mechanisms including mitophagy and proteolysis. Other processes not discussed here in-depth include mitochondrial dynamics (Liesa and Shirihai, 2013; Pernas and Scorrano, 2015), as well as the biosynthetic properties of mitochondria. The best evidence for the relevance of the latter property comes from yeast, where mitochondrial regulation of iron-sulfur cluster biogenesis clearly modulates nuclear genomic integrity (Veatch et al., 2009). In many of the early studies, the association between mitochondria and the aging process was mostly correlative. Increasingly, however, causative connections are being established. This suggests that attempts to rejuvenate mitochondrial function or improve mitochondrial quality control might be an effective strategy to combat aging. Towards this goal, there are a number of ongoing efforts to develop small molecules to therapeutically augment mitochondrial biogenesis (Suliman and Piantadosi, 2016). Similarly, raising NAD+ levels in older mice appears to restore mitochondrial function (Gomes et al., 2013). As such, there is considerable enthusiasm to develop methods to increase NAD+ levels, either through direct supplementation, or by altering NAD+ metabolism (Canto et al., 2015). Pharmacologic activation of mitophagy is another approach that might be widely beneficial in patients with age-related neurodegenerative disorders, or to combat aspects of normal aging. With the relatively detailed molecular understanding of PINK1 and Parkin activation, efforts are underway in academia and industry to directly or indirectly modulate the activity of PINK1, (Hertz et al., 2013) Parkin or USP30 (Bingol et al., 2014; Hasson et al., 2015) in order to promote mitophagic flux. As such, the next decade appears to hold considerable promise for developing a wide range of effective mitochondria-targeted therapies. With such agents, clinical trial can ultimately test the very tenable hypothesis that reversing the decline in mitochondrial function will slow, or even reverse, the rate which we age.
Figure 5.
Mitochondria as regulators of organismal aging. The contribution of mitochondria to the aging process occurs through multiple distinct pathways. Although depicted as separate pathways, clear intersections occur as is evident between the connection between activation of the UPRmt and the induction of the inflammatory response (see text for details).
Acknowledgement
We are grateful to members of the Finkel and Youle Labs for helpful comments and for Ilsa Rovira for help with the preparation of the manuscript. This work was supported by NIH Intramural Funds and a Leducq FoundationTransatlantic Network Award.
Footnotes
Author Contributions: N.S., R.J.Y. and T.F. all particpated in the writing of this manuscript.
Competing Financial Interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahlqvist KJ, Hamalainen RH, Yatsuga S, Uutela M, Terzioglu M, Gotz A, Forsstrom S, Salven P, Angers-Loustau A, Kopra OH, et al. Somatic progenitor cell vulnerability to mitochondrial DNA mutagenesis underlies progeroid phenotypes in Polg mutator mice. Cell Metab. 2012;15:100–109. doi: 10.1016/j.cmet.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, Ahmad F, Matsui T, Chin S, Wu PH, et al. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metab. 2005;1:259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Argiles JM, Busquets S, Stemmler B, Lopez-Soriano FJ. Cachexia and sarcopenia: mechanisms and potential targets for intervention. Curr Opin Pharmacol. 2015;22:100–106. doi: 10.1016/j.coph.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Austin S, St-Pierre J. PGC1alpha and mitochondrial metabolism--emerging concepts and relevance in ageing and neurodegenerative disorders. J Cell Sci. 2012;125:4963–4971. doi: 10.1242/jcs.113662. [DOI] [PubMed] [Google Scholar]
