SUMMARY
During mitochondrial dysfunction or the accumulation of unfolded proteins within mitochondria, cells employ a transcriptional response known as the mitochondrial unfolded protein response (UPRmt) to promote cell survival along with the repair and recovery of defective mitochondria. Considerable progress has been made in understanding how cells monitor mitochondrial function and activate the response, as well as in identifying scenarios where the UPRmt plays a protective role such as during bacterial infection, hematopoietic stem cell maintenance or general aging. To date, much of the focus has been on the role of the UPRmt in maintaining or re-establishing protein homeostasis within mitochondria by transcriptionally inducing mitochondrial molecular chaperone and protease genes. In this review, we focus on the metabolic adaptations or rewiring mediated by the UPRmt and how this may contribute to the resolution of mitochondrial unfolded protein stress and cell type-specific physiology.
Mitochondrial recovery via the UPRmt
Mitochondria are a dynamic network of double membrane bound organelles responsible for the vast majority of ATP generation in non-dividing differentiated cells. In addition to housing the tricarboxylic acid (TCA) cycle and oxidative phosphorylation (OxPhos) machinery, mitochondria are also important contributors to amino acid, lipid, and nucleotide metabolism. Because mitochondrial function impacts numerous essential cellular and organismal functions, it is perhaps not surprising that mechanisms or pathways have evolved to monitor mitochondrial function and rapidly respond to mitochondrial stress to recover organelle activity. Such pathways are typically referred to as retrograde responses as the upstream signal initiates at mitochondria and communicates the status to the cytosol and nucleus to impact gene transcription and protein synthesis in a protective manner (Liu and Butow, 2006). Here, we focus on the mitochondrial unfolded protein response (UPRmt), which is a transcriptional response originally discovered to increase mitochondrial localized molecular chaperones and proteases to promote the recovery of organellar protein homeostasis (proteostasis) (Yoneda et al., 2004; Zhao et al., 2002). However, the UPRmt also promotes a rewiring of cellular metabolism that includes an increase in glycolysis and amino acid catabolism genes with a simultaneous repression of TCA cycle and OxPhos-encoding genes potentially to relieve mitochondrial stress and/or alter cellular metabolism to promote survival (Nargund et al., 2015).
Many potential mechanisms exist to evaluate mitochondrial activity or function including monitoring mitochondrial metabolites or products such as ATP or iron-sulfur clusters (Hardie et al., 2015). However, a number of recent studies demonstrate that an effective strategy is to monitor mitochondrial protein import efficiency (Harbauer et al., 2014). Mitochondria are composed of over 1000 proteins of which ~99 percent are encoded by nuclear genes and translated on cytosolic ribosomes. Thus, in order for mitochondria to function properly, these proteins must be imported into mitochondria where they are appropriately folded and assembled. Transit across the mitochondrial inner membrane not only requires the Tim23 complex, but also an intact TCA cycle and OxPhos system that maintains the membrane potential, as well as mitochondrial chaperones located within the matrix (Chacinska et al., 2009; Shariff et al., 2004; van der Laan et al., 2006). Thus, the efficiency of the protein import process reflects diverse aspects of mitochondrial function.
