Abstract
The enthusiasm for phenotypic screening as an approach for small-molecule discovery has increased dramatically over the last several years. The recent increase in phenotype-based discoveries is in part due to advancements in phenotypic readouts in improved disease models that recapitulate clinically relevant biology in cell culture. Of course, a major historical barrier to using phenotypic assays in chemical biology has been the challenge in determining the mechanism of action (MoA) for compounds of interest. With the combination of medically inspired phenotypic screening and the development of modern MoA methods, we can now start implementing this approach in chemical probe and drug discovery. In this Perspective, we highlight recent advances in phenotypic readouts and MoA determination by discussing several case studies in which both activities were required for understanding the chemical biology involved and, in some cases, advancing toward clinical development.
A resurgence in phenotypic screening
Many drug-discovery efforts in the past several decades have focused on target-based discoveries, in which disease modeling and pathway analysis generate candidate proteins, generally leading to high-throughput biochemical screening. A less-biased approach, however, has the advantage of allowing the cell (or sometimes the organism) to reveal a target necessary to achieve a desired phenotype (Eggert, 2013). It also addresses the possibility that we may not know as much about a disease pathway as we think. An important analysis of the means by which drugs were discovered between 1999 and 2008 was recently published (Swinney and Anthony, 2011). Of 50 first-in-class new molecular entities (NMEs) approved clinically, 28 (56%) were phenotype-based discoveries, while 17 (34%) were target-based. The cancer field has had more success in targeted approaches for drug discovery, perhaps due to the success of kinase inhibition (Moffat et al., 2014), but it seems likely that cancer drug discovery will ultimately witness the same trends, with phenotype-based screening and discovery playing a more major role.
Advancement in the sophistication of phenotypic assays is important to success in small-molecule discovery. Recently, development of a phenotypic “rule of 3”, similar to Lipinski's “rule of 5”, was proposed (Vincent et al., 2015). These rules involve developing highly disease-relevant assay systems, maintaining the disease relevance of cell stimuli, and assay readouts that are close to the clinically desired outcome. Such reports reveal the increasing intellectual commitment to the importance of phenotypic screening, and determining how best to improve the medical applicability of these efforts.
Mechanism-of-action (MoA) studies are a critical complement to phenotypic screening. Importantly, MoA can be developed at the level of the molecule (e.g., target identification, or the cellular entity that the compound binds), at the level of the biochemical pathway, at the level of the cell, or even at the level of the whole organism (Swinney, 2013). Perhaps most attractive to the chemical biology community are compounds that have a novel (previously unknown) mechanism of action (nMoA). Rather than discovering yet another microtubule inhibitor, for example, expanding our chemical repertoire to new cellular components and targets with nMoA compounds will enhance our understanding of human disease and our ability to modulate cellular processes. We have organized this Perspective by the MoA method used in each case: affinity chromatography, gene-expression analyses, genetic modifier screening, resistance mutation selection, and computational approaches (see Table). We hope that each of the methods described here will help embolden the scientific community to apply phenotypic approaches to small-molecule discovery.
Table
| Mechanism-of-action method Process | Strengths | |
|---|---|---|
| Affinity-based | Western blotting SILAC LC/MS |
identifies direct targets |
| Gene expression-based | array-based profiling RNA-Seq reporter-gene assays |
can uncover dependencies identifies modulated pathways |
| Genetic modifier screening | shRNA CRISPR ORFs |
loss of target can be identified enables chemical genetic epistasis |
| Resistance selection | low dose + sequencing | identifies bypass mechanisms largely used in infectious disease best for death phenotype |
| Computational approaches | profiling-based methods inferential approaches | hypothesis generation by small-molecule similarity |
Affinity-based methods to determine MoA
A small-molecule inducer of chondrocyte differentiation
In an effort to identify novel therapeutic candidates for osteoarthritis, Johnson et al. (2012) developed an image-based assay to identify small molecules that induce chondrocyte differentiation (Johnson et al., 2012). Treatments for osteoarthritis are limited to pain management and surgical intervention, so new methods for stimulating the production of cartilage in affected joints would have the potential to provide, for the first time, a disease-modifying strategy. Since mesenchymal stem cells (MSCs) in the bone marrow can differentiate into bone, fat, and cartilage, the team used primary human bone marrow MSCs, isolated by virtue of having the right cell-surface marker profile, and rhodamine B, which can highlight cells that contain cartilage-specific components, such as proteoglycans and type II collagen. The authors then screened over 20,000 heterocyclic compounds for differentiation, and identified kartogenin (KGN) as a top-ranked hit (Figure 1A).
