Abstract
Background. Severe H1N1 influenza can be lethal in otherwise healthy individuals and can have features of reactive hemophagocytic lymphohistiocytosis (HLH). HLH is associated with mutations in lymphocyte cytolytic pathway genes, which have not been previously explored in H1N1 influenza.
Methods. Sixteen cases of fatal influenza A(H1N1) infection, 81% with histopathologic hemophagocytosis, were identified and analyzed for clinical and laboratory features of HLH, using modified HLH-2004 and macrophage activation syndrome (MAS) criteria. Fourteen specimens were subject to whole-exome sequencing. Sequence alignment and variant filtering detected HLH gene mutations and potential disease-causing variants. Cytolytic function of the PRF1 p.A91V mutation was tested in lentiviral-transduced NK-92 natural killer (NK) cells.
Results. Despite several lacking variables, cases of influenza A(H1N1) infection met 44% and 81% of modified HLH-2004 and MAS criteria, respectively. Five subjects (36%) carried one of 3 heterozygous LYST mutations, 2 of whom also possessed the p.A91V PRF1 mutation, which was shown to decrease NK cell cytolytic function. Several patients also carried rare variants in other genes previously observed in MAS.
Conclusions. This cohort of fatal influenza A(H1N1) infections confirms the presence of hemophagocytosis and HLH pathology. Moreover, the high percentage of HLH gene mutations suggests they are risk factors for mortality among individuals with influenza A(H1N1) infection.
Keywords: hemophagocytosis, macrophage activation syndrome, H1N1 influenza, perforin, cytolytic pathway, LYST
A novel influenza A(H1N1) virus emerged in spring 2009 and rapidly reached global pandemic status [1]. Subsequently, this virus has largely replaced previously circulating viruses and become the predominant agent of seasonal influenza [2]. Although the 2009 H1N1 influenza outbreak was associated with a relatively low case-fatality rate, H1N1 influenza disproportionately affected children and younger adults, with a significantly higher number of reported pediatric deaths than had been seen in prior seasonal epidemics [1, 3]. Indeed, significant numbers of children and younger adults required critical care, including extracorporeal membrane oxygenation [3]. Several reports of severe and often fatal H1N1 influenza have demonstrated liver dysfunction, cytopenias, coagulopathy, hyperferritinemia, and hemophagocytosis on bone marrow examination [4–11]. These features are consistent with reactive hemophagocytic lymphohistiocytosis (rHLH; also known as “secondary HLH”), also referred to as macrophage activation syndrome (MAS) [12–14], and indeed several patients were successfully treated with HLH-specific therapy [5, 8, 15]. There is some evidence that rHLH/MAS may be a common complication in fatal H1N1 influenza. An autopsy series found evidence of hemophagocytosis (a hallmark of HLH) in all cases examined [16, 17], and a prospective study of all critically ill patients with influenza found that 36% satisfied diagnostic criteria for rHLH, of whom 89% died, compared with 25% without rHLH [18].
Familial HLH (fHLH; also known as “primary HLH”) is a rare autosomal recessive disorder of multisystem inflammation, typically fatal in the absence of hematopoietic stem cell transplantation in infancy. fHLH is caused by mutations affecting the cytolytic pathway of natural killer (NK) cells and CD8+ T lymphocytes. Normally, these cells recognize and kill virus-infected cells through the exocytosis of cytotoxic effector molecules, such as perforin (PRF1). This function requires the coordinated movement, docking, and fusion of lytic granules toward the target cell. Thus, a majority of patients with fHLH have identified mutations in PRF1, which encodes perforin itself, or in genes encoding proteins required for the docking and fusion of these granules: UNC13D (encoding MUNC13–4), STX11 (encoding syntaxin 11), and STXBP2 (encoding syntaxin-binding protein 2) [13, 19]. Additional rare genetic disorders also lead to HLH and have cytolytic dysfunction, including Chediak–Higashi syndrome (caused by LYST mutations) and Griscelli syndrome (caused by RAB27A mutations) [13].
In contrast, while rHLH/MAS share clinical and pathological hallmarks with fHLH, their etiology is not fully understood. However, MAS occurring in the setting of systemic juvenile idiopathic arthritis (SJIA) shares with HLH defective NK cell function [20]. Moreover, polymorphisms in the HLH-associated gene UNC13D were soon identified [21]. Subsequently, heterozygous uncommon-to-rare protein-altering mutations in HLH-associated genes have been identified in approximately one third of patients with MAS in the setting of SJIA [22–25] and were significantly more common than in patients with SJIA without MAS [22, 25]. This suggests that the distinction between fHLH and rHLH may be less clear and, rather, that HLH exists as a spectrum of disease [24]. However, genetic contributions to infection-triggered HLH have not been well examined. Similarly, the diagnosis of infection-associated HLH/MAS is challenging. Diagnostic criteria for fHLH include results of tests, such as those measuring NK cell function and soluble interleukin 2 (IL-2) receptor α levels, not routinely performed or readily available in the care of critically ill patients [26]. While criteria have been proposed for MAS complicating SJIA, these have not been validated in the setting of acute infections [27].
In this study, 16 patients with fatal H1N1 influenza were examined. We hypothesized that patients with H1N1 influenza and features of MAS would have a high frequency of protein-altering variants in genes implicated in HLH/MAS. Using whole-exome sequencing (WES), we found mutations in the gene encoding perforin, the granule trafficking protein LYST, and other genes associated with MAS. We also show that a perforin variant caused decreased NK cell cytoxicity, suggesting that these variants contribute to MAS etiology in fatal H1N1 influenza.
METHODS
Patient Identification
All autopsy cases between June 2009 and January 2014 with laboratory-confirmed influenza A(H1N1) infection were identified from a single institution, either prospectively or retrospectively, by query of the University of Michigan Hospital Department of Pathology database. No fatal cases were excluded. A subset of autopsies involving cases of H1N1 influenza in this study have been previously described [16]. The diagnosis of H1N1 influenza was made by antemortem polymerase chain reaction testing, except for patient 1, for whom postmortem testing was performed. The presence or absence of hemophagocytosis in each case was confirmed by a board-certified hematopathologist (L. B. S.).