- Bennett CF, Vander Wende H, Simko M, Klum S, Barfield S, Choi H, Pineda VV, Kaeberlein M. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nat Commun. 2014;5:3483. doi: 10.1038/ncomms4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezawork-Geleta A, Brodie EJ, Dougan DA, Truscott KN. LON is the master protease that protects against protein aggregation in human mitochondria through direct degradation of misfolded proteins. Sci Rep. 2015;5:17397. doi: 10.1038/srep17397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigarella CL, Liang R, Ghaffari S. Stem cells and the impact of ROS signaling. Development. 2014;141:4206–4218. doi: 10.1242/dev.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, Sheng M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510:370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- Brown K, Xie S, Qiu X, Mohrin M, Shin J, Liu Y, Zhang D, Scadden DT, Chen D. SIRT3 reverses aging-associated degeneration. Cell Rep. 2013;3:319–327. doi: 10.1016/j.celrep.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Menzies KJ, Auwerx J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell metabolism. 2015;22:31–53. doi: 10.1016/j.cmet.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty EF, Hubert HB, Lingala VB, Fries JF. Reduced disability and mortality among aging runners: a 21-year longitudinal study. Arch Intern Med. 2008;168:1638–1646. doi: 10.1001/archinte.168.15.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ML, Logan TD, Hochberg ML, Shelat SG, Yu X, Wilding GE, Tan W, Kujoth GC, Prolla TA, Selak MA, et al. Erythroid dysplasia, megaloblastic anemia, and impaired lymphopoiesis arising from mitochondrial dysfunction. Blood. 2009;114:4045–4053. doi: 10.1182/blood-2008-08-169474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay L, Caudron F, Denoth-Lippuner A, Boettcher B, Buvelot Frei S, Snapp EL, Barral Y. A sphingolipid-dependent diffusion barrier confines ER stress to the yeast mother cell. Elife. 2014;3:e01883. doi: 10.7554/eLife.01883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy MJ, Conboy IM, Rando TA. Heterochronic parabiosis: historical perspective and methodological considerations for studies of aging and longevity. Aging Cell. 2013;12:525–530. doi: 10.1111/acel.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortopassi GA, Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 1990;18:6927–6933. doi: 10.1093/nar/18.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Morvan F, Jourde B, Meier V, Kahle P, Brebbia P, Toussaint G, Glass DJ, Fornaro M. ATP citrate lyase improves mitochondrial function in skeletal muscle. Cell Metab. 2015;21:868–876. doi: 10.1016/j.cmet.2015.05.006. [DOI] [PubMed] [Google Scholar]
- Dell'agnello C, Leo S, Agostino A, Szabadkai G, Tiveron C, Zulian A, Prelle A, Roubertoux P, Rizzuto R, Zeviani M. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum Mol Genet. 2007;16:431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Durcan TM, Fon EA. The three 'P's of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev. 2015;29:989–999. doi: 10.1101/gad.262758.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easlon E, Tsang F, Skinner C, Wang C, Lin SJ. The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev. 2008;22:931–944. doi: 10.1101/gad.1648308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013;17:162–184. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Egerman MA, Glass DJ. Signaling pathways controlling skeletal muscle mass. Crit Rev Biochem Mol Biol. 2014;49:59–68. doi: 10.3109/10409238.2013.857291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Shan C, Kang HB, Elf S, Xie J, Tucker M, Gu TL, Aguiar M, Lonning S, Chen H, et al. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol Cell. 2014;53:534–548. doi: 10.1016/j.molcel.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, Mitchell JR, Croteau DL, Bohr VA. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014;157:882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felkai S, Ewbank JJ, Lemieux J, Labbe JC, Brown GG, Hekimi S. CLK-1 controls respiration, behavior and aging in the nematode Caenorhabditis elegans. Embo J. 1999;18:1783–1792. doi: 10.1093/emboj/18.7.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Bonafe M, Valensin S, Olivieri F, De Luca M, Ottaviani E, De Benedictis G. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- Giblin W, Skinner ME, Lombard DB. Sirtuins: guardians of mammalian healthspan. Trends Genet. 2014;30:271–286. doi: 10.1016/j.tig.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodpaster BH, Park SW, Harris TB, Kritchevsky SB, Nevitt M, Schwartz AV, Simonsick EM, Tylavsky FA, Visser M, Newman AB. The loss of skeletal muscle strength, mass, and quality in older adults: the health, aging and body composition study. J Gerontol A Biol Sci Med Sci. 2006;61:1059–1064. doi: 10.1093/gerona/61.10.1059. [DOI] [PubMed] [Google Scholar]
- Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamalainen RH, Ahlqvist KJ, Ellonen P, Lepisto M, Logan A, Otonkoski T, Murphy MP, Suomalainen A. mtDNA Mutagenesis Disrupts Pluripotent Stem Cell Function by Altering Redox Signaling. Cell Rep. 2015;11:1614–1624. doi: 10.1016/j.celrep.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Spiegelman BM. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature. 2008;454:463–469. doi: 10.1038/nature07206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Hasson SA, Fogel AI, Wang C, MacArthur R, Guha R, Heman-Ackah S, Martin S, Youle RJ, Inglese J. Chemogenomic profiling of endogenous PARK2 expression using a genome-edited coincidence reporter. ACS Chem Biol. 2015;10:1188–1197. doi: 10.1021/cb5010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, Yang Y, Blais SP, Neubert TA, Ron D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol Cell. 2010;37:529–540. doi: 10.1016/j.molcel.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepple RT. Mitochondrial involvement and impact in aging skeletal muscle. Front Aging Neurosci. 2014;6:211. doi: 10.3389/fnagi.2014.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz NT, Berthet A, Sos ML, Thorn KS, Burlingame AL, Nakamura K, Shokat KM. A neo-substrate that amplifies catalytic activity of parkinson's-disease-related kinase PINK1. Cell. 2013;154:737–747. doi: 10.1016/j.cell.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Gottschling DE. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature. 2012;492:261–265. doi: 10.1038/nature11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes BG, Hekimi S. A mild impairment of mitochondrial electron transport has sex-specific effects on lifespan and aging in mice. PLoS One. 2011;6:e26116. doi: 10.1371/journal.pone.0026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KA, Reynolds RM. Evolutionary and mechanistic theories of aging. Annu Rev Entomol. 2005;50:421–445. doi: 10.1146/annurev.ento.50.071803.130409. [DOI] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Koyama-Honda I, Mizushima N. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. Journal of cell science. 2012;125:1488–1499. doi: 10.1242/jcs.094110. [DOI] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- Jensen MB, Jasper H. Mitochondrial proteostasis in the control of aging and longevity. Cell Metab. 2014;20:214–225. doi: 10.1016/j.cmet.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Du W, Mancuso A, Wellen KE, Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493:689–693. doi: 10.1038/nature11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9:1750–1757. doi: 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG., Jr. Cancer and altered metabolism: potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb Symp Quant Biol. 2011;76:335–345. doi: 10.1101/sqb.2011.76.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, van der Burg SH, Verdegaal EM, Cascante M, Shlomi T, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498:109–112. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- Katajisto P, Dohla J, Chaffer CL, Pentinmikko N, Marjanovic N, Iqbal S, Zoncu R, Chen W, Weinberg RA, Sabatini DM. Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science. 2015;348:340–343. doi: 10.1126/science.1260384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrapko K, Vijg J. Mitochondrial DNA mutations and aging: devils in the details? Trends Genet. 2009;25:91–98. doi: 10.1016/j.tig.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmadakis GC, Bevington A, Smith AC, Clapp EL, Viana JL, Bishop NC, Feehally J. Physical exercise in patients with severe kidney disease. Nephron Clin Pract. 2010;115:c7–c16. doi: 10.1159/000286344. [DOI] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AC, Fenster BE, Ito H, Takeda K, Bae NS, Hirai T, Yu ZX, Ferrans VJ, Howard BH, Finkel T. Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species. J Biol Chem. 1999;274:7936–7940. doi: 10.1074/jbc.274.12.7936. [DOI] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell metabolism. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnane AW, Marzuki S, Ozawa T, Tanaka M. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;1:642–645. doi: 10.1016/s0140-6736(89)92145-4. [DOI] [PubMed] [Google Scholar]