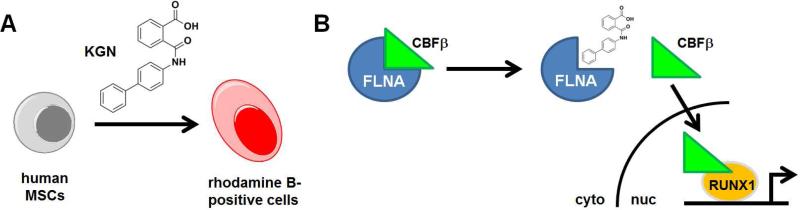
Figure 1. Discovery and MoA of kartogenin (KGN).
(A) Schematic of the phenotypic assay for rhodamine-positive chondrocyte-like cells derived from human MSCs. (B) Affinity-based pull-down experiments revealed filamin A (FLNA) as a cellular binder of KGN. KGN disrupts the cytoplasmic interaction of FLNA and CBFβ, enabling CBFβ nuclear translocation and RUNX1-dependent transcription of cartilage-specific genes.
Follow-up analysis of KGN revealed a potent compound (EC50 100 nM) that induced multiple chondrocyte markers, including SOX9, aggrecan, and lubricin, as measured by mRNA and protein expression. Upon treatment of a three-dimensional culture of human MSCs with KGN for 21 days, cells retain a chondrocyte-cell state, without evidence of matrix breakdown by matrix metalloproteinases. These results led to the study of KGN under pathophysiological conditions; the compound was able to suppress the release of nitric oxide and glycosaminoglycans from primary bovine chondrocytes treated with TNFα and oncostatin M, which mimics cartilage damage in vitro. Two mouse models of cartilage damage, representing chronic destruction (collagenase VII-induced) or acute injury (surgical ligament transection) showed that KGN has reasonable in vivo efficacy as well. Weekly intra-articular injection of KGN reduced inflammation and pain, and induced cartilage regeneration over 1-2 months of treatment.
These data demonstrate that a well-designed phenotypic screen can result in efficacious compounds with eventual clinical potential. The next step in such a process is to uncover the MoA by which KGN induces chondrocyte differentiation. Here, the authors synthesized an analog of KGN with similar activity, conjugated to biotin and with a phenyl azide moiety for photo-crosslinking. After small-molecule treatment of cells and ultraviolet exposure, cell lysates were evaluated by Western blot for biotin. The authors identified two bands, at 90- and 280-kDa, both of which decreased in intensity after treatment with 100 μM free KGN. Remarkably, both bands corresponded to filamin A (FLNA), a 280-kDa protein that can be cleaved in certain cell types to a 90-kDa form. Knock-down of FLNA by shRNA in human MSCs increased chondrocyte formation, recapitulating the effect of KGN in cells. Filamin A is an actin-binding protein, but KGN had no effect on G-actin or F-actin, suggesting that the compound binds distally to the actin-binding site; protein truncations revealed that only the distal FC-1 end of the protein could bind KGN. Fortuitously, it was already known that FC-1 is bound by the core-binding factor beta subunit (CBFβ), and pull-down experiments in MSCs showed that KGN disrupts the interaction between filamin A and CBFβ.
CBFβ is a regulatory subunit that binds to the runt-related transcription factor members (RUNX). In the cytoplasm, CBFβ remains bound to FLNA; upon activation, it dissociates and translocates to nucleus, where it binds RUNX members and controls transcription (Figure 1B). Accordingly, CBFβ was found in the nucleus upon KGM treatment, while shRNA of CBFβ reduced the effects of KGM on chondrocyte differentiation. Gene-expression profiling of KGN-treated human MSCs resulted in only 39 genes changing 1.5-fold after six hours; five of those genes were involved in RUNX transcriptional pathways. Based on cell-specific expression, the authors speculated that RUNX1 is likely the family member most responsible for activity, and shRNA of RUNX1 caused a loss of KGN differentiation activity.
This story provides an important lesson in the value of an inspired phenotypic screen coupled with modern methods of MoA determination. The use of primary human cells to perform high-throughput screening is important to identifying small molecules with the potential for human medical relevance. To the authors’ credit, the report is careful to call this approach an image-based high-throughput screen, and not a high-content screen. Although the terms are frequently used interchangeably, the majority of image-based screens still use single-point readouts, similar to more conventional plate reader-based assays. Further, the use of photo-crosslinking and protein analysis revealed that KGN disrupts a key protein–protein interaction (PPI); such PPI inhibitors have long been sought after in numerous indications (Laraia et al., 2015). In this case, the use of Western blotting to separate and identify binders was quite fortuitous. More typically, mass spectrometry has been successful in identifying binders of immobilized compounds (Larrieu et al., 2014). In the few years since this paper was published, other groups have been using KGN to evaluate the induction of cartilage tissue in tendon-bone junction injury (Zhang and Wang, 2014), limb and joint development in mice (Decker et al., 2014), and microfracture-induced repair of cartilage defects in rabbits (Xu et al., 2015), supporting the conclusions derived in the original studies.
