Abstract
The selectivity with which a biomolecule can bind its cognate ligand when confronted by the vast array of structurally similar, competing ligands that are present in the cell underlies the fidelity of some of the most fundamental processes in biology. Because they collectively comprise one of only a few methods that can sensitively detect the ‘encounter’ complexes and subsequent intermediate states that regulate the selectivity of ligand binding, single-molecule fluorescence, and particularly single-molecule fluorescence resonance energy transfer (smFRET), approaches have revolutionized studies of ligand-binding reactions. Here, we describe a widely used smFRET strategy that enables investigations of a large variety of ligand-binding reactions, and discuss two such reactions, aminoacyl-tRNA selection during translation elongation and splice site selection during spliceosome assembly, that highlight both the successes and challenges of smFRET studies of ligand-binding reactions. We conclude by reviewing a number of emerging experimental and computational approaches that are expanding the capabilities of smFRET approaches for studies of ligand-binding reactions and that promise to reveal the mechanisms that control the selectivity of ligand binding with unprecedented resolution.
Keywords: Encounter Complex, Fidelity, Single-Molecule Fluorescence Resonance Energy Transfer, Ribosome, Spliceosome
1. Introduction
The ability of a biomolecule to selectively bind its cognate ligand when faced with the vast array of structurally similar near- and non-cognate ligands present in the cellular environment is critical for maintaining the fidelity of biological processes. Specific examples include critical processes spanning all of biology: a transcription factor binding a DNA promoter site, a small nuclear ribonucleoprotein (snRNP) binding a precursor mRNA (pre-mRNA) splice site, a microRNA binding a target mRNA, or a ribosome binding an aminoacyl-tRNA (aa-tRNA) substrate. Such specificity of ligand binding is typically achieved through multistep, ligand-binding reactions in which the first step encompasses the reversible formation of a nearly non-specific, weakly interacting, transient, intermediate ‘encounter’ complex between the biomolecule and a potential ligand [1]. Subsequent steps lead to the formation of a final biomolecule-ligand complex [1,2]. Multistep ligand-binding reactions such as these are governed by free-energy landscapes such as the one shown in Figure 1, which depicts a minimal, two-step, ligand-binding reaction. Regardless of the number of steps, formation of an encounter complex reduces the search that is required to form the final biomolecule-ligand complex from three spatial dimensions to two [1]. This results in a lowering of the considerably large entropic barrier(s) that would otherwise separate the unbound state of the biomolecule from the bound state and potentially allows the search for the bound state to be guided along a primarily enthalpic energy funnel [1–4]. As a consequence, formation of an encounter complex significantly increases the rate with which biomolecules can conformationally screen potential ligands. In addition, the weakly interacting, transient, and reversible nature of an encounter complex allows it to rapidly dissociate into its component biomolecule and potential ligand. The reversibility and selectivity of ligand-binding reactions can therefore be precisely regulated by coupling the identity of the ligand to the probability of dissociating versus the probability of proceeding along the reaction pathway. Near- and non-cognate ligands will have a higher probability of dissociating while cognate ligands will have a higher probability of proceeding along the reaction pathway and forming the final biomolecule-ligand complex. We note here that more complex ligand-binding reactions encompassing three or more steps allow additional opportunities for the ligand to dissociate from the post-transition state (‡EC-B in Figure 1) intermediate states [2]. This allows for editing mechanisms, such as kinetic proofreading [5], kinetic enhancement [6], or double-feature selection [4], all of which can greatly increase the specificity of ligand binding without significantly decreasing the rate of the ligand-binding reaction.
Figure 1.