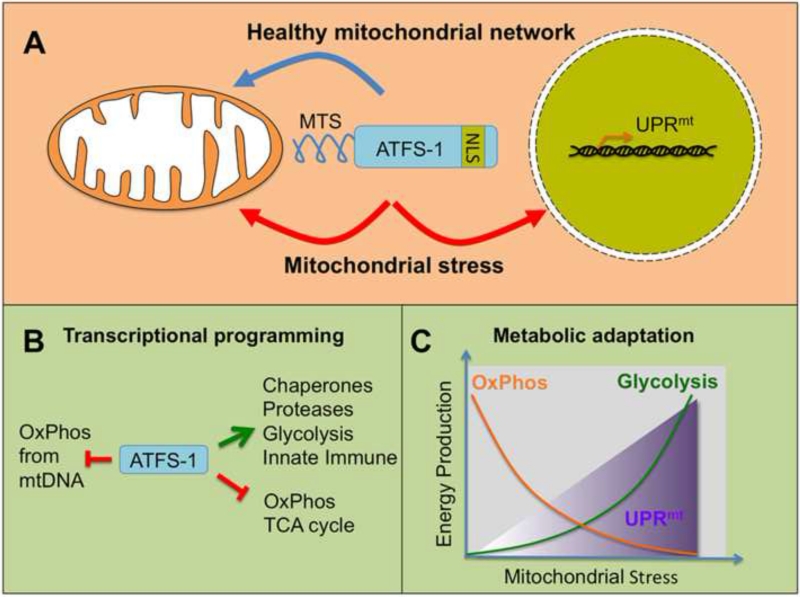
Indeed, signaling molecules have been identified that are activated by mitochondrial dysfunction or stresses that reduce mitochondrial import efficiency. For example, the transcription factor ATFS-1, which regulates the UPRmt in C. elegans has both a mitochondrial targeting sequence (MTS) and a nuclear localization sequence (NLS). Normally, ATFS-1 is efficiently imported into mitochondria where it is rapidly degraded. However during mitochondrial stress, reduced import efficiency causes many mitochondrial-targeted proteins to accumulate in the cytosol. Most are recognized and targeted for degradation by the proteasome (Wrobel et al., 2015) so as to prevent toxicity of mislocalized protein accumulation (Wang and Chen, 2015). But, because ATFS-1 has a NLS it traffics to the nucleus to activate the UPRmt (Nargund et al., 2012) (Figure 1A). Included in the stresses that perturb import and activate the UPRmt are depletion of mtDNA (Martinus et al., 1996; Yoneda et al., 2004), accumulation of misfolded proteins within mitochondria (Papa and Germain, 2011; Zhao et al., 2002), mitochondrial ribosome impairment (Houtkooper et al., 2013; Moullan et al., 2015), mitochondrial chaperone and protease inhibition (Yoneda et al., 2004), OxPhos perturbation (Liu et al., 2014; Nargund et al., 2012), high glucose consumption (Tauffenberger et al., 2016) and high levels of reactive oxygen species (Runkel et al., 2013; Yoneda et al., 2004). Thus, the mitochondrial import capacity of the entire organellar network controls expression of a mitochondrial recovery program. Of note, while many of the conditions outlined above have been reported to deplete membrane potential, none of them cause complete membrane depolarization. To our knowledge the relationship between the inner membrane potential and UPRmt activation has not been explicitly evaluated. Regardless, it is clear that depletion of the mitochondrial inner membrane potential is not necessary to activate the UPRmt (Jin and Youle, 2013).
Figure 1. ATFS-1 represses OxPhos and TCA cycle gene expression while inducing mitochondrial proteostasis and glycolysis genes during mitochondrial dysfunction.
(A) Beause ATFS-1 contains a mitochondrial targeting sequence (MTS), it is efficiently imported into mitochondria and degraded. However during mitochondrial stress, import efficiency of ATFS-1 is reduced causing a percentage of it to traffic to the nucleus via its nuclear localization sequence (NLS) where it regulates the UPRmt.
(B) In the nucleus, ATFS-1 regulates expression of over 400 genes including the induction of mitochondrial proteostasis genes (chaperones, proteases) as well as glyolysis and innate immune genes. Simultaneously, ATFS-1 limits the accumulation of the highly expressed TCA cycle and oxidative phosphorylation (OxPhos) transcripts. During stress, a percentage of ATFS-1 is also stabilized within mitochondria where it limits the accumulation of mtDNA-encoded OxPhos transcripts.
(C) UPRmt activation results in a reduction of nuclear and mtDNA-encoded OxPhos transcripts to promote stoichiometric complex assembly and reduce the substrate burden on the overwhelmed proteostasis environment in stressed mitochondria. Together, ATFS-1 promotes the regeneration of OxPhos complexes while increasing glycolytic capacity to maintain ATP levels.