Gene-expression profiling to uncover MoA
A small-molecule activator of stem cell proliferation
In addition to differentiating stem cells, there is also a need to expand and keep them in a multipotent state. For example, in autologous and allogeneic bone-marrow transplant in cancer, a limitation in the number of hematopoietic stem cells (HSCs) available for transplant prevents taking full advantage of this strategy to treat certain cancers. Current methods to expand HSCs using a cytokine cocktail induce proliferation, but the cells then differentiate and lose their activity, accompanied by a loss of CD34 and CD133 expression. To identify small molecules capable of expanding HSCs, Boitano et al. (2010) measured CD34 and CD133 expression in primary human CD34+ cells isolated from human blood (Boitano et al., 2010). After 5-day treatment with a collection of ~100,000 heterocycles, CD34 and CD133 expression were evaluated using confocal microscopy. StemRegenin 1 (SR1) emerged as a top screening hit, with an EC50 ~120 nM. SR1 induces massive expansion of HSCs over three weeks in culture, with over 1000-fold increase in CD34+ cells over the input material. HSCs treated with SR1 were able to engraft over long-term conditions in NSG mice, showing the in vivo relevance of the compound. Further characterization of SR1 revealed some fascinating properties of the compound. First, SR1 has no effect in the absence of the cytokine cocktail used in culture, suggesting an enhancement of stimulation. Second, HSCs could differentiate when SR1 was removed, indicating a reversible effect that may make the compound more suitable in a clinical setting. Third, SR1 does not affect the rate of cell division, suggesting a direct mechanism outside of the cell cycle. Finally, and most intriguingly, SR1 had no effect on murine HSCs, but could expand human, monkey, and dog HSCs, underscoring the importance of using primary human culture in this case.
In this case, the authors relied on gene-expression analysis of compound-treated cells to uncover MoA for SR1. Comparison between SR1 and LGC006, a much less potent analog, revealed two genes repressed by SR1, but not by LGC006: cytochrome P450 1B1 (CYP1B1) and the aryl hydrocarbon receptor repressor (AHRR). Remarkably, both genes are transcriptionally regulated by the aryl hydrocarbon receptor (AHR), suggesting that SR1 could be an AHR antagonist. Accordingly, the AHR agonist dioxin upregulated CYP1B1 expression, while SR1 dose-dependently downregulated this transcript. Further, two other known AHR antagonists also expanded CD34+ cells. In keeping with the phenotypic observations, SR1 had no effect on either rat or mouse AHR, underscoring the remarkable species specificity of SR1 for HSC expansion.
To test SR1 binding, a photoaffinity ligand (PAL) was used to show binding to human AHR isolated from hAHR transgenic mice. SR1 inhibited PAL binding with an IC50 of 40nM, while the less active analog LGC006 was much less effective in this assay. Knock-down of AHR by shRNA resulted in sustained CD34 expression throughout culture, in a similar manner to SR1 treatment. Finally, a constitutively active AHR construct that lacks the ligand-binding domain was used to show that SR1 could not inhibit AHR-dependent luciferase activity in this context, further cementing AHR antagonism as the MoA for SR1-induced expansion of human HSCs.