DNA Extraction and WES
Prior to DNA extraction, paraffin was removed using 1 mL of xylene per 20-μm scroll of formalin-fixed paraffin-embedded (FFPE) tissue. DNA was extracted from these FFPE samples, using the Recover all Total Nuclei Acid isolation kit (Life Technologies, Grand Island, New York). Recovered DNA was treated with RNase A to remove contaminating RNA. The DNA quantity was assessed on the Qubit Fluorometer (Life Technologies) and analyzed on a 1% agarose gel to evaluate sample quality. Two DNA samples (from patients 15 and 16) were of inadequate quality for WES or Sanger sequencing. The library preparation, postcapture steps, and sequencing were performed by PerkinElmer (Branford, Connecticut). The Agilent SureSelectXT Target enrichment work flow was used to generate the library, followed by sequencing with the Illumina HiSeq 2000, using 100–base pair paired end sequencing.
Sequencing Alignment and Variant Filtering
Sequencing reads in FASTQ format were filtered by quality scores. Quality reads were aligned to the human genome reference sequence GRCh37 with NextGene software v 2.3 (SoftGenetics, State College, Pennsylvania). Ninety-three percent of reads were successfully matched to the human genome reference sequence (GRCh37). Coverage statistics for the library (Agilent V4 51Mb) were calculated using NextGene Software (SoftGenetics). The coverage sequence depth was 118-fold, with >95% of the target bases covered at least 10-fold (Supplementary Table 1). On average, 46 million paired reads were generated for each sample, with a minimum of 10-fold coverage for >90% of exomes (Supplementary Table 1). Sequence variants were filtered by Ingenuity Variant Analysis. All protein-altering variants in HLH-associated genes were identified. Sequence alignments supporting variant calls were then visually inspected for quality to rule out common sequence alignment errors. Variant calls due to mismatches primarily occurring at ends of reads were classified as “likely false positive.” All putative variants in HLH-linked genes were confirmed by Sanger sequencing (Supplementary Table 2). For variants in MAS-associated genes, all previously reported rare protein-altering variants (defined as those with a minor allele frequency of <0.5% in 1000 Genomes and Exome Aggregation Consortium) were identified.
DNA Constructs
Complementary DNA (cDNA) encoding wild-type (WT) human PRF1 was generated by reverse transcription from RNA of the human NK-92 NK cell line, which expresses endogenous human WT PRF1. The cDNA was cloned into an expression vector, and the WT sequence was confirmed by DNA sequencing. The patient-derived PRF1 mutant cDNA (mutation site, c.272C > T) was generated from the WT PRF1 cDNA by site-directed mutagenesis as described elsewhere [28] and confirmed by DNA sequencing. The lentiviral expression vector z-368-ΔNP and the packaging plasmid Δ8.91 were kindly provided by Dr Philip Zoltick (The Children's Hospital of Philadelphia, Pennsylvania) [29]. Both WT and mutant PRF1 cDNAs were independently subcloned into z-368-ΔNP to help generate recombinant lentiviruses. These viruses were separately transduced into NK-92 cells as detailed below. Transduction efficiencies were monitored by coexpression of green fluorescent protein (GFP), detected by flow cytometry. There was no decrease in perforin-1 levels noted (data not shown).
Lentiviral Preparation and Transduction
HEK293T cells were transfected with the respective z-368-ΔNP expression constructs, along with Δ8.91 and pVSV-G, using the FuGENE HD transfection reagent (Roche, Branford, Connecticut). Lentiviral production was concentrated using the lenti-X concentrator reagent (Clontech, Mountain View, California). NK-92 cells were infected with lentiviruses overnight at a multiplicity of infection of 50:1. Transduction efficiency was 45%–55% on day 5 and afterward for up to 4 weeks. All assays were performed 2–3 weeks following lentiviral infection.
Cytotoxicity Assays
The NK-sensitive K562 erythroleukemia target cells were labeled by the cell tracer dye eFluor450 (eBioscience, San Diego, California) 12 hours prior to the cytotoxicity assay. The GFP-expressing, lentiviral-transduced NK-92 cells and labeled K562 target cells were mixed together at different effector to target (E:T) cell ratios and incubated at 37°C for 4 hours. The cells were then stained with live/dead near-IR dye (Invitrogen, Carlsbad, California) and analyzed by flow cytometry (LSRII; BD Biosciences, San Jose, California).
RESULTS
Clinical and Laboratory Features of HLH/MAS
Sixteen adult patients were identified with fatal influenza A(H1N1) infection (Table 1). Their clinical presentations were initially typical for influenza, with fever, chills, cough, dyspnea, malaise, myalgias, and diarrhea (Supplementary Table 3). None of the patients had prior histories suggestive of immunodeficiency, but all required intensive care with mechanical ventilation and circulatory support (Supplementary Table 3). Their hospital courses were complicated by a variety of comorbid infections (Supplementary Table 3). Eighty-one percent (13) had evidence of hemophagocytosis in their bone marrow, spleen, and/or lymph nodes (Figure 1). The majority of patients also had thrombocytopenia (for 69%, platelet counts were <100 × 109 platelets/L; for 94%, counts were <150 × 109 platelets/L) and elevated liver enzyme levels (75%), both of which are commonly reported in HLH/MAS [31]. Several patients also had abnormalities in serum ferritin, fibrinogen, or triglyceride levels, although these were not measured in most patients. Taken together, 31% (5) met ≥4 HLH-2004 diagnostic criteria, which is considered consistent with rHLH (Supplementary Table 4) [32]. Notably, none of the patients had testing performed for 2 of 8 diagnostic criteria (NK cell functional testing and soluble IL-2 receptor α level). When the 2009 modification of these criteria are applied, 44% (7) met diagnostic criteria for HLH (Supplementary Table 4) [13]. A diagnostic scoring system for adults with rHLH, termed the HScore, has recently been reported, and, using the optimal cutoff reported, 44% of patients (7) scored positive (Supplementary Table 4) [30]. Finally, recently proposed consensus classification criteria for MAS require a serum ferritin measurement, which was only performed for 4 patients; excluding the ferritin requirement, 81% (13) met modified MAS classification criteria (Supplementary Table 4) [33].