Schematic of a ligand-binding reaction studied by smFRET. The minimal, three-state, energy landscape describes the energy of a ligand-binding reaction along the reaction coordinate. The free biomolecule and ligand begin in the unbound state, and may collide and cross an initial energy barrier (‡U-EC) to form an encounter complex. The encounter complex may then either dissociate to reestablish the free biomolecule and ligand or cross a second energy barrier (‡EC-B) to form the final bound state. The energy landscape is projected onto a plane below it, where the hypothetical trajectory of a single ligand-binding reaction is depicted diffusing along the reaction coordinate (gray curved line). Below this trajectory, a hypothetical signal versus time trajectory corresponding to the ligand-binding reaction trajectory is shown, where the signal of the unbound state, encounter complex, and bound state are denoted (red horizontal lines). The reaction is initiated at t = 0, cross ‡U-EC several times to transiently sample the encounter complex, and ultimately crosses ‡EC-B to form the bound state.
Although they play a decisive role in ensuring that biomolecules can rapidly and selectively bind their cognate ligands, the encounter complexes and, if present, subsequent intermediate states that form during ligand-binding reactions have traditionally been very difficult to experimentally observe and characterize [1]. This is because the stochastic nature of ligand-binding kinetics and the transient nature of the encounter complexes and subsequent intermediate states that are formed give rise to a situation in which such complexes and states comprise exceedingly low-population states that are extremely difficult to detect using traditional, ensemble biophysical techniques that report only on the average behavior of the entire ensemble. Further exacerbating this situation is the fact that, due to their partially non-specific nature, encounter complexes and, possibly, subsequent early intermediate states, can exhibit significant heterogeneity in their structures and other physical properties. Despite these challenges, the emergence in the early 2000s of nuclear magnetic resonance (NMR) spectroscopic techniques, particularly paramagnetic relaxation enhancement, that enable the detection of low-population states have allowed the partial characterization of several encounter complexes and/or subsequent intermediate states formed during ligand-binding reactions [7–9]. Almost simultaneously, the advent of single-molecule biophysical techniques, particularly single-molecule fluorescence approaches, has provided a powerful complement to NMR spectroscopic methods. Single-molecule techniques allow stochastic transitions to rarely and transiently sampled states to be directly observed, thereby allowing those states that were difficult to detect in ensemble biophysical experiments to be sensitively detected and comprehensively characterized [10]. Moreover, the observation of individual transitions to and from a state of interest allows heterogeneities in the physical properties of that state to be identified, sorted, and analyzed [11,12].
Despite the tremendous promise that single-molecule fluorescence approaches hold for studies of encounter complexes and subsequent intermediate states formed during ligand-binding reactions, these approaches are often limited by (i) data collection throughput that can be too low for sufficient statistical analysis, (ii) time resolutions that can be too slow to capture exceedingly transient states, (iii) limitations in spatial resolution that can make it difficult to determine when a ligand is co-localized to a biomolecule, and (iv) the ‘concentration barrier’ generated by the background fluorescence from the relatively high concentrations of fluorophore-labeled ligands that are required to observe ligand binding events within the experimental observation time that compromises the high signal-to-background ratio that is required to sensitively observe the fluorescence from a single molecule [13,14]. Notably, single-molecule fluorescence resonance energy transfer (smFRET) approaches are particularly powerful for studies of ligand-binding reactions [15–17], as they ameliorate several of these limitations, and are able to directly report on both the spatial localization as well as the conformational dynamics of biomolecules and/or ligands [18]. We begin this article by providing a brief description of the basic experimental platform that is typically used to investigate ligand-binding reactions using smFRET and then highlight two representative examples of ligand-binding reactions that have been studied using smFRET. We then close by reviewing emerging experimental and computational developments that are expanding the capabilities of smFRET experiments to characterize the encounter complexes and subsequent intermediate states that regulate the rate and specificity of ligand-binding reactions at an ever-increasing level of detail. Given the large number of ligand-binding reactions that are the subject of ongoing smFRET studies and of emerging techniques that are currently being developed, we wish to apologize to those colleagues whose work we were unable to review due to space constraints.