A small-molecule activator of fatty acid desaturase-2
The case of SR1 provides further support for using gene-expression profiling to develop a model of the MoA for a compound discovered by phenotypic screening. Other inference-based methods, such as those using the NCI60 cancer cell line (CCL) collection (Weinstein et al., 1997) and the Connectivity Map (Lamb et al., 2006), also rely upon cellular perturbation to induce gene-expression profiles that can be compared. Recently, however, an analysis of the basal gene expression in hundreds of CCLs proved a useful method to illuminate the MoA for a compound with unknown activity (Rees et al., 2015). Here, the phenotypic assay involved profiling 842 CCLs against 481 small molecules of interest to cancer therapy. Correlating small-molecule sensitivity with basal levels of transcripts in CCLs provided numerous examples where the MoA for particular compounds could be inferred, and the pattern of sensitivity made sense in the context of the uncovered MoA. CCL sensitivity to ML239, a small molecule first identified in a phenotypic screen for compounds that selectively kill a model of breast cancer stem cells (Germain et al., 2012), was highly correlated with high expression of FADS2, encoding delta(6) fatty-acid desaturase. Both co-treatment with a small-molecule FADS2 inhibitor and FADS2 knock-down reduced the toxicity of ML239 in a large-cell lung carcinoma cell line, indicating that ML239 actually activates this enzyme. FADS2 is a key enzyme in polyunsaturated fatty acid (PUFA) metabolism, and the accumulation of polyunsaturated plasmalogens in ML239-treated cells suggested that increased oxidative lipid stress in high FADS2-expressing cells may render them more sensitive to the effects of ML239. This unexpected finding helps to emphasize the importance of phenotypic approaches to identifying novel MoA; by letting the cell reveal relevant MoAs necessary to arrive at phenotypes of interest, we remove bias introduced by selecting candidate targets based on literature. Further pathway analysis and structure-based studies are necessary to gain a more precise understanding of the MoA of ML239.
Genetic modifiers of small-molecule activity to reveal MoA
A small-molecule inhibitor of cell stress responses
Screening for genes whose knock-down or overexpression modulates small-molecule activity is an attractive approach to determining MoA (Mohr et al., 2014; Palchaudhuri and Hergenrother, 2011). A good example of this tactic involves a compound that inhibits the integrated stress response (ISR) in cells. The ISR is activated by eIF2α phosphorylation, which regulates rate of translation initiation; four kinases in turn control eIF2α (PERK during the unfolded protein response, GCN2 in response to starvation, PKR in response to viral infection, and HRI in response to heme deficiency. Sidrauski et al. (2013) sought to identify inhibitors of PERK signaling through a cell-based assay in which the 5’ untranslated region of ATF4 was fused to luciferase in human 293T cells (Sidrauski et al., 2013). Unlike most reporter genes, this system allows for monitoring activity in live cells, and the researchers screened over 100,000 compounds for suppressors of thapsigargin-induced stress. Through an iterative process of secondary assays, orthogonal readouts, and tertiary analysis by Western blot, 28 compounds were identified to pass every filter. ISRIB, a symmetric bisglycolamide, was the most potent compound (IC50 40 nM; Figure 2A). Mice in which eIF2α cannot be phosphorylated have improved memory, so the authors tested ISRIB in a variety of mouse models of memory. To the authors’ credit, they first measured the pharmacokinetic properties of ISRIB before testing in animals, a step that is often not routinely observed in literature reports. In each case, intraperitoneal injection of ISRIB improved hippocampal learning, hippocampal contextual fear conditioning, and amygdala-based fear conditioning. Thus, this phenotypic approach arrived at a small molecule with interesting and potentially clinically applicable effects in the brain.
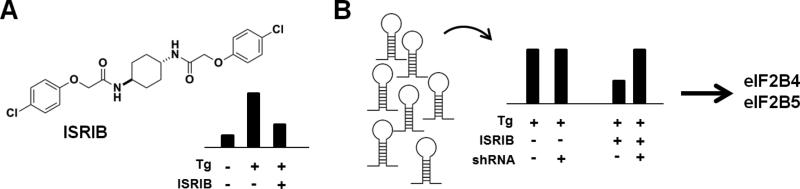
Figure 2. Discovery and MoA of ISRIB.
(A) An ATF4-based luciferase assay identified ISRIB as a small-molecule suppressor of thapsigargin (Tg). (B) An shRNA library was used to screen cells for genes whose knock-down selectively abolished ISRIB suppression. eIF2B4 and eIF2B5 were identified as key genes and followed up for MoA (see main text).
Surprisingly, ISRIB had effects on neither PERK phosphorylation nor eIF2α phosphorylation, suggesting that ISRIB affects signaling events downstream of these proteins. eIF2α binds GTP, and its guanine exchange factor (GEF) is eIF2B; the existence of yeast mutations in eIF2B that, like ISRIB, render cells resistant to eIF2α phosphorylation led the authors to hypothesize eIF2B may be a target of ISRIB. To identify the precise genes involved, a genetic screen was performed for knock-downs that modulate sensitivity to ISRIB (Sidrauski et al., 2015), using an array-based shRNA library targeting 2933 genes involved in proteostasis. Such a targeted approach is appropriate when a solid hypothesis has been formulated. Using a K562 cell line containing an ATF4-venus reporter, whose fluorescence increases in response to eIF2α phosphorylation, the authors induced ER stress with thapsigargin in the presence or absence of ISRIB, and FACS-sorted cells in the top and bottom third of the population. Focusing on hairpins that decreased fluorescence only in the presence of ISRIB, they determined that knock-down of eIF2B4 and eIF2B5 (two subunits of the eIF2B complex) significantly reduced ISRIB sensitivity, suggesting that ISRIB activates the complex (Figure 2B).