Table 1.
Pathologic and Laboratory Findings in Patients With Fatal H1N1 Influenza
| Patient ID | HP | Fever | Cytopenia (Criteria Satisfied)a | Nadir Plt Count, × 109 Plt/L | HSMb | AST Level, U/L | Ferritin Level, mg/L | Triglycerides Level, mg/dL | Fibrinogen Level, mg/L | HLH Criteriac | MAS Criteriad | Mod HLHe | H-Scoref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Sp, BM | Yes | No | 111 | Liver | 5373 | ND | 397 | ND | 3 | Yes–F | No | 154 |
| 2 | Sp, BM, LN | Yes | No (Hb) | 106 | Sp, Liver | 276 | ND | 308 | ND | 4 | Yes–F | Yes | 209 |
| 3 | BM | Yes | Yes (ANC, Plt) | 71 | Liver | 53 | ND | ND | ND | 3 | Yes–F | Yes | 144 |
| 4 | Sp, BM | Yes | No (Plt) | 92 | Sp, Liver | 15479 | ND | 789 | ND | 4 | Yes–F | Yes | 213 |
| 5 | BM, LN | Yes | No | 116 | Liver | ND | Normal | ND | ND | 2 | No | No | 107 |
| 6 | Sp, BM | No | No | 206 | Liver | 454 | 443 | 314 | ND | 2 | No | No | 121 |
| 7 | BM | Yes | No (Plt) | 69 | No | 200 | >3000 | Normal | Normal | 3 | Yes | No | 146 |
| 8 | None | Yes | Yes (Hb, Plt) | 62 | Sp, Liver | 1137 | ND | ND | ND | 3 | Yes–F | No | 140 |
| 9 | BM | Yes | No (Plt) | 44 | No | 7640 | ND | 574 | ND | 3 | Yes–F | No | 191 |
| 10 | None | Yes | Yes (Hb, Plt) | 38 | Sp, Liver | 49 | ND | ND | ND | 3 | Yes–F | No | 140 |
| 11 | Sp, BM, LN | Yes | Yes (Hb, Plt) | 25 | Liver | 397 | ND | 368 | 91 | 4 | Yes–F | Yes | 234 |
| 12 | LN | Yes | Yes (Hb, Plt) | 18 | No | 401 | 1214 | 453 | ND | 5 | Yes | Yes | 201 |
| 13 | None | Yes | Yes (Hb, Plt) | 54 | Sp, Liver | 2931 | ND | ND | ND | 3 | Yes–F | No | 140 |
| 14 | Sp, BM | Yes | No (Plt) | 15 | Liver | ND | ND | ND | ND | 2 | Yes–F | No | 115 |
| 15 | BM | Yes | Yes (Hb, Plt) | 99 | Sp, Liver | ND | ND | 237 | ND | 4 | Yes–F | Yes | 200 |
| 16 | BM, LN | Yes | Yes (ANC, Plt) | 27 | Liver | 1086 | ND | ND | ND | 3 | Yes–F | Yes | 160 |
Abbreviations: ANC, absolute neutrophil count; BM, bone marrow; Hb, hemoglobin; HLH, hemophagocytic lymphohistiocytosis; HP, hemophagocytosis; HSM, hepatosplenomegaly; LN, lymph node; MAS, macrophage activation syndrome; ND, no data; Plt, platelet; Sp, spleen.
a Cytopenia was defined as satisfaction of ≥2 HLH-2004 diagnostic criteria: Hb level, <90 g/L; platelet count, <100 × 109 platelets/L; and neutrophil count, <1.0 × 109 neutrophils/L.
b As determined by organ size at autopsy.
c Number of HLH-2004 diagnostic criteria met.
d Based on proposed MAS classification criteria. Yes–F indicates that the ferritin level was not measured but patients otherwise satisfied classification criteria.
e Based on HLH-2004 criteria modified by Filipovich [13].
f As described by Fardet et al [30]; optimal cutoff, 169.
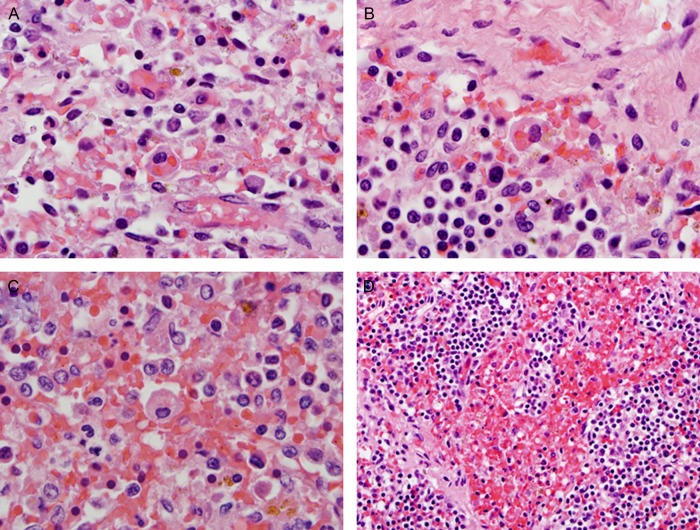
Figure 1.
Representative examples of hemophagocytosis characterized by macrophages with engulfed red blood cells in lymph node (A and B) and spleen (C and D) in fatal cases of H1N1 influenza. Hematoxylin-eosin stain was used (1000× [oil; A–C] or 400× [D] original magnification).