A second pathway regulated by mitochondrial protein import efficiency is a form of organelle quality control known as mitophagy (Narendra et al., 2008). Similar to ATFS-1, the kinase PINK1 has a MTS, which allows it to be efficiently imported into healthy organelles and processed leading to its degradation in the cytosol (Tanaka et al., 2010). However, when import efficiency is severely impaired, PINK1 integrates into the outer membrane of the defective compartment where it serves several functions. The most well-characterized function involves the subsequent phosphorylation of ubiquitin (Kane et al., 2014; Kazlauskaite et al., 2014; Koyano et al., 2014) and the recruitment of the ubiquitin ligase Parkin (Matsuda et al., 2010; Narendra et al., 2010; Okatsu et al., 2015; Vives-Bauza et al., 2010) that ultimately results in isolation of the defective organelle, or subcompartment, and targeting to the lysosome for degradation (Heo et al., 2015; Lazarou et al., 2015; McLelland et al., 2014; Yang and Yang, 2013). Thus, the mitochondrial network is self-monitoring as import deficiency directly initiates the downstream activation of at least two protective responses. By activating a protective transcriptional response and eliminating the most defective mitochondria, the UPRmt and mitophagy pathways function to improve or recover the health of the mitochondrial network.
Regulated expression of TCA cycle and OxPhos components
In flies, worms and mammals, the UPRmt includes the induction of mitochondrial proteostasis machinery such as mitochondrial molecular chaperones and proteases as well as anti-oxidant genes to limit damage due to increased generation of reactive oxygen species (Nargund et al., 2012; Owusu-Ansah et al., 2013; Wu et al., 2014; Zhao et al., 2002). Additionally, the UPRmt includes the induction of multiple metabolic pathways including genes required for glycolysis and amino acid catabolism. Interestingly, the UPRmt also limits the accumulation of transcripts that encode the highly expressed TCA cycle and OxPhos components (Nargund et al., 2015) (Figure 1B).
The OxPhos complexes are large multi-subunit structures located within the mitochondrial inner membrane. With the exception of the succinate dehydrogenase complex (Complex II), the other three respiratory chain complexes and the ATP synthase are composed of proteins encoded by both the nuclear and mitochondrial genomes (mtDNA). 100s of mtDNA copies exist per cell, with multiple copies per mitochondrion. They encode 13 OxPhos subunits as well as tRNAs and rRNAs required for their synthesis. Therefore, to insure efficient complex assembly and biogenesis, transcription from both genomes must be coordinated to promote stoichiometric expression and prevent the accumulation of OxPhos subunits that fail to integrate or assemble into specific complexes (Jovaisaite and Auwerx, 2015).
In addition to trafficking to the nucleus during mitochondrial dysfunction and limiting the expression of nuclear-encoded OxPhos components, a percentage of ATFS-1 also accumulates within mitochondria where it limits the accumulation of the OxPhos transcripts encoded by mtDNA (Figure 1A). ATFS-1-dependent repression of mtDNA-encoded transcripts appears to be direct as ATFS-1 binds the mtDNA promoter region, which contains the same sequence motif that ATFS-1 binds in the nuclear genome to activate mitochondrial proteostasis gene transcription. Combined, these observations suggest that ATFS-1 and the UPRmt simultaneously limit expression of OxPhos components from both genomes to promote efficient stoichiometric expression and assembly of the OxPhos complexes (Nargund et al., 2015). Thus, in addition to increasing the expression of the machinery required to assemble the OxPhos complexes, the UPRmt limits the influx of nascent OxPhos components so as to not overwhelm the protein folding and complex assembly capacity of the defective organelle. Concomitantly, the deficit in ATP production is maintained by increased glycolysis gene expression (Fig. 1C). Supporting the role of matching substrate load and mitochondrial proteostasis capacity, additional pathways are also in place to reduce the burden on the potentially deficient mitochondrial protein-folding environment during stress by reducing cytosolic protein synthesis (Baker et al., 2012; Wang and Chen, 2015).