2. smFRET studies of ligand-binding reactions
2.1 General experimental platform
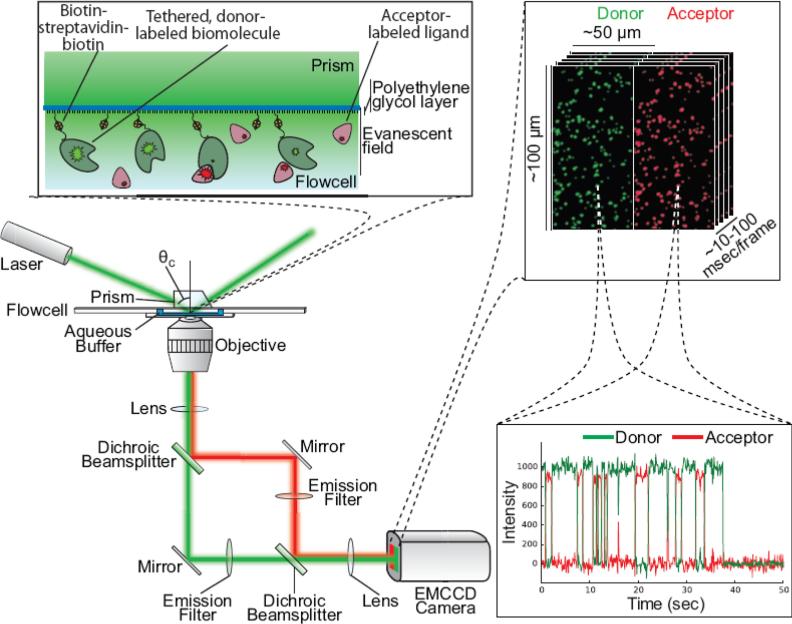
Fluorescence resonance energy transfer (FRET) is a dipole-dipole interaction in which the excitation of a donor fluorophore (e.g., Cy3) can be transferred to an acceptor fluorophore (e.g., Cy5) with an efficiency (termed the FRET efficiency, or EFRET) that, among other variables, depends monotonically on the distance between the fluorophores [19,20]. Thus, EFRET can be interpreted as a “spectroscopic ruler” (over a fluorophore pair-specific range of distances that is typically in the tens of Å) [21]. The EFRET of an energy-transfer event can be quantified by directly exciting the donor; measuring the fluorescence intensities of both fluorophores; and calculating the EFRET using the relationship EFRET = IA / (IA + ID), where IA and ID are the fluorescence intensities of the acceptor and the donor, respectively. Perhaps the most common experimental platform for studying ligand-binding reactions using smFRET (Fig. 2) involves tethering a biotin-derivatized, donor-labeled biomolecule of interest to the surface of a microfluidic, observation flowcell via a biotin-streptavidin-biotin bridge formed with biotin-derivatized polyethylene glycol (PEG) that has been used to functionalize the surface of the flowcell [22,23]. Introduction of an acceptor-labeled ligand into the imaging buffer within the flowcell and imaging using wide-field total internal reflection fluorescence (TIRF) microscopy [24–26] enables direct excitation of the surface-localized donors and simultaneous observation of both the donor- and acceptor fluorescence intensities originating from hundreds of individual biomolecules [23,27]. In such experiments, an anti-correlated donor- and acceptor fluorescence intensities versus time trajectory that exhibits single-step fluorophore photobleaching originating from diffraction-limited donor and acceptor spots serves as unambiguous evidence for resonance energy transfer arising from the encounter of a single, acceptor-labeled ligand with a single, surface-tethered, donor-labeled biomolecule. Importantly, the acceptor will only fluoresce when it is within tens of Å of the donor (i.e., when the ligand is bound to the biomolecule). Thus, smFRET enables the detection of biomolecule-ligand encounters in the presence of relatively higher, tens to one hundred nM, concentrations of fluorophore-labeled ligand in the imaging buffer than is possible with other single-molecule fluorescence microscopy approaches – thereby partially alleviating the limitations imposed by the concentration barrier described in Section 1 [28]. In addition to reporting on ligand-biomolecule encounters, smFRET can also report on conformational changes of the biomolecule and/or ligand that result in changes in the distance between the positions of the donor and acceptor. In summary, because it is able to simultaneously and sensitively report on hundreds of single biomolecule-ligand encounters at fluorophore-labeled ligand concentrations that are higher than is possible with other single-molecule fluorescence approaches, as well as because it can report on conformational changes that take place during the ligand-binding reaction, the TIRF-based smFRET experimental platform has been particularly successful for investigating the kinetic and thermodynamic properties of ligand-binding reactions [10].