Structure-activity relationship (SAR) analysis suggested that, as a compound with twofold rotational symmetry, ISRIB may bind and stabilize eIF2B as a dimer. Sucrose gradient sedimentation and mass spectrometry analyses, aided by the very valuable tool of an inactive analog, indicated that the entire complex was dimerized by ISRIB. The authors went a step further and, employing CETSA (cell-based thermal shift assay), a recently developed technique to measure protein thermal stability in cells (Martinez Molina et al., 2013), determined that eIF2B4 was slightly but reproducibly stabilized by ISRIB. Finally, radioactive analysis of nucleotide release showed that ISRIB enhances the GEF activity of eIF2B. This exhaustive study shows the importance of taking advantage of multiple modern MoA methods to make maximal value of small molecules discovered during phenotypic screening.
Using resistance mutation selection to discover MoA
The continuous advancement in sequencing technologies has made the selection of clones of cells resistant to a small molecule of interest a very attractive strategy. This approach, however, is generally limited to compounds with toxicity, as the selection pressure of cell death is necessary to generate these clones. Thus, resistance clone selection has been most highly used by the infectious disease community. The MoA for a variety of antimalarial compounds have been determined by several groups (Baragana et al., 2015; Rottmann et al., 2010). Here, we focus on a story involving a highly novel approach to discovering antibacterial compounds.
A small-molecule inhibitor of bacterial riboswitches
Because bacterial riboswitches, structural elements in the 5’-UTR of mRNAs, bind metabolites and regulate biosynthetic pathways essential for growth, a novel therapeutic strategy could be to mimic the activity of a natural ligand, downregulating such growth pathways. Howe et al. (2015) focused on riboflavin biosynthesis by targeting flavin mononucleotide (FMN) riboswitches in E. coli, which regulate riboflavin uptake and synthesis (Howe et al., 2015). Importantly, such riboswitches are absent in vertebrates, and humans must consume riboflavin as part of their diet. Although a number of in vitro screening approaches have been taken to target riboswitches, no successes have been reported. Here, the authors took a phenotypic approach by screening 57,000 compounds with antibacterial activity to look for compounds whose inhibition of E. coli viability was dependent on riboflavin. The compound ribocil was the top-screening hit, as its activity was completely suppressed by riboflavin (Figure 3A).
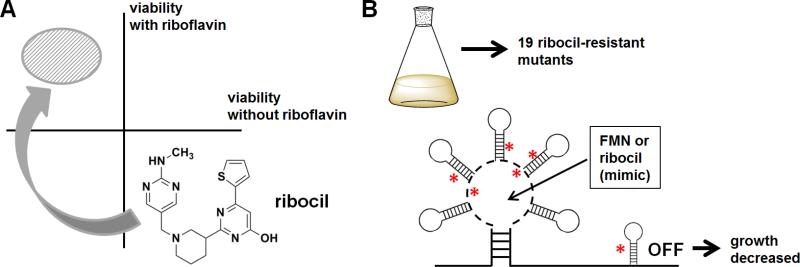
Figure 3. Discovery and MoA of ribocil.
(A) Phenotypic screening in E. coli for small molecules that cause loss of viability in the absence of riboflavin led to the discovery of ribocil. (B) Resistant mutant selection followed by whole-genome sequencing revealed that ribocil targets the FMN riboswitch as an FMN mimic. Asterisks indicate general sites of mutation.
To expand on the MoA of ribocil, the authors found 19 independent ribocil-resistant E.coli mutants and performed whole-genome sequencing to determine where mutants were generated. Remarkably, all drug-resistant mutants had base-pair changes in the FMN riboswitch within ribB gene (Figure 3B). None of these mutants just increased riboflavin levels, which would have bypassed ribocil's effects in a trivial way. A comprehensive biophysical analysis was then made, using the X-ray crystal structure of an FMN riboswitch in Fusobacterium nucleatum. Electron density mapping suggested that only the (S) enantiomer (ribocil-B) is bound; this enantiomer was superior to the (R) enantiomer in antibacterial activity, inhibition of riboflavin synthesis, and in binding to the FMN aptamer. To fully drive home the mechanism, structure-based analysis suggested that deleting part of the FMN aptamer would abolish binding, which was borne out by experimentation. This remarkable paper, which also confirmed in vivo activity of ribocil in a septicemia model, showed that bacterial riboswitches are therefore druggable targets for new antibiotic development.