Variants in Known fHLH-Associated Genes
To examine genetic variants associated with fatal H1N1 influenza, WES was performed on DNA extracted from FFPE tissue obtained at autopsy. For 2 (patients 15 and 16), insufficient DNA was obtained to perform WES. Initial efforts were focused on identifying rare, nonsynonymous variants in genes associated with fHLH. Rare variants in HLH-associated genes were identified in 36% patients (5 of 14) with fatal H1N1 influenza who underwent sequencing (Table 2), all of whom had evidence of hemophagocytosis. Two patients were heterozygous for the p.A91V PRF1 mutation. This perforin gene variant, with a MAF of 1%–4%, has been reported in patients with fHLH, including late-onset HLH, as well as in patients with SJIA who developed MAS [22, 34]. Additionally, 5 patients carried rare (MAF, < 0.6%), protein-altering variants in LYST, the causative gene of Chediak–Higashi syndrome and HLH [35]. Three patients were heterozygous carriers for the same LYST p.T1982I (rs146591126, 0.6%) missense mutation. This amino acid position is moderately conserved, and, although this mutation is predicted to have modest effects on protein function, it was detected significantly more frequently in our population than predicted (P < .0001, by the χ2 test). Additionally, 2 other patients were heterozygous carriers for LYST (c.8913T > G/p.N2971K; rs34702903; 0.2%) and LYST (c.11086G > A/p.V3696I; rs147221131; 0.08%). The functional significance of these variants is unknown. Interestingly the 2 patients who carried the PRF1 p.A91V mutation described above were also carriers of LYST mutations (p.T1892I and p.V3696I). There were no rare protein-altering variants identified in UNC13D, STX11, STXBP2, or RAB27A.
Table 2.
Variants in Hemophagocytic Lymphohistiocytosis (HLH) Genes
| Sample Source | Gene | Variant | Identifier | Variant Type | Prediction SIFT/Polyphen | 1000 Genomes MAF | ExAC MAF | Conservation (PhyloP) |
|---|---|---|---|---|---|---|---|---|
| Patients 7 and 11 | PRF1 | c.272C > T(p.A91V) | rs35947132 | Missense | Damaging/Probably damaging | 0.013 | 0.031 | 3.6 |
| Patient 4 | LYST | c.8913T > G(p.N2971K) | rs34702903 | Missense | Tolerated/Possibly damaging | 0.0006 | 0.002 | 0.77 |
| Patients 1, 11, and 14 | LYST | c.5945C > T(p.T1982I) | rs146591126 | Missense | Tolerated/Benign | 0.0016 | 0.006 | 3.03 |
| Patient 7 | LYST | c.11086G > A(p.V3696I) | rs147221131 | Missense | Tolerated/Benign | 0.0002 | 0.0008 | 2.87 |
Transcript accession numbers were as follows: PRF1 NM_001083116.1, LYST NM_000081.3.
Abbreviation: MAF, minor allele frequency.
Perforin p.A91V Variant Confers Decreased NK Cell Cytotoxicity
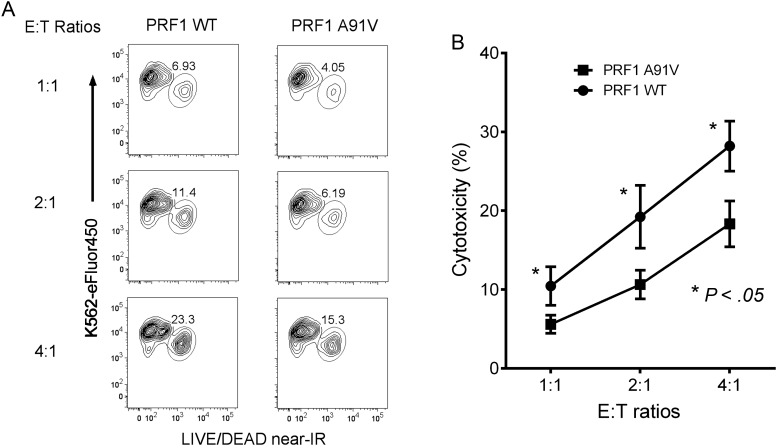
Controversy exists as to whether the PRF1 p.A91V mutation results in decreased cytolytic activity [36–38]. It has even been suggested that the p.A91V change helps to stabilize the perforin molecule [39]. To test the functional significance of the p.A91V PRF1 mutation in a controlled fashion without in vivo influences [40], we cloned the PRF1 WT and p.A91V mutant cDNA constructs into lentiviral expression vectors (which coexpress GFP) for introduction into a highly relevant cell type, the human NK cell line NK-92. GFP-expressing transduced NK-92 cells were incubated with dye-loaded K562 target cells at graded E:T ratios for 4 hours prior to analysis of cell death by flow cytometry. The PRF1 p.A91V mutant demonstrated 35%–45% reduction in NK cell lysis of K562 cells at all E:T ratios in comparison to WT PRF1-transduced NK-92 cells (Figure 2A). Multiple repeated experiments revealed statistically significant decreased NK cell cytolytic activity all E:T ratios examined (Figure 2B). Thus, consistent with a recent report analyzing patient-derived heterozygous PRF1 p.A91V-expressing NK cells [41], the p.A91V mutant notably impaired NK cell cytolytic function and likely contributed to MAS pathophysiology.
Figure 2.