- Liu J, Cao L, Chen J, Song S, Lee IH, Quijano C, Liu H, Keyvanfar K, Chen H, Cao LY, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459:387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Samuel BS, Breen PC, Ruvkun G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 2014;508:406–410. doi: 10.1038/nature13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luce K, Osiewacz HD. Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nature cell biology. 2009;11:852–858. doi: 10.1038/ncb1893. [DOI] [PubMed] [Google Scholar]
- Macip S, Igarashi M, Fang L, Chen A, Pan ZQ, Lee SW, Aaronson SA. Inhibition of p21-mediated ROS accumulation can rescue p21-induced senescence. Embo J. 2002;21:2180–2188. doi: 10.1093/emboj/21.9.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinus RD, Garth GP, Webster TL, Cartwright P, Naylor DJ, Hoj PB, Hoogenraad NJ. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur J Biochem. 1996;240:98–103. doi: 10.1111/j.1432-1033.1996.0098h.x. [DOI] [PubMed] [Google Scholar]
- Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, Kang Y, Shenk T, Cristea IM. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159:1615–1625. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick MA, Delaney JR, Tsuchiya M, Tsuchiyama S, Shemorry A, Sim S, Chou AC, Ahmed U, Carr D, Murakami CJ, et al. A Comprehensive Analysis of Replicative Lifespan in 4,698 Single-Gene Deletion Strains Uncovers Conserved Mechanisms of Aging. Cell Metab. 2015;22:895–906. doi: 10.1016/j.cmet.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFaline-Figueroa JR, Vevea J, Swayne TC, Zhou C, Liu C, Leung G, Boldogh IR, Pon LA. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell. 2011;10:885–895. doi: 10.1111/j.1474-9726.2011.00731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, Haynes CM, Chen D. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science. 2015;347:1374–1377. doi: 10.1126/science.aaa2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan RM, Barnes RG, Fisher K, Andreou T, Rooney N, Poulin GB, Whitmarsh AJ. A nuclear role for the respiratory enzyme CLK-1 in regulating mitochondrial stress responses and longevity. Nat Cell Biol. 2015;17:782–792. doi: 10.1038/ncb3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt) Mol Cell. 2015;58:123–133. doi: 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norddahl GL, Pronk CJ, Wahlestedt M, Sten G, Nygren JM, Ugale A, Sigvardsson M, Bryder D. Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. Cell Stem Cell. 2011;8:499–510. doi: 10.1016/j.stem.2011.03.009. [DOI] [PubMed] [Google Scholar]
- O'Connor KG, Tobin JD, Harman SM, Plato CC, Roy TA, Sherman SS, Blackman MR. Serum levels of insulin-like growth factor-I are related to age and not to body composition in healthy women and men. J Gerontol A Biol Sci Med Sci. 1998;53:M176–182. doi: 10.1093/gerona/53a.3.m176. [DOI] [PubMed] [Google Scholar]
- Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer L, Fuehr K. Low oxygen concentration extends the lifespan of cultured human diploid cells. Nature. 1977;267:423–425. doi: 10.1038/267423a0. [DOI] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 2015;521:525–528. doi: 10.1038/nature14300. [DOI] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos JF, Nelson G, Wang C, Richter T, Simillion C, Proctor CJ, Miwa S, Olijslagers S, Hallinan J, Wipat A, et al. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol Syst Biol. 2010;6:347. doi: 10.1038/msb.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM, Kirienko NV, Gillis R, Fiorese CJ, Haynes CM. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature. 2014;516:414–417. doi: 10.1038/nature13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernas L, Scorrano L. Mito-Morphosis: Mitochondrial Fusion, Fission, and Cristae Remodeling as Key Mediators of Cellular Function. Annual review of physiology. 2015 doi: 10.1146/annurev-physiol-021115-105011. [DOI] [PubMed] [Google Scholar]
- Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW, Shulman GI. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300:1140–1142. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell AM, Huang CH, Kennedy SR, Ordureau A, Sideris DP, Hoekstra JG, Harper JW, Youle RJ. Endogenous Parkin Preserves Dopaminergic Substantia Nigral Neurons following Mitochondrial DNA Mutagenic Stress. Neuron. 2015;87:371–381. doi: 10.1016/j.neuron.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piko L, Hougham AJ, Bulpitt KJ. Studies of sequence heterogeneity of mitochondrial DNA from rat and mouse tissues: evidence for an increased frequency of deletions/additions with aging. Mech Ageing Dev. 1988;43:279–293. doi: 10.1016/0047-6374(88)90037-1. [DOI] [PubMed] [Google Scholar]