Computational approaches to triangulate on small-molecule MoA
Numerous methods have been developed to analyze large-scale high-throughput screening or small-molecule profiling data to generate hypotheses regarding MoA for compounds discovered phenotypically (Dancik et al., 2014; Schirle and Jenkins, 2015). Such inferential methods are particularly useful, and in fact possibly necessary, when dealing with the large amount of data generated by cancer cell line (CCL) profiling.
Small-molecule profiling based on cell viability
Seashore-Ludlow et al. (2015) developed a new analytical method, termed ACME (annotated cluster multidimensional enrichment), to find new associations between small-molecule MoA and particular CCL dependencies (Seashore-Ludlow et al., 2015). This approach started with hierarchical clustering of data collected on 664 CCLs treated individually with 481 small molecules, some with known MoA. The ensuing matrix was then mined for “hot spots”, subclusters in which CCLs sharing genetic features were enriched for sensitivity to small molecules sharing MoA (Figure 4A). In cases where one or more compounds in the hot spot have a known MoA, it is possible to then hypothesize that unknown compounds in that same cluster share a similar MoA. Using this analysis, the authors found a hot spot containing neuroblastoma CCLs that clustered with four IGF1R inhibitors and one ALK kinase inhibitor, NVP-TAE684. This last compound, though, was also known to inhibit IGF1R phosphorylation, and we determined that IGF1R inhibition also contributes to neuroblastoma sensitivity. Overall, dual ALK/IGF1R inhibition may be a clinically actionable combination that overcomes resistance in some neuroblastomas.
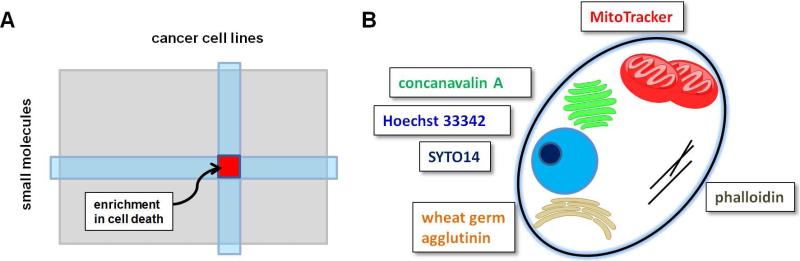
Figure 4. Computational approaches to inferential MoA.
(A) ACME (annotated cluster multidimensional enrichment) mines cancer cell line profiling data to identify clusters in which cell lines are enriched for genetic features, and small molecules are enriched for protein targets. (B) Cell painting uses six dyes to stain cellular components, and automated feature extraction is used to cluster small molecules with similar phenotypic outcomes.
This approach used cell viability across hundreds of CCLs as the phenotypic approach. A newer elegant strategy is to leverage the rich morphological data generated by cell-based imaging to generate small-molecule profiles that, like gene-expression profiles, can be used to develop similarity models and hypotheses regarding MoA (Singh et al., 2014).
Small-molecule profiling based on cell morphology
One of the first reports on this approach looked at several cell-based stains and clustered compounds based on morphological profiles, and was able to successfully bin compounds with similar MoA (Perlman et al., 2004). More recently, Gustafsdottir et al. (2013) developed a method called cell painting (Figure 4B), in which six fluorescent dyes annotate seven cellular components over five microscope channels (Gustafsdottir et al., 2013). CellProfiler (Carpenter et al., 2006) analysis generates over 800 cellular features, some of which are not detectable by the human eye, that can be clustered and analyzed in a similar fashion to gene-expression or CCL viability profiles. Many compounds with similar MoA co-clustered, validating the approach for testing nMoA compounds. A retrospective analysis of 30,000 compounds tested by cell painting and in various HTS campaigns revealed that, somewhat surprisingly, biological performance diversity is better represented by selecting for diverse activity in cell painting than it is by selecting for chemical diversity (Wawer et al., 2014). Thus, the assumption that chemical diversity leads to biological performance diversity is not always valid. This result suggests that methods such as cell painting may be appropriate up-front experiments to perform on a new set of compounds to select the most performance-diverse set for later HTS campaigns.