PRF1 p.A91V mutation decreases natural killer (NK) cell cytolytic function. A, eFluor450-labeled K562 target cells were mixed at increasing effector to target (E:T) cell ratios with NK-92 cells transduced with lentiviruses expressing PRF1 wild type (WT; left), or PRF1 p.A91V (right) and were incubated for 4 hours prior to flow cytometry of K562 cell death. Flow cytometry plots were gated on eFluor450-expressing cells, with eFluor450 depicted on the y-axis and near–infrared (IR) staining (dead cells) along the x-axis. One representative experiment is shown, with percentages of lysed K562 cells enumerated. B, Percentage cytotoxicity at 3 different E:T cell ratios for lentivirus-transduced NK-92 cells (PRF1 WT [circles] and PRF1 p.A91V [squares]). Results are plotted as means ± standard error of the mean for 5 independent experiments. Two-way analysis of variance revealed statistically significant differences (P < .05) between the PRF1 WT– and PRF1 p.A91V mutant–transduced NK-92 cells at all 3 E:T cell ratios.
Variants in Other MAS-Associated Genes
Our recent analysis of WES from patients with MAS in SJIA revealed potentially causative variants in numerous genes not previously implicated in the pathogenesis of HLH [25]. Filtering specifically for these genes, we identified rare protein-altering variants in 36% patients (5 of 14; Table 3), of whom 2 (patients 1 and 11) also had variants in LYST and/or PRF1 described above. Two patients were carriers of rare variants in Xin actin-binding protein 2 (XIRP2), which stabilizes the actin cytoskeleton, and 1 patient was a carrier of a variant in leucine-rich repeats and guanylate kinase-domain containing protein (LRGUK), which is involved in microtubule structure. Intriguingly, biallelic mutations in both XIRP2 and LRGUK were previously identified in children with MAS and SJIA [25]. Patient 11, who carried the PRF1 p.A91V allele, also carried 2 protein-altering mutations in MEFV, the causative gene of familial Mediterranean fever. These 2 variants, p.P369S and p.R408Q, are typically found in cis and associated with clinical features consistent with autoinflammatory disease [42]. Interestingly, we have observed these same MEFV mutations in a pediatric patient with periodic fevers and MAS; this child also carried a single disease-associated RAB27A mutation (A. A. Grom, unpublished data). This specific RAB27A mutation (c.259G > C/p.A87P) has also recently been found to delay NK cell lytic activity in 2 unrelated teenagers with MAS (Cron et al, unpublished data). Finally, we also identified mutations in nipped-B homolog (NIPBL) and FAM220A, all of which were found as de novo mutations in patients with MAS [25]. It is interesting to speculate on the potential contributions to HLH pathophysiology of these variants in proteins that might disrupt lymphocyte cytolytic function.
Table 3.
Other Variants of Interest
| Sample Source | Gene | Variant | Variant Type | Prediction SIFT/Polyphen | 1000 Genomes MAF | ExAC MAF | Conservation (PhyloP) |
|---|---|---|---|---|---|---|---|
| Patient 13 | XIRP2 | c.462del (p.S155fs) | Frameshift | NA | Not listed | 0.0005 | NA |
| Patient 10 | XIRP2 | c.7856T > C (p.R2619Q) | Missense | Tolerated/benign | 0.0026 | 0.0030 | 1.34 |
| Patient 6 | NIPBL | c.1965G > T (p.E655D) | Missense | Tolerated/benign | 0.0022 | 0.0010 | 0.53 |
| Patient 9 | FAM220A | c.437C > T (p.P146L) | Missense | Tolerated/benign | 0.0014 | 0.0010 | −0.52 |
| Patient 11 | LRGUK | c.2227T > C (p.C743R) | Missense | Tolerated/benign | 0.0014 | 0.0030 | −0.52 |
| Patient 11 | MEFV | c.1105C > T (p.P369S) + c.1223G > A (p.R408Q) | Missense | Deleterious/probably damaging + tolerated/benign | 0.02 + 0.02 | 0.01423 + 0.01295 | 0.21 + −0.28 |
Transcript accession numbers were as follows: XIRP2 NM_152381.5, NIPBL NM_133133.3, FAM220A NM_001037163.1, LRGUK NM_144648.1, MEFV NM_000243.
Abbreviations: MAF, minor allele frequency; NA, not applicable.
DISCUSSION
Patients with life-threatening H1N1 influenza can manifest features of HLH/MAS, including hyperinflammation, pancytopenia, coagulopathy, and liver dysfunction; however, the frequency of these findings and their pathologic basis remains undefined. Here, we describe 16 patients with fatal H1N1 influenza, of whom 81% had evidence of hemophagocytosis at autopsy, and 31%–81% satisfied modified diagnostic criteria for HLH/MAS. We also found that 36% of these patients were carriers of rare, protein-altering variants in genes associated with fHLH. Furthermore, patients carried mutations previously reported in MAS. This work also shows that the PRF1 variant found in 2 patients with H1N1 influenza impairs NK cell cytotoxicity, which provides a link between these rare mutations and rHLH/MAS pathophysiology. Together, these data demonstrate that a rHLH/MAS process occurs in a sizable number of fatal influenza A(H1N1) virus infections. Furthermore, the high frequency of mutations in HLH-associated genes suggests a genetic predisposition to developing a hyperinflammatory syndrome in the setting of severe influenza A(H1N1) infection.
There are no widely accepted and validated diagnostic criteria for rHLH. fHLH is diagnosed using the HLH-2004 criteria, by either defined genetic lesions or a combination of clinical and laboratory information, including NK cell functional testing, soluble IL-2 receptor levels, and bone marrow examination [26]. These criteria are limited by their reliance on esoteric testing and are widely viewed as insensitive for the diagnosis of rHLH. Indeed, in children with SJIA, the presence of at least 4 of these criteria was 100% specific but only 35% sensitive for the diagnosis of MAS [43]. Two other proposed systems, a modification of the HLH-2004 criteria [13] and the HScore [30], together classify 50% of patients in the present study with fatal H1N1 influenza as having rHLH, despite lacking results of several laboratory tests. Finally, a recent international consortium used expert consensus plus analysis of actual patient data to define classification criteria for MAS in the setting of SJIA [33]. The proposed criteria require a measurement of the serum ferritin level, which is lacking in the majority of the patients with H1N1 influenza in the present study, but absent its requirement these criteria classify as many as 81% as having MAS. Regardless of criteria, it seems clear that a sizable number of patients with fatal influenza A(H1N1) infection develop rHLH and that critically ill patients with H1N1 influenza should be carefully evaluated for rHLH/MAS.