Metabolic adaptation in physiology and diseases
The cellular benefits of what appears to be a metabolic shift similar to that observed in rapidly growing cells (Vander Heiden et al., 2009) are potentially numerous during mitochondrial dysfunction. An increase in glycolysis is potentially a means to maintain ATP production in the presence of compromised OxPhos, which would promote cell survival as well as the regeneration of defective mitochondria and the OxPhos complexes. However, the induction of glycolysis and repression of TCA cycle and OxPhos transcripts could also serve to rewire cellular metabolism to effect cellular proliferation, growth or differentiation. Here, we explore the role of UPRmt-mediated metabolic adaptations in several physiologic scenarios where the UPRmt is known to play a role.
Innate immunity during bacterial infection
In addition to inducing genes that promote mitochondrial recovery, the UPRmt also includes xenobiotic detoxification genes as well as innate immunity genes (Liu et al., 2014; Pellegrino et al., 2014) (Figure 1B). And perhaps not surprisingly, a number of bacterial-produced OxPhos inhibitors such as antimycin and oligomycin, as well as mitochondrial ribosome inhibitors (Moullan et al., 2015) activate the UPRmt in C. elegans and mammals (Liu et al., 2014; Pellegrino et al., 2014; Runkel et al., 2013). These observations suggest a means to detect pathogenic, or toxic, bacteria by monitoring mitochondrial function and initiating an innate immune response accordingly. In support of this idea, pathogenic strains of Pseudomonas aeruginosa cause mitochondrial dysfunction and activate the UPRmt. Activation of the UPRmt requires the P. aeruginosa virulence response, which includes the production of cyanide (a respiratory chain inhibitor) and iron chelators (Kirienko et al., 2013). Worms lacking ATFS-1 are sensitive to P. aeruginosa infection while worms with a constitutively active UPRmt are resistant to the pathogen and limit the intestinal accumulation of the bacteria. Thus, it appears that cells perceive mitochondrial stress or damage as a potential bacterial infection, which may be an important strategy to detect toxic bacteria in non-sterile environments such as the intestine or skin. Increased intestinal clearance of P. aeruginosa suggests the UPRmt provides anti-bacterial activity, but the role of the UPRmt-mediated metabolic adaptations is less clear. Perhaps the simplest explanation is that in response to the cytochrome c oxidase (Complex IV) inhibitor cyanide, the UPRmt increases glycolysis genes to maintain energy levels and limits OxPhos biogenesis so as not to exacerbate the accumulating mitochondrial damage until the animal clears the infection (Melo and Ruvkun, 2012; Pellegrino et al., 2014).
Aging
A decline in mitochondria function is a hallmark of aging (Lopez-Otin et al., 2013), and multiple studies suggest that increased UPRmt activation can recover or prolong mitochondrial function in a variety of tissues and promote longevity. Modest OxPhos dysfunction in worms, flies and mice results in lifespan extension and causes UPRmt activation (Durieux et al., 2011; Houtkooper et al., 2013; Lapointe et al., 2009; Owusu-Ansah et al., 2013). In worms it has been demonstrated that ATFS-1 (Schieber and Chandel, 2014), and other UPRmt signaling components are required for this form of lifespan extension (Durieux et al., 2011; Houtkooper et al., 2013). However, it should be noted that UPRmt activation alone is not sufficient to prolong lifespan (Bennett et al., 2014; Rauthan et al., 2013), suggesting multiple pathways are in place to respond to mitochondrial stress in addition to the UPRmt. It is unclear which UPRmt-mediated activities specifically contribute to longevity, however recent work points towards a role in the recovery of mitochondrial function via mitochondrial biogenesis (Houtkooper et al., 2013; Mouchiroud et al., 2013; Nargund et al., 2015).