Figure 2.
Schematic of a ligand-binding reaction studied by smFRET using a typical TIRF microscope. A laser excitation source is totally internally reflected (TIR) at the interface formed between the surface of the quartz, microfluidic, observation flowcell to which the donor-labeled biomolecules are tethered and the aqueous imaging buffer in the flowcell. The evanescent field that is generated by TIR propagates into the imaging buffer and decays exponentially as a function of increasing distance from the quartz–buffer interface, thereby selectively exciting only those donors that are localized within ~300 nm of the quartz–buffer interface (top left inset). Donor and acceptor fluorescence emission is collected by an objective, separated by wavelength using dichroic mirrors, and detected using an electron-multiplying charge-coupled device (EMCCD) camera (bottom left inset). The separated donor and acceptor fluorescence intensities reporting on the binding of acceptor-labeled ligands to individual, spatially resolved, donor-labeled biomolecules can then be quantified and plotted as a function of time (bottom right inset). Figure and caption adapted from ref. [174].
2.2 aa-tRNA selection during translation elongation
In all organisms, mRNAs are translated into proteins by the ribosome, a 2.5-4.3 MDa [29,30] molecular machine that is composed of two ribonucleoprotein subunits. During translation, aa-tRNAs are delivered to the ribosomal aa-tRNA binding (A) site by the guanine triphosphatase (GTPase) elongation factor Tu (EF-Tu) as part of an EFTu(GTP)aa-tRNA ternary complex (TC) (Fig. 3A) [31–35]. Despite the pool of 41-55 different aa-tRNA isoacceptors that is found in cells [36], the ribosome is able to accurately select the aa-tRNA whose anticodon correctly basepairs to the mRNA codon at the A site (i.e., the cognate aa-tRNA), only misincorporating aa-tRNAs with one-base mismatches (i.e., near-cognate aa-tRNAs) or with two-base or greater mismatches (i.e., non-cognate aa-tRNAs) with a frequency of 1 in 103-104 [37–40]. Notably, this level of fidelity greatly exceeds the maximum misincorporation frequency of 1 in 102 that would be expected from just the thermodynamic stability differences between the anticodoncodon interactions formed by a cognate aa-tRNA and those formed by the corresponding near-cognate aa-tRNAs [41–43]. Extensive studies of the mechanisms through which the ribosome achieves the observed high fidelity of aa-tRNA selection have led to the development of a widely accepted, multistep mechanism [44,45] in which kinetic proofreading [5,6,46–48] and induced fit [48,49] strategies are employed to increase the otherwise relatively low fidelity of aa-tRNA selection that would be expected [33].
Figure 3.
(A) Cartoon schematic of the mechanism of aa-tRNA selection during translation elongation. The EFRET values for the states that are observed by smFRET are denoted below the corresponding states. Dashed arrows represent steps believed to have exceedingly low probabilities of occurring. smFRET experiments have utilized both tRNA-tRNA and tRNA-ribosome donor-acceptor labeling schemes. This mechanistic scheme is based on a similar scheme appearing in ref. [35]. (B) Cartoon schematic of the mechanism of splice site selection during spliceosome assembly and the subsequent splicing reaction that converts pre-mRNA into mRNA. Single-molecule fluorescence experiments have utilized numerous fluorophore labeling schemes involving fluorophore-labeled U1, U2, U5, NTC, and/or pre-mRNA. Again, dashed arrows represent steps believed to have exceedingly low probabilities of occurring. This mechanistic scheme is based on a similar scheme appearing in ref. [175].