fHLH has been linked to mutations in the cytolytic pathway, notably the genes encoding perforin and proteins essential for the trafficking and fusion of perforin-containing granules. While the pathogenesis of rHLH is incompletely understood, several recent reports have shown that children who develop MAS in the setting of SJIA have a high frequency of mutations in these same genes [22–25]. Here, we found that 36% of adult patients with fatal H1N1 influenza similarly carried rare, protein-altering mutations in genes implicated in fHLH, namely PRF1 and LYST. Two patients were heterozygous carriers of the p.A91V mutation in PRF1. Homozygous p.A91V mutations and heterozygotes who carry a second cytolytic pathway mutation have been reported among children with fHLH [44, 45]; single mutations have also been found in cases of adult-onset HLH [33] and in children with SJIA who developed MAS [22]. However, p.A91V occurs in at least 3% of the US population, with conflicting reports as to its effect on cytolytic activity [46]. Here, we found a clear reduction in human NK cell cytotoxicity when the p.A91V variant was expressed in vitro. This is in agreement with in vitro data from Voskoboinik and colleagues [36], as well as with primary NK cell data from patients heterozygous for this mutation [41]. As perforin forms a multimolecular (19–24 perforin molecules)[47] channel into the target cells, it is possible that heterozygous mutants function as hypomorphic alleles or even partial dominant-negative proteins, as has been shown for other heterozygous mutations in fHLH-associated genes involved in perforin-mediated cytolysis [24, 48].
By comparison, there is comparably little known as to how LYST mutations impact NK cell cytotoxicity, although one report highlights that patients with Chediak–Higashi syndrome have severely diminished NK cell function but relatively preserved CD8+ T-cell function [49]. Interestingly, the adult patients identified in this study, as well as in our prior study of MAS, had missense mutations largely affecting the C-terminal region of the protein; these types of mutations are largely associated with milder clinical phenotypes [50]. Although many of the LYST variants are predicted to be benign mutations, their identification here and in prior studies supports the need for further study of the impact of these rare mutations on NK cell function.
This is the first demonstration of mutations in HLH-associated genes in patients with H1N1-associated rHLH/MAS. We have recently reported a rare PRF1 mutation in a child with the autoinflammatory disease mevalonate kinase deficiency and recurrent MAS [51]. This may indicate that genetic polymorphisms in the cytolytic pathway predispose patients to development of HLH in the setting of hyperinflammation, whether caused by infection, SJIA, or other autoinflammatory disorders. This is highly consistent with a model of HLH pathogenesis proposed by Canna and Behrens and by De Benedetti et al, in which children with severe and/or biallelic HLH-associated mutations present early in childhood with fHLH, while those with single or less severe mutations develop rHLH/MAS only in the setting of significant environmental triggers. This also suggests a genetic disposition to development of severe influenza A(H1N1) infection. Crucially, this also raises the intriguing question of whether patients who develop life-threatening H1N1 influenza will benefit from immunosuppressive therapy for rHLH/MAS [14]. Indeed, there are several reports of severe influenza A(H1N1) infection successfully treated with these approaches [5, 8, 15], indicating that this is likely a fruitful avenue for further prospective trials.
In addition to H1N1, there are a variety of infectious triggers known to be associated with rHLH/MAS, most notably members of the herpes virus family (eg, Epstein-Barr virus, cytomegalovirus, and human herpesvirus 6) [13]. Perhaps viruses that trigger robust immune responses, including hemorrhagic fever viruses, are more likely to trigger rHLH in genetically susceptible individuals. Ultimately, prenatal screening for mutations in common HLH-associated genes may identify a substantial portion (as high as 10%) of the general population at risk for developing MAS/HLH when a threshold of inflammation has been reached from a variety of potential triggers, including H1N1 influenza. The identification of novel genes involved in HLH pathogenesis may even broaden this screening approach.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Financial support. This work was supported by the National Institutes of Health (R01-AR059049 to A. A. G. and K12-HL119986 to G. S. S.), the Kaul Pediatric Research Institute (to R. Q. C.), and the American College of Rheumatology Rheumatology Research Foundation (scientist development award to G. S. S.).