In aged cells where mitochondria are potentially damaged, recovery of dysfunctional organelles may require a different, perhaps more tightly regulated, program than mitochondrial biogenesis in an otherwise healthy network. Studies in mice and worms have shown that NAD is reduced in aged tissues such as muscle (Canto et al., 2015; Pirinen et al., 2014), as well as in livers of mice fed a high fat and high sugar diet (Gariani et al., 2015). NAD is converted to NADH by the TCA cycle within mitochondria, which acts as an electron donor for the respiratory chain, which maintains the membrane potential across the mitochondrial inner membrane and can be used to generate ATP. Impressively, increasing NAD levels by genetic or pharmacologic means promotes mitochondrial function and prolongs lifespan (Andreux et al., 2013; Gariani et al., 2015; Mouchiroud et al., 2013). Increased NAD leads to mitochondrial recovery via sirtuin-mediated activation of the transcriptional co-activator PGC-1α, FOXO as well as UPRmt activation, which combined yields efficient recovery of mitochondrial activity (Mouchiroud et al., 2013). In this context, we anticipate that UPRmt activation orchestrates a very coordinated mitochondrial recovery program by fine-tuning OxPhos and TCA cycle expression to match the protein folding and complex assembly of the defective organelles while simultaneously increasing the mitochondrial proteostasis capacity. Interestingly, the UPRmt may also play a role in maintaining a high NAD/NADH ratio by limiting TCA activity until mitochondrial activity has been recovered when normal TCA cycle gene transcription resumes.
Stem cell maintenance
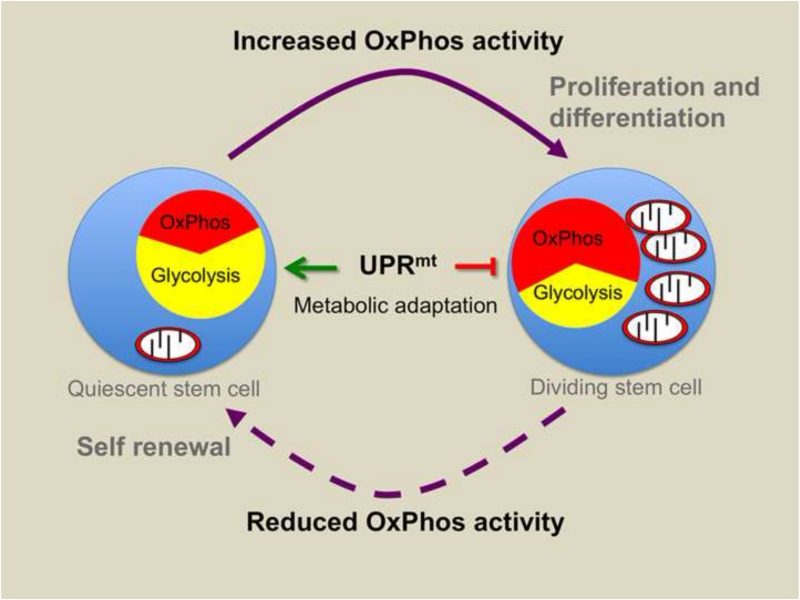
Aging can also be attributed to deterioration of tissue-specific stem cells (Lopez-Otin et al., 2013). However, unlike somatic cells, quiescent stem cells maintain few mitochondria with relatively low metabolic activity (Kohli and Passegue, 2014). Interestingly, a recent study found that hematopoietic stem cells (HSC) utilize a signaling pathway involving a UPRmt to repress mitochondrial biogenesis and OxPhos to coordinate the metabolism required for stem cell maintenance (Mohrin et al., 2015). Hematopoietic stem cell maintenance requires an interaction between the histone deacetylase SIRT7 and the transcription factor NRF1, which regulates the expression of genes that encode mitochondrial ribosome components (Scarpulla et al., 2012). In this context, SIRT7 expression is increased by mitochondrial protein folding stress associated with the burst of mitochondria biogenesis that occurs during stem cell proliferation. By inhibiting NRF1 activity, SIRT7 limits mitochondrial biogenesis and OxPhos preserving stem cell metabolism (Figure 2). Therefore, SIRT7 promotes the maintenance of a pristine mitochondrial protein-folding environment keeping the organelles and the stem cells in a “youthful” state. Consistent with this idea, HSCs lacking SIRT7 have increased mitochondrial stress and an increased propensity to proliferate. Thus, HSCs require SIRT7 to limit mitochondrial stress and proliferation (Mohrin et al., 2015), which maintains their regenerative function (Miyamoto et al., 2007). Indeed, SIRT7 expression decreases in aged mice where HSC maintenance fails leading to a reduction in white blood cell number (Mohrin et al., 2015) suggesting that by simply limiting OxPhos complex biogenesis and unfolded protein accumulation, stem cell function can be preserved during aging.