Over the past decade, the mechanism of aa-tRNA selection has been the subject of numerous smFRET studies [50–55]. Several aspects of the experimental systems available for studying aa-tRNA selection provide important advantages for designing, implementing, and interpreting smFRET experiments. These advantages include: (i) the involvement of interactions between only two molecular subcomplexes, the TC and a ribosomal elongation complex (REC); (ii) the availability of a reconstituted in vitro translation system composed of a full set of purified components that allows individual components and steps of the reaction to be manipulated (reviewed in ref. [56]); (iii) the existence of a large number of small-molecule inhibitors that enable inhibition of specific and well-defined steps of the reaction [57–59]; and, (iv) the availability of a series of cryogenic electron microscopy (cryo-EM) and X-ray crystallography structures that approximate the structures of the initial and final states of the reaction, as well as those of several intermediate states (reviewed in refs. [31,35]).
smFRET studies of aa-tRNA selection are typically performed using RECs that are biotinylated at the 5’ end of the mRNA and that have been labeled with a donor fluorophore either within the fMet-tRNAfMet that is bound at the ribosomal peptidyl-tRNA binding (P) site [50,60] or within ribosomal protein L11 [52,61]. RECs are then tethered to the surface of a microfluidic, observation flowcell via their 5’-biotinylated mRNA such that they can be imaged with single-molecule resolution using TIRF microscopy. Stopped-flow delivery of a TC carrying an acceptor-labeled aa-tRNA to an REC carrying a donor-labeled P-site tRNA and a cognate A-site codon then yields acceptor- and donor intensities versus time trajectories that are used to calculate pre-steady-state EFRET versus time trajectories. EFRET versus time trajectories initiate at zero-EFRET and evolve through transiently sampled low- and mid-EFRET states before arriving at a high-EFRET final state that is consistent with structures approximating the final state of the reaction in which the aa-tRNA has been accommodated into the A site (Fig. 3A) [50–55]. The mid-EFRET state has been assigned to a mixture of at least two intermediate states that had been previously observed in biochemical [48] and structural studies [62,63], and that correspond to the conformations of the TC-bound REC that immediately precede and immediately follow ribosome-catalyzed GTP hydrolysis by EF-Tu (Fig. 3A) [50]. The state that precedes GTP hydrolysis can be biochemically ‘captured’ and stabilized using a non-hydrolyzable GTP analog [64] or a GTP hydrolysis-deficient EF-Tu mutant [65]. Likewise, the state that immediately follows GTP hydrolysis can be captured and stabilized using the EF-Tu-targeting antibiotic kirromycin [66]. Such approaches allow the populations of these ordinarily transient and low-population states to be increased such that they can be easily studied using ensemble biochemical and structural methods [31–33,35].
In contrast to the mid-FRET state, the low-EFRET state has been assigned to a structurally novel intermediate state that has thus far eluded capture and stabilization using mutations, biochemical analogs, or small-molecule inhibitors, thereby precluding its direct detection using ensemble biochemical or structural studies (Fig. 3A). Nonetheless, the conformation of the TC-bound REC corresponding to the low-EFRET state is a critical, codon-dependent intermediate state during aa-tRNA selection. Experiments in which the TC is delivered to an REC carrying a non-cognate A-site codon, for example, do not result in any detectable smFRET signals, including even the detection of highly transient sampling of the low-EFRET state. Analogous experiments using RECs carrying a near-cognate A-site codon, on the other hand, result in the detection of a highly transient low-EFRET state that corresponds to the formation of a weakly interacting, transient TC-bound REC from which TC has a much higher probability of dissociating from the REC than of progressing along the reaction pathway. In contrast, experiments using RECs carrying a cognate A-site codon result the detection of a slightly longer-lived low-EFRET state that corresponds to the formation of a slightly more stably interacting, less transient TC-bound REC from which the TC has a much higher probability of progressing along the reaction pathway than of dissociating from the REC. Interestingly, the precise EFRET values of the low-EFRET state differ for cognate and near-cognate TC-bound RECs, indicating that the TC in a cognate TC-bound REC is positioned in a manner that differs slightly from how it is positioned in a near-cognate TC-bound REC. This suggests that positioning of the TC within the low-EFRET TC-bound REC state is an important, yet transient, structural response to the recognition of a cognate codon at the A site of the REC.