Conclusion and Future Perspectives
Increasingly sophisticated technologies have made MoA determination more straightforward, resulting in less uncertainty involved in undertaking a phenotypic screening campaign. By combining assay technologies and cell sources that emphasize in vivo relevance with modern MoA methods, researchers are identifying novel compounds, as well as their MoA, at an increasing rate. Although affinity-based methods are most well known for identifying intracellular targets, the success of MoA methods based on gene expression, genetic modifier screening, resistance mutation, or computational inference shows that complementary approaches can be equally effective. In fact, no one single approach is sufficient to identify MoA in all cases. For example, resistance selection is best performed when cell death is the outcome, but is currently less suitable for more subtle phenotypes. Computational approaches enable hypothesis generation, but do not usually give a definitive answer as to MoA. None of these approaches, aside from affinity-based methods, can identify a direct biochemical target of a small molecule. Instead of relying upon one method to solve all problems, MoA campaigns should take a combination of approaches. Because the phenotypes being used for screening are becoming increasingly sophisticated, there is greater incentive to fully understand the MoA for small molecules discovered in this fashion.
These approaches also highlight the fact that MoA is not equivalent to target identification; rather, target ID is an important, but not sufficient, component of an MoA study. An early understanding of the MoA for small molecules identified phenotypically should contribute not only to more rapid translational development, but also to more likely outcomes of success. We anticipate a surge in the advancement of our knowledge of cell biology and human disease enabled by these tools developed by the chemical biology community.
Highlights.
Phenotypic screening enables medically relevant small-molecule discovery
Modern mechanism-of-action methods fuels phenotypic approaches
Integrating these activities will help discover new chemistry and biology
Acknowledgements
We thank S.Gill, T.Sundberg, A.Choudhary, A.Kramm, M.Bray, M.Palmer, and A.Carpenter for helpful suggestions and comments. S.L.S. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baragana B, Hallyburton I, Lee MC, Norcross NR, Grimaldi R, Otto TD, Proto WR, Blagborough AM, Meister S, Wirjanata G, et al. A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature. 2015;522:315–320. doi: 10.1038/nature14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitano AE, Wang J, Romeo R, Bouchez LC, Parker AE, Sutton SE, Walker JR, Flaveny CA, Perdew GH, Denison MS, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancik V, Carrel H, Bodycombe NE, Seiler KP, Fomina-Yadlin D, Kubicek ST, Hartwell K, Shamji AF, Wagner BK, Clemons PA. Connecting Small Molecules with Similar Assay Performance Profiles Leads to New Biological Hypotheses. Journal of biomolecular screening. 2014;19:771–781. doi: 10.1177/1087057113520226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker RS, Koyama E, Enomoto-Iwamoto M, Maye P, Rowe D, Zhu S, Schultz PG, Pacifici M. Mouse limb skeletal growth and synovial joint development are coordinately enhanced by Kartogenin. Developmental biology. 2014;395:255–267. doi: 10.1016/j.ydbio.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert US. The why and how of phenotypic small-molecule screens. Nat Chem Biol. 2013;9:206–209. doi: 10.1038/nchembio.1206. [DOI] [PubMed] [Google Scholar]
- Germain AR, Carmody LC, Morgan B, Fernandez C, Forbeck E, Lewis TA, Nag PP, Ting A, VerPlank L, Feng Y, et al. Identification of a selective small molecule inhibitor of breast cancer stem cells. Bioorganic & medicinal chemistry letters. 2012;22:3571–3574. doi: 10.1016/j.bmcl.2012.01.035. [DOI] [PubMed] [Google Scholar]
- Gustafsdottir SM, Ljosa V, Sokolnicki KL, Anthony Wilson J, Walpita D, Kemp MM, Petri Seiler K, Carrel HA, Golub TR, Schreiber SL, et al. Multiplex cytological profiling assay to measure diverse cellular states. PloS one. 2013;8:e80999. doi: 10.1371/journal.pone.0080999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe JA, Wang H, Fischmann TO, Balibar CJ, Xiao L, Galgoci AM, Malinverni JC, Mayhood T, Villafania A, Nahvi A, et al. Selective small-molecule inhibition of an RNA structural element. Nature. 2015 doi: 10.1038/nature15542. [DOI] [PubMed] [Google Scholar]