Figure 2. A metabolic checkpoint involving a UPRmt via SIRT7 is required for stem cell maintainance.
Quiescent stem cells rely heavily on glycolysis. However, stem cell proliferation and differentiation is accompanied by increased mitochondrial biogenesis and a shift to oxidative phosphorylation (OxPhos). SIRT7 is a histone deacetylase transcriptionally induced during the mitochondrial stress associated with mitochondrial biogenesis in proliferating hematopoietic stem cells (HSCs). SIRT7 expression is induced during mitochondrial unfolded protein stress potentially to reduce the number of OxPhos proteins expressed so as to not further perturb a dysfunctional protein folding environment. In the context of HSCs, SIRT7 promotes a pristine mitochondrial proteostasis environment, which also limits mitochondrial biogenesis, cell proliferation and differentiation.
CONCLUSIONS AND PERSPECTIVES
We have highlighted scenarios identified in multiple organisms where the UPRmt has been documented to play a protective role, focusing on the potential protective outputs of UPRmt-mediated metabolic regulation rather than the maintenance of mitochondrial proteostasis. However, it should be noted that while the regulation of the UPRmt in C. elegans relies on the transcription factor ATFS-1, less is known regarding the regulation of the UPRmt in mammals or flies. A pressing question in the field is whether the UPRmt-related observations in flies or mammals are regulated via mitochondrial protein import efficiency or through an alternative means. Unfortunately homology searches do not yield candidates with especially significant homology beyond the DNA binding domain. Interestingly, the yeast transcription factor Hap1 was recently found to harbor both a MTS and a NLS (Williams et al., 2014), suggesting it is regulated similarly to ATFS-1. Consistent with Hap1 responding to mitochondrial protein import impairment, it is activated when oxygen and heme are limiting and activates genes that promote heme biogenesis (Kwast et al., 1998). Because, multiple mammalian transcription factors have putative MTSs (Claros and Vincens, 1996) and have been found to localize to mitochondria (Marinov et al., 2014), we are optimistic a similar mode of regulation will be uncovered. However, other means of sensing and responding to mitochondrial stress should not be excluded.
While roles for the UPRmt in adapting metabolism to promote recovery of the mitochondrial network are only beginning to emerge, considerable data suggests multiple pathologic scenarios where improved mitochondrial health may be beneficial. Interestingly, a dichotomy may be emerging where the UPRmt affects proliferating cells differently than post-mitotic cells such as muscles or neurons, which likely reflects each cell types preferred means of glucose utilization. In response to mitochondrial dysfunction, mitotic and post-mitotic cells employ aerobic glycolysis (or Warburg metabolism) by increasing glycolytic while impairing TCA cycle and OxPhos gene expression. In proliferating cells, perpetual or prolonged UPRmt activation may promote glycolysis while maintaining or stabilizing mitochondrial function (Mohrin et al., 2015). Promoting aerobic glycolysis in stem or cancer cells is potentially a concern, but recent work demonstrates that these cells require mitochondrial function and maintenance of the membrane potential in order to proliferate (Martinez-Reyes et al., 2015). Presumably muscle cells or neurons employ the UPRmt over shorter periods of time to promote repair or recovery of mitochondrial function without permanently rewiring cellular metabolism (Lamech and Haynes, 2015). As additional physiologic roles of the UPRmt are identified, coupled with a better understanding of the underlying regulatory mechanisms in mammals (Arnould et al., 2015), we are optimistic that strategies to engage this pathway therapeutically will develop. In addition to neurodegenerative diseases such as Parkinson’s where loss of mitochondrial function contributes to dopaminergic neuronal loss, diverse metabolic diseases may also be considered. For example, increased muscle mitochondrial biogenesis is a proposed strategy to limit or prevent insulin resistance, a precursor to type 2 diabetes (Egan and Zierath, 2013).