Despite the success of smFRET in identifying and characterizing the highly transient low-EFRET TC-bound REC state, it is important to note that there are one or more states preceding this state in the reaction pathway that play important roles during aa-tRNA selection and that remain to be identified and characterized. For example, the very first state resulting from the codon-independent, initial binding of the TC to the REC, which is likely to have properties that are similar or identical to those of an encounter complex, has yet to be observed (Fig. 3A). In addition, the fact that the low-EFRET state is not detected in smFRET experiments in which a TC is delivered to an REC carrying a non-cognate A-site codon suggests that, either: (i) the low-EFRET state is sampled too rarely to be detected due to current experimental limitations (e.g., the low concentrations of acceptor-labeled TC that must be used to maintain the signal-to-background required to observe single fluorophores, the rate with which the donors on the RECs photobleach, etc.); (ii) the low-EFRET state is sampled too transiently to be detected due to current limitations on the time resolution of the experiments; (iii) non-cognate TCs are recognized and discriminated against within a state that precedes and is physically distinct from the low-EFRET state, but in which the distance between the donors and acceptors in the currently available fluorophore labeling schemes are too far away to generate a detectable EFRET; or (iv) a combination of these possibilities. Thus, weakly interacting, highly transient states that are critical for fully understanding the physical and molecular mechanisms underlying aa-tRNA selection during translation remain to be identified and characterized – representing an ongoing challenge for the field.
Towards these goals, emerging technologies hold great promise for ongoing investigations of the mechanism of aa-tRNA selection. Recent single-molecule fluorescence co-localization microscopy experiments that use next-generation, nanofabricated, microfluidic, observation flowcells to overcome the concentration barrier (see Sec. 3.3), for example, have demonstrated that fluorophore-labeled TCs do indeed transiently co-localize with fluorophore-labeled RECs carrying non-cognate A-site codons. Although these were not smFRET experiments in which the measured EFRET could be compared to what is observed in RECs carrying near-cognate or cognate A-site codons, they represent an important step towards understanding where in the pathway and how RECs discriminate against non-cognate TCs [67].
2.3 Splice site selection during spliceosome assembly
In eukaryotes, transcription of the genomic DNA often produces pre-mRNAs that must be spliced in order to excise the non-coding intronic sequences and ligate the coding exonic sequences, ultimately yielding mature mRNAs that can be translated by the ribosome. Pre-mRNA splicing is carried out by the spliceosome, a 2-3 MDa molecular machine that is composed of five RNA-protein complexes known as snRNP complexes and dozens of additional protein factors (Fig. 3B) [68–72]. During splicing, the spliceosome must assemble de novo across each pre-mRNA intron. Assembly often begins with binding of the U1 snRNP to the junction of the intron with the 5’ exon (i.e., the 5’ splice site) and one or more proteins to the short sequence that specifies the 2’-OH for nucleophilic attack of the phosphodiester bond at the 5’ splice site during the first chemical step of splicing (i.e., the branchsite). Some of these branchsite-binding proteins (Branchpoint Bridging Protein in yeast or SF1 in humans) are then displaced by binding of the U2 snRNP. This is then followed by binding of the U4/U6.U5 tri-snRNP and the protein-only nineteen complex (NTC), an event that ultimately triggers the release of the U1 and U4 snRNPs and activates the spliceosome for catalysis [68,69,72,73]. Despite the fact that the 5’ splice site, the junction of the intron with the 3’ exon (i.e., the 3’ splice site), and the branchsite are comprised of short consensus sequence elements that, at least in metazoans, are very poorly conserved [73–75], the spliceosome is able to accurately recognize and select these sequence elements in order to generate the proper mRNA with an error rate for splicing of 1 in 102 to only 1 in 105 [73,76–78]. Similar to aa-tRNA selection, the unexpectedly high fidelity of splice site selection by the spliceosome has been attributed to multistep mechanisms for spliceosome assembly, activation, and catalysis in which kinetic proofreading [5,6] strategies are employed to increase the low fidelity of splice site selection that would otherwise be predicted [73,75].