Potential conflicts of interest. A. A. G. has received consulting fees from NovImmune and Roche and has research collaborations with Novartis and NovImmune. R. Q. C. has received consulting fees from AbbVie and Novartis and has a clinical trial planned with SOBI. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Tang JW, Shetty N, Lam TT-Y. Features of the new pandemic influenza A/H1N1/2009 virus: virology, epidemiology, clinical and public health aspects. Curr Opin Pulm Med 2010; 16:235–41. [DOI] [PubMed] [Google Scholar]
- 2.York I, Donis RO. The 2009 pandemic influenza virus: where did it come from, where is it now, and where is it going? Curr Top Microbiol Immunol 2013; 370:241–57. [DOI] [PubMed] [Google Scholar]
- 3.Randolph AG, Vaughn F, Sullivan R et al. . Critically ill children during the 2009–2010 influenza pandemic in the United States. Pediatrics 2011; 128:e1450–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casciaro R, Cresta F, Favilli F, Naselli A, De Alessandri A, Minicucci L. Macrophage activation syndrome induced by A/H1N1 influenza in cystic fibrosis. Pediatr Pulmonol 2014; 49:E10–2. [DOI] [PubMed] [Google Scholar]
- 5.Yöntem Y, Ilker D, Yeşim O et al. . Analysis of fatal cases of pandemic influenza A (H1N1) virus infections in pediatric patients with leukemia. Pediatr Hematol Oncol 2013; 30:437–44. [DOI] [PubMed] [Google Scholar]
- 6.Shrestha B, Omran A, Rong P, Wang W. Report of a Fatal Pediatric Case of Hemophagocytic Lymphohistiocytosis Associated with Pandemic Influenza A (H1N1) Infection in 2009. Pediatr Neonatol 2015; 56:189–92. [DOI] [PubMed] [Google Scholar]
- 7.Ozdemir H, Çiftçi E, Ince EU, Ertem M, Ince E, Doğru U. Hemophagocytic lymphohistiocytosis associated with 2009 pandemic influenza A (H1N1) virus infection. J Pediatr Hematol Oncol 2011; 33:135–7. [DOI] [PubMed] [Google Scholar]
- 8.Unal S, Gökçe M, Aytaç-Elmas S et al. . Hematological consequences of pandemic influenza H1N1 infection: a single center experience. Turk J Pediatr 52:570–5. [PubMed] [Google Scholar]
- 9.To KKW, Hung IFN, Li IWS et al. . Delayed clearance of viral load and marked cytokine activation in severe cases of pandemic H1N1 2009 influenza virus infection. Clin Infect Dis 2010; 50:850–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X-Y, Ye X-W, Feng D-X, Han J, Li D, Zhang C. Hemophagocytic lymphohistiocytosis induced by severe pandemic influenza A (H1N1) 2009 virus infection: a case report. Case Rep Med 2011; doi:10.1155/2011/951910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Y, Yang Y, Zhao W, Wang H. Novel swine-origin influenza A (H1N1) virus-associated hemophagocytic syndrome--a first case report. Am J Trop Med Hyg 2010; 82:743–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravelli A, Grom AA, Behrens EM, Cron RQ. Macrophage activation syndrome as part of systemic juvenile idiopathic arthritis: diagnosis, genetics, pathophysiology and treatment. Genes Immun 2012; 13:289–98. [DOI] [PubMed] [Google Scholar]
- 13.Filipovich AH. Hemophagocytic lymphohistiocytosis (HLH) and related disorders. Hematology Am Soc Hematol Educ Program 2009:127–31. [DOI] [PubMed] [Google Scholar]
- 14.Schulert GS, Grom AA. Pathogenesis of macrophage activation syndrome and potential for cytokine- directed therapies. Annu Rev Med 2015; 66:145–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henter J-I, Palmkvist-Kaijser K, Holzgraefe B, Bryceson YT, Palmér K. Cytotoxic therapy for severe swine flu A/H1N1. Lancet (London, England) 2010; 376:2116. [DOI] [PubMed] [Google Scholar]
- 16.Harms PW, Schmidt LA, Smith LB et al. . Autopsy findings in eight patients with fatal H1N1 influenza. Am J Clin Pathol 2010; 134:27–35. [DOI] [PubMed] [Google Scholar]
- 17.Mauad T, Hajjar LA, Callegari GD et al. . Lung pathology in fatal novel human influenza A (H1N1) infection. Am J Respir Crit Care Med 2010; 181:72–9. [DOI] [PubMed] [Google Scholar]
- 18.Beutel G, Wiesner O, Eder M et al. . Virus-associated hemophagocytic syndrome as a major contributor to death in patients with 2009 influenza A (H1N1) infection. Crit Care 2011; 15:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bode SF, Lehmberg K, Maul-Pavicic A et al. . Recent advances in the diagnosis and treatment of hemophagocytic lymphohistiocytosis. Arthritis Res Ther 2012; 14:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grom AA, Villanueva J, Lee S, Goldmuntz EA, Passo MH, Filipovich A. Natural killer cell dysfunction in patients with systemic-onset juvenile rheumatoid arthritis and macrophage activation syndrome. J Pediatr 2003; 142:292–6. [DOI] [PubMed] [Google Scholar]
- 21.Zhang K, Biroschak J, Glass DN et al. . Macrophage activation syndrome in patients with systemic juvenile idiopathic arthritis is associated with MUNC13–4 polymorphisms. Arthritis Rheum 2008; 58:2892–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vastert SJ, van Wijk R, D'Urbano LE et al. . Mutations in the perforin gene can be linked to macrophage activation syndrome in patients with systemic onset juvenile idiopathic arthritis. Rheumatol 2010; 49:441–9. [DOI] [PubMed] [Google Scholar]
- 23.Bracaglia C, Sieni E, Da Ros M et al. . Mutations of familial hemophagocytic lymphohistiocytosis (FHL) related genes and abnormalities of cytotoxicity function tests in patients with macrophage activation syndrome (MAS) occuring in systemic juvenile idiopathic arthritis (sJIA). Pediatr Rheumatol Online J 2014; 12(suppl 1):P53. [Google Scholar]
- 24.Zhang M, Behrens EM, Atkinson TP, Shakoory B, Grom AA, Cron RQ. Genetic defects in cytolysis in macrophage activation syndrome. Curr Rheumatol Rep 2014; 16:439. [DOI] [PubMed] [Google Scholar]
- 25.Kaufman KM, Linghu B, Szustakowski JD et al. . Whole exome sequencing reveals overlap between macrophage activation syndrome in systemic juvenile idiopathic arthritis and familial hemophagocytic lymphohistiocytosis. Arthritis Rheumatol 2014; 66:3486–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henter JI, Horne A, Arico M et al. . HLH-2004: Diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer 2007; 48:124–31. [DOI] [PubMed] [Google Scholar]