- Johnson K, Zhu S, Tremblay MS, Payette JN, Wang J, Bouchez LC, Meeusen S, Althage A, Cho CY, Wu X, et al. A stem cell-based approach to cartilage repair. Science. 2012;336:717–721. doi: 10.1126/science.1215157. [DOI] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene- expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Laraia L, McKenzie G, Spring DR, Venkitaraman AR, Huggins DJ. Overcoming Chemical, Biological, and Computational Challenges in the Development of Inhibitors Targeting Protein-Protein Interactions. Chemistry & biology. 2015;22:689–703. doi: 10.1016/j.chembiol.2015.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrieu D, Britton S, Demir M, Rodriguez R, Jackson SP. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science. 2014;344:527–532. doi: 10.1126/science.1252651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, Nordlund P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- Moffat JG, Rudolph J, Bailey D. Phenotypic screening in cancer drug discovery - past, present and future. Nature reviews Drug discovery. 2014;13:588–602. doi: 10.1038/nrd4366. [DOI] [PubMed] [Google Scholar]
- Mohr SE, Smith JA, Shamu CE, Neumuller RA, Perrimon N. RNAi screening comes of age: improved techniques and complementary approaches. Nature reviews Molecular cell biology. 2014;15:591–600. doi: 10.1038/nrm3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palchaudhuri R, Hergenrother PJ. Transcript profiling and RNA interference as tools to identify small molecule mechanisms and therapeutic potential. ACS chemical biology. 2011;6:21–33. doi: 10.1021/cb100310h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. Multidimensional drug profiling by automated microscopy. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S, Javaid S, Coletti ME, Jones VL, Bodycombe NE, et al. Correlating small-molecule sensitivity with basal gene expression to illuminate mechanism of action. Nat Chem Biol. 2015 doi: 10.1038/nchembio.1986. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottmann M, McNamara C, Yeung BK, Lee MC, Zou B, Russell B, Seitz P, Plouffe DM, Dharia NV, Tan J, et al. Spiroindolones, a potent compound class for the treatment of malaria. Science. 2010;329:1175–1180. doi: 10.1126/science.1193225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle M, Jenkins JL. Identifying compound efficacy targets in phenotypic drug discovery. Drug discovery today. 2015 doi: 10.1016/j.drudis.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Seashore-Ludlow B, Rees MG, Cheah JH, Cokol M, Price EV, Coletti ME, Jones V, Bodycombe NE, Soule CK, Gould J, et al. Harnessing Connectivity in a Large- Scale Small-Molecule Sensitivity Dataset. Cancer discovery. 2015 doi: 10.1158/2159-8290.CD-15-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H, Gamache K, Gallagher CM, Ang KK, Wilson C, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife. 2013;2:e00498. doi: 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Tsai JC, Kampmann M, Hearn BR, Vedantham P, Jaishankar P, Sokabe M, Mendez AS, Newton BW, Tang EL, et al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife. 2015;4:e07314. doi: 10.7554/eLife.07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Carpenter AE, Genovesio A. Increasing the Content of High-Content Screening: An Overview. Journal of biomolecular screening. 2014;19:640–650. doi: 10.1177/1087057114528537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinney DC. The contribution of mechanistic understanding to phenotypic screening for first-in-class medicines. Journal of biomolecular screening. 2013;18:1186–1192. doi: 10.1177/1087057113501199. [DOI] [PubMed] [Google Scholar]
- Swinney DC, Anthony J. How were new medicines discovered? Nature reviews Drug discovery. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- Vincent F, Loria P, Pregel M, Stanton R, Kitching L, Nocka K, Doyonnas R, Steppan C, Gilbert A, Schroeter T, et al. Developing predictive assays: the phenotypic screening “rule of 3”. Science translational medicine. 2015;7:293ps215. doi: 10.1126/scitranslmed.aab1201. [DOI] [PubMed] [Google Scholar]
- Wawer MJ, Li K, Gustafsdottir SM, Ljosa V, Bodycombe NE, Marton MA, Sokolnicki KL, Bray MA, Kemp MM, Winchester E, et al. Toward performance-diverse small-molecule libraries for cell-based phenotypic screening using multiplexed high- dimensional profiling. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:10911–10916. doi: 10.1073/pnas.1410933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JN, Myers TG, O'Connor PM, Friend SH, Fornace AJ, Jr., Kohn KW, Fojo T, Bates SE, Rubinstein LV, Anderson NL, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- Xu X, Shi D, Shen Y, Xu Z, Dai J, Chen D, Teng H, Jiang Q. Full-thickness cartilage defects are repaired via a microfracture technique and intraarticular injection of the small-molecule compound kartogenin. Arthritis research & therapy. 2015;17:20. doi: 10.1186/s13075-015-0537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang JH. Kartogenin induces cartilage-like tissue formation in tendon-bone junction. Bone research. 2014;2 doi: 10.1038/boneres.2014.8. [DOI] [PMC free article] [PubMed] [Google Scholar]