- Pulliam DA, Deepa SS, Liu Y, Hill S, Lin AL, Bhattacharya A, Shi Y, Sloane L, Viscomi C, Zeviani M, et al. Complex IV-deficient Surf1(−/−) mice initiate mitochondrial stress responses. Biochem J. 2014;462:359–371. doi: 10.1042/BJ20140291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijano C, Cao L, Fergusson MM, Romero H, Liu J, Gutkind S, Rovira II, Mohney RP, Karoly ED, Finkel T. Oncogene-induced senescence results in marked metabolic and bioenergetic alterations. Cell Cycle. 2012;11:1383–1392. doi: 10.4161/cc.19800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros PM, Langer T, Lopez-Otin C. New roles for mitochondrial proteases in health, ageing and disease. Nat Rev Mol Cell Biol. 2015;16:345–359. doi: 10.1038/nrm3984. [DOI] [PubMed] [Google Scholar]
- Rana A, Rera M, Walker DW. Parkin overexpression during aging reduces proteotoxicity, alters mitochondrial dynamics, and extends lifespan. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8638–8643. doi: 10.1073/pnas.1216197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Zarse K, Oberbach A, Kloting N, Birringer M, Kiehntopf M, Stumvoll M, Kahn CR, Bluher M. Antioxidants prevent health-promoting effects of physical exercise in humans. Proc Natl Acad Sci U S A. 2009;106:8665–8670. doi: 10.1073/pnas.0903485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockstein M, Brandt KF. Enzyme changes in flight muscle correlated with aging and flight ability in the male housefly. Science. 1963;139:1049–1051. doi: 10.1126/science.139.3559.1049. [DOI] [PubMed] [Google Scholar]
- Rongvaux A, Jackson R, Harman CC, Li T, West AP, de Zoete MR, Wu Y, Yordy B, Lakhani SA, Kuan CY, et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159:1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP, Letellier T. Mitochondrial threshold effects. Biochem J. 2003;370:751–762. doi: 10.1042/BJ20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe GC, Safdar A, Arany Z. Running forward: new frontiers in endurance exercise biology. Circulation. 2014;129:798–810. doi: 10.1161/CIRCULATIONAHA.113.001590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruas JL, White JP, Rao RR, Kleiner S, Brannan KT, Harrison BC, Greene NP, Wu J, Estall JL, Irving BA, et al. A PGC-1alpha isoform induced by resistance training regulates skeletal muscle hypertrophy. Cell. 2012;151:1319–1331. doi: 10.1016/j.cell.2012.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safdar A, Bourgeois JM, Ogborn DI, Little JP, Hettinga BP, Akhtar M, Thompson JE, Melov S, Mocellin NJ, Kujoth GC, et al. Endurance exercise rescues progeroid aging and induces systemic mitochondrial rejuvenation in mtDNA mutator mice. Proc Natl Acad Sci U S A. 2011;108:4135–4140. doi: 10.1073/pnas.1019581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz A, Fernandez-Ayala DJ, Stefanatos RK, Jacobs HT. Mitochondrial ROS production correlates with, but does not directly regulate lifespan in Drosophila. Aging (Albany NY) 2010;2:200–223. doi: 10.18632/aging.100137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Mitchell SJ, Fang EF, Iyama T, Ward T, Wang J, Dunn CA, Singh N, Veith S, Hasan-Olive MM, et al. A high-fat diet and NAD(+) activate Sirt1 to rescue premature aging in cockayne syndrome. Cell metabolism. 2014;20:840–855. doi: 10.1016/j.cmet.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. [DOI] [PubMed] [Google Scholar]
- Schulz AM, Haynes CM. UPR(mt)-mediated cytoprotection and organismal aging. Biochim Biophys Acta. 2015;1847:1448–1456. doi: 10.1016/j.bbabio.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, Ramanujan VK, Wolf AJ, Vergnes L, Ojcius DM, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soong NW, Hinton DR, Cortopassi G, Arnheim N. Mosaicism for a specific somatic mitochondrial DNA mutation in adult human brain. Nat Genet. 1992;2:318–323. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- Stein LR, Imai S. Specific ablation of Nampt in adult neural stem cells recapitulates their functional defects during aging. Embo J. 2014;33:1321–1340. doi: 10.1002/embj.201386917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner JL, Murphy EA, McClellan JL, Carmichael MD, Davis JM. Exercise training increases mitochondrial biogenesis in the brain. J Appl Physiol (1985) 2011;111:1066–1071. doi: 10.1152/japplphysiol.00343.2011. [DOI] [PubMed] [Google Scholar]