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants (R01AG040061 and R01AG047182) to CMH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andreux PA, Houtkooper RH, Auwerx J. Pharmacological approaches to restore mitochondrial function. Nature reviews. Drug discovery. 2013;12:465–483. doi: 10.1038/nrd4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnould T, Michel S, Renard P. Mitochondria Retrograde Signaling and the UPR mt: Where Are We in Mammals? International journal of molecular sciences. 2015;16:18224–18251. doi: 10.3390/ijms160818224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BM, Nargund AM, Sun T, Haynes CM. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS genetics. 2012;8:e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CF, Vander Wende H, Simko M, Klum S, Barfield S, Choi H, Pineda VV, Kaeberlein M. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nature communications. 2014;5:3483. doi: 10.1038/ncomms4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Menzies KJ, Auwerx J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell metabolism. 2015;22:31–53. doi: 10.1016/j.cmet.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. European Journal Of Biochemistry / Febs. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell metabolism. 2013;17:162–184. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Gariani K, Menzies KJ, Ryu D, Wegner CJ, Wang X, Ropelle ER, Moullan N, Zhang H, Perino A, Lemos V, et al. Eliciting the mitochondrial unfolded protein response via NAD repletion reverses fatty liver disease. Hepatology. 2015 doi: 10.1002/hep.28245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbauer AB, Zahedi RP, Sickmann A, Pfanner N, Meisinger C. The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell metabolism. 2014;19:357–372. doi: 10.1016/j.cmet.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Schaffer BE, Brunet A. AMPK: An Energy-Sensing Pathway with Multiple Inputs and Outputs. Trends in cell biology. 2015 doi: 10.1016/j.tcb.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Molecular cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9:1750–1757. doi: 10.4161/auto.26122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovaisaite V, Auwerx J. The mitochondrial unfolded protein response-synchronizing genomes. Current opinion in cell biology. 2015;33:74–81. doi: 10.1016/j.ceb.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S, Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. The Journal of cell biology. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M, Muqit MM. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. The Biochemical journal. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirienko NV, Kirienko DR, Larkins-Ford J, Wahlby C, Ruvkun G, Ausubel FM. Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell host & microbe. 2013;13:406–416. doi: 10.1016/j.chom.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli L, Passegue E. Surviving change: the metabolic journey of hematopoietic stem cells. Trends in cell biology. 2014;24:479–487. doi: 10.1016/j.tcb.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- Kwast KE, Burke PV, Poyton RO. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. The Journal of experimental biology. 1998;201:1177–1195. doi: 10.1242/jeb.201.8.1177. [DOI] [PubMed] [Google Scholar]
- Lamech LT, Haynes CM. The unpredictability of prolonged activation of stress response pathways. The Journal of cell biology. 2015;209:781–787. doi: 10.1083/jcb.201503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe J, Stepanyan Z, Bigras E, Hekimi S. Reversal of the mitochondrial phenotype and slow development of oxidative biomarkers of aging in long-lived Mclk1+/− mice. The Journal of biological chemistry. 2009;284:20364–20374. doi: 10.1074/jbc.M109.006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Samuel BS, Breen PC, Ruvkun G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 2014;508:406–410. doi: 10.1038/nature13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA. Mitochondrial retrograde signaling. Annual review of genetics. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinov GK, Wang YE, Chan D, Wold BJ. Evidence for site-specific occupancy of the mitochondrial genome by nuclear transcription factors. PloS one. 2014;9:e84713. doi: 10.1371/journal.pone.0084713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Reyes I, Diebold LP, Kong H, Schieber M, Huang H, Hensley CT, Mehta MM, Wang T, Santos JH, Woychik R, et al. TCA Cycle and Mitochondrial Membrane Potential Are Necessary for Diverse Biological Functions. Molecular cell. 2015 doi: 10.1016/j.molcel.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinus RD, Garth GP, Webster TL, Cartwright P, Naylor DJ, Hoj PB, Hoogenraad NJ. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. European journal of biochemistry / FEBS. 1996;240:98–103. doi: 10.1111/j.1432-1033.1996.0098h.x. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. The Journal of cell biology. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. The EMBO journal. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JA, Ruvkun G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell. 2012;149:452–466. doi: 10.1016/j.cell.2012.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell stem cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, Haynes CM, Chen D. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science. 2015;347:1374–1377. doi: 10.1126/science.aaa2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moullan N, Mouchiroud L, Wang X, Ryu D, Williams EG, Mottis A, Jovaisaite V, Frochaux MV, Quiros PM, Deplancke B, et al. Tetracyclines Disturb Mitochondrial Function across Eukaryotic Models: A Call for Caution in Biomedical Research. Cell reports. 2015 doi: 10.1016/j.celrep.2015.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. The Journal of cell biology. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS biology. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt) Molecular cell. 2015;58:123–133. doi: 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K, Koyano F, Kimura M, Kosako H, Saeki Y, Tanaka K, Matsuda N. Phosphorylated ubiquitin chain is the genuine Parkin receptor. The Journal of cell biology. 2015;209:111–128. doi: 10.1083/jcb.201410050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa L, Germain D. Estrogen receptor mediates a distinct mitochondrial unfolded protein response. Journal of cell science. 2011;124:1396–1402. doi: 10.1242/jcs.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM, Kirienko NV, Gillis R, Fiorese CJ, Haynes CM. Mitochondrial UPR-regulated innate immunity provides resistance to pathogen infection. Nature. 2014;516:414–417. doi: 10.1038/nature13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirinen E, Canto C, Jo YS, Morato L, Zhang H, Menzies KJ, Williams EG, Mouchiroud L, Moullan N, Hagberg C, et al. Pharmacological Inhibition of poly(ADP-ribose) polymerases improves fitness and mitochondrial function in skeletal muscle. Cell metabolism. 2014;19:1034–1041. doi: 10.1016/j.cmet.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauthan M, Ranji P, Aguilera Pradenas N, Pitot C, Pilon M. The mitochondrial unfolded protein response activator ATFS-1 protects cells from inhibition of the mevalonate pathway. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5981–5986. doi: 10.1073/pnas.1218778110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runkel ED, Liu S, Baumeister R, Schulze E. Surveillance-activated defenses block the ROS-induced mitochondrial unfolded protein response. PLoS genetics. 2013;9:e1003346. doi: 10.1371/journal.pgen.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends in endocrinology and metabolism: TEM. 2012;23:459–466. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber M, Chandel NS. TOR signaling couples oxygen sensing to lifespan in C. elegans. Cell reports. 2014;9:9–15. doi: 10.1016/j.celrep.2014.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shariff K, Ghosal S, Matouschek A. The force exerted by the membrane potential during protein import into the mitochondrial matrix. Biophysical journal. 2004;86:3647–3652. doi: 10.1529/biophysj.104.040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. The Journal of cell biology. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauffenberger A, Vaccaro A, Parker JA. Fragile lifespan expansion by dietary mitohormesis in C. elegans. Aging. 2016 doi: 10.18632/aging.100863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M, Wiedemann N, Mick DU, Guiard B, Rehling P, Pfanner N. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Current biology: CB. 2006;16:2271–2276. doi: 10.1016/j.cub.2006.10.025. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen XJ. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature. 2015;524:481–484. doi: 10.1038/nature14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L, Topf U, Bragoszewski P, Wiese S, Sztolsztener ME, Oeljeklaus S, Varabyova A, Lirski M, Chroscicki P, Mroczek S, et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature. 2015;524:485–488. doi: 10.1038/nature14951. [DOI] [PubMed] [Google Scholar]
- Wu Y, Williams EG, Dubuis S, Mottis A, Jovaisaite V, Houten SM, Argmann CA, Faridi P, Wolski W, Kutalik Z, et al. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158:1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Yang WY. Bit-by-bit autophagic removal of parkin-labelled mitochondria. Nature communications. 2013;4:2428. doi: 10.1038/ncomms3428. [DOI] [PubMed] [Google Scholar]
- Yoneda T, Benedetti C, Urano F, Clark SG, Harding HP, Ron D. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. Journal of cell science. 2004;117:4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. The EMBO journal. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]