- 27.Ravelli A, Magni-Manzoni S, Pistorio A et al. . Preliminary diagnostic guidelines for macrophage activation syndrome complicating systemic juvenile idiopathic arthritis. J Pediatr 2005; 146:598–604. [DOI] [PubMed] [Google Scholar]
- 28.Selliah N, Zhang M, White S et al. . FOXP3 inhibits HIV-1 infection of CD4 T-cells via inhibition of LTR transcriptional activity. Virology 2008; 381:161–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endo M, Zoltick PW, Peranteau WH et al. . Efficient in vivo targeting of epidermal stem cells by early gestational intraamniotic injection of lentiviral vector driven by the keratin 5 promoter. Mol Ther 2008; 16:131–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fardet L, Galicier L, Lambotte O et al. . Development and validation of the HScore, a score for the diagnosis of reactive hemophagocytic syndrome. Arthritis Rheumatol 2014; 66:2613–20. [DOI] [PubMed] [Google Scholar]
- 31.Minoia F, Davì S, Horne A et al. . Clinical features, treatment, and outcome of macrophage activation syndrome complicating systemic juvenile idiopathic arthritis: a multinational, multicenter study of 362 patients. Arthritis Rheumatol 2014; 66:3160–9. [DOI] [PubMed] [Google Scholar]
- 32.Townsend JL, Shanbhag S, Hancock J, Bowman K, Nijhawan AE. Histoplasmosis-Induced Hemophagocytic Syndrome: A Case Series and Review of the Literature. Open Forum Infect Dis 2015; doi:10.1093/ofid/ofv055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravelli A, Minoia F, Davì S et al. ; Pediatric Rheumatology International Trials Organization; Childhood Arthritis & Rheumatology Research Alliance; Pediatric Rheumatology Collaborative Study Group; Histiocyte Society. Development and initial validation of classification criteria for macrophage activation syndrome complicating systemic juvenile idiopathic arthritis. Arthritis Rheumatol 2015; doi:10.1002/art.39332. [Google Scholar]
- 34.Zhang K, Jordan MB, Marsh RA et al. . Hypomorphic mutations in PRF1, MUNC13–4, and STXBP2 are associated with adult-onset familial HLH. Blood 2011; 118:5794–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbosa MD, Nguyen QA, Tchernev VT et al. . Identification of the homologous beige and Chediak-Higashi syndrome genes. Nature 1996; 382:262–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voskoboinik I, Sutton VR, Ciccone A et al. . Perforin activity and immune homeostasis: the common A91V polymorphism in perforin results in both presynaptic and postsynaptic defects in function. Blood 2007; 110:1184–90. [DOI] [PubMed] [Google Scholar]
- 37.Risma KA, Frayer RW, Filipovich AH, Sumegi J. Aberrant maturation of mutant perforin underlies the clinical diversity of hemophagocytic lymphohistiocytosis. J Clin Invest 2006; 116:182–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zur Stadt U, Beutel K, Weber B, Kabisch H, Schneppenheim R, Janka G. A91V is a polymorphism in the perforin gene not causative of an FHLH phenotype. Blood 2004; 104:1909. [DOI] [PubMed] [Google Scholar]
- 39.An O, Gursoy A, Gurgey A, Keskin O. Structural and functional analysis of perforin mutations in association with clinical data of familial hemophagocytic lymphohistiocytosis type 2 (FHL2) patients. Protein Sci 2013; 22:823–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cifaldi L, Prencipe G, Caiello I et al. . Inhibition of Natural Killer Cell Cytotoxicity by Interleukin-6: Implications for the Pathogenesis of Macrophage Activation Syndrome. Arthritis Rheumatol 2015; 67:3037–46. [DOI] [PubMed] [Google Scholar]
- 41.House IG, Thia K, Brennan AJ et al. . Heterozygosity for the common perforin mutation, p.A91V, impairs the cytotoxicity of primary natural killer cells from healthy individuals. Immunol Cell Biol 2015; 93:575–80. [DOI] [PubMed] [Google Scholar]
- 42.Ryan JG, Masters SL, Booty MG et al. . Clinical features and functional significance of the P369S/R408Q variant in pyrin, the familial Mediterranean fever protein. Ann Rheum Dis 2010; 69:1383–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davì S, Minoia F, Pistorio A et al. . Performance of current guidelines for diagnosis of macrophage activation syndrome complicating systemic juvenile idiopathic arthritis. Arthritis Rheumatol 2014; 66:2871–80. [DOI] [PubMed] [Google Scholar]
- 44.Zhang K, Chandrakasan S, Chapman H et al. . Synergistic defects of different molecules in the cytotoxic pathway lead to clinical familial hemophagocytic lymphohistiocytosis. Blood 2014; 124:1331–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cetica V, Sieni E, Pende D et al. . Genetic predisposition to hemophagocytic lymphohistiocytosis: Report on 500 patients from the Italian registry. J Allergy Clin Immunol 2015; doi:10.1016/j.jaci.2015.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voskoboinik I, Trapani JA. Perforinopathy: a spectrum of human immune disease caused by defective perforin delivery or function. Front Immunol 2013; 4:441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Law RHP, Lukoyanova N, Voskoboinik I et al. . The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature 2010; 468:447–51. [DOI] [PubMed] [Google Scholar]
- 48.Spessott WA, Sanmillan ML, McCormick ME et al. . Hemophagocytic lymphohistiocytosis caused by dominant-negative mutations in STXBP2 that inhibit SNARE-mediated membrane fusion. Blood 2015; 125:1566–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jessen B, Maul-Pavicic A, Ufheil H et al. . Subtle differences in CTL cytotoxicity determine susceptibility to hemophagocytic lymphohistiocytosis in mice and humans with Chediak-Higashi syndrome. Blood 2011; 118:4620–9. [DOI] [PubMed] [Google Scholar]
- 50.Karim MA, Suzuki K, Fukai K et al. . Apparent genotype-phenotype correlation in childhood, adolescent, and adult Chediak-Higashi syndrome. Am J Med Genet 2002; 108:16–22. [PubMed] [Google Scholar]
- 51.Schulert GS, Bove K, McMasters R, Campbell K, Leslie N, Grom AA. Mevalonate kinase deficiency associated with recurrent liver dysfunction, macrophage activation syndrome and perforin gene polymorphism. Arthritis Care Res 2014; 67:1173–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.