- Stessman J, Hammerman-Rozenberg R, Cohen A, Ein-Mor E, Jacobs JM. Physical activity, function, and longevity among the very old. Arch Intern Med. 2009;169:1476–1483. doi: 10.1001/archinternmed.2009.248. [DOI] [PubMed] [Google Scholar]
- Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9:298–310. doi: 10.1016/j.stem.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Sugiura A, McLelland GL, Fon EA, McBride HM. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. Embo J. 2014;33:2142–2156. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suliman HB, Piantadosi CA. Mitochondrial Quality Control as a Therapeutic Target. Pharmacol Rev. 2016;68:20–48. doi: 10.1124/pr.115.011502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, Holmstrom KM, Fergusson MM, Yoo YH, Combs CA, et al. Measuring In Vivo Mitophagy. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RE, Andrews LA, Burman JL, Lin WY, Pallanck LJ. PINK1-Parkin pathway activity is regulated by degradation of PINK1 in the mitochondrial matrix. PLoS Genet. 2014;10:e1004279. doi: 10.1371/journal.pgen.1004279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ, Pallanck LJ. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6400–6405. doi: 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen XJ. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature. 2015;524:481–484. doi: 10.1038/nature14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci U S A. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, van Delft MF, Bedoui S, Lessene G, Ritchie ME, et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams George C. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- Williams SL, Huang J, Edwards YJ, Ulloa RH, Dillon LM, Prolla TA, Vance JM, Moraes CT, Zuchner S. The mtDNA mutation spectrum of the progeroid Polg mutator mouse includes abundant control region multimers. Cell metabolism. 2010;12:675–682. doi: 10.1016/j.cmet.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis BL, Gao A, Leonard D, Defina LF, Berry JD. Midlife fitness and the development of chronic conditions in later life. Arch Intern Med. 2012;172:1333–1340. doi: 10.1001/archinternmed.2012.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L, Topf U, Bragoszewski P, Wiese S, Sztolsztener ME, Oeljeklaus S, Varabyova A, Lirski M, Chroscicki P, Mroczek S, et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature. 2015;524:485–488. doi: 10.1038/nature14951. [DOI] [PubMed] [Google Scholar]
- Wu CY, Chen YF, Wang CH, Kao CH, Zhuang HW, Chen CC, Chen LK, Kirby R, Wei YH, Tsai SF, et al. A persistent level of Cisd2 extends healthy lifespan and delays aging in mice. Hum Mol Genet. 2012;21:3956–3968. doi: 10.1093/hmg/dds210. [DOI] [PubMed] [Google Scholar]
- Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- Wu Y, Williams EG, Dubuis S, Mottis A, Jovaisaite V, Houten SM, Argmann CA, Faridi P, Wolski W, Kutalik Z, et al. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158:1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Hekimi S. A mitochondrial superoxide signal triggers increased longevity in Caenorhabditis elegans. PLoS Biol. 2010;8:e1000556. doi: 10.1371/journal.pbio.1000556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Li J, Hekimi S. A Measurable increase in oxidative damage due to reduction in superoxide detoxification fails to shorten the life span of long-lived mitochondrial mutants of Caenorhabditis elegans. Genetics. 2007;177:2063–2074. doi: 10.1534/genetics.107.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee C, Yang W, Hekimi S. The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans. Cell. 2014;157:897–909. doi: 10.1016/j.cell.2014.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youm YH, Grant RW, McCabe LR, Albarado DC, Nguyen KY, Ravussin A, Pistell P, Newman S, Carter R, Laque A, et al. Canonical Nlrp3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab. 2013;18:519–532. doi: 10.1016/j.cmet.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR, Ristow M. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 2012;15:451–465. doi: 10.1016/j.cmet.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 2010;464:104–107. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. Embo J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]