Although studies of splice site selection during spliceosome assembly would ideally be performed using smFRET [69], the experimental systems available for studying splice-site selection during spliceosome assembly present several important challenges for designing, implementing, and interpreting smFRET experiments. These challenges include, (i) the fact that even a minimal version of the reaction involves the interaction of numerous molecular subcomplexes; (ii) the unavailability of a reconstituted in vitro splicing system such that experiments must be performed in whole cell or nuclear cell extracts, making it difficult to manipulate individual components and steps of the reaction [79]; (iii) the existence of only a few small-molecule inhibitors that enable inhibition of specific and well-defined steps of the reaction [80,81]; and (iv) the availability of only a very limited number of cryo-EM and/or X-ray crystallography structures (reviewed in ref. [82]) for designing donor-acceptor labeling schemes. Due to these challenges, few smFRET studies of the spliceosome have been performed, and these either focus on structural rearrangements of a donor- and acceptor-labeled premRNA upon binding of the indirectly detected, fluorophore-free splicesomal components [83,84] or on structural rearrangements of protein-free, RNA components of the snRNPs [85,86]. Notably, smFRET studies reporting on the dynamics of spliceosome assembly and how these dynamics are modulated by the identities of the 5’- and 3’ splice sites and the branch point sequence have not been reported. Recently, however, a combination of chemical biology approaches for systematically and orthogonally fluorophore-labeling sets of spliceosomal subcomplexes within Saccharomyces cerevisiae (S. cerevisiae) whole cell extracts (WCEs) and single-molecule fluorescence co-localization microscopy experiments have been used to investigate the dynamics of spliceosome assembly. Notably, this approach enabled the authors to demonstrate how these dynamics are modulated by the unique splice-site features of specific pre-mRNA introns [79,84].
WCEs for single-molecule fluorescence co-localization microscopy experiments were prepared from S. cerevisiae strains in which the C-termini of specific proteins that are associated with sets of well-defined spliceosomal subcomplexes had been fused to either a ‘DHFR’ tag (a variant of Escherichia coli dihydrofolate reductase) or a ‘SNAP’ tag (a variant of human alkylguanine S-transferase) [79,84]. DHFR- and SNAP fusions are then differentially fluorophore labeled using fluorophore-labeled trimethoprim analogs that bind DHFR very tightly, with an equilibrium dissociation constant of <1 nM [87], or benzyl-guanine derivatives that covalently modify the active site cysteine of SNAP [88]. Single intron-containing pre-mRNAs to be used for single-molecule fluorescence colocalization microscopy were fluorophore labeled near the 3’ end, biotinylated at the 3’ end, and then tethered to the surface of a microfluidic, observation flowcell via their 3’ biotin. In this manner, they could be imaged using TIRF microscopy and their spatial location in the field-of-view could be determined to within single-molecule resolution [89]. Introduction of a WCE containing a set of fluorophore-labeled spliceosomal subcomplexes into such a flowcell then allows binding of each subcomplex to the fluorophore-labeled and surface-tethered pre-mRNA to be monitored via their spatial co-localization with the pre-mRNA.
Collectively, these studies have revealed that, depending on the identity of the pre-mRNA, spliceosome assembly preferentially follows one of two pathways that differ in the order in which the U1 and U2 snRNPs are observed to co-localize to the premRNA (Fig. 3B). Thus, the first steps of the assembly reaction may be either U1→U2→U4/U6.U5→NTC or U2→U1→U4/U6.U5→NTC. In addition, these studies revealed that no single step is rate limiting for the overall assembly reaction. Perhaps most interesting, these experiments have demonstrated that, for all pre-mRNAs tested thus far, binding of each subcomplex is highly reversible and transient on the timescale of the experiments. This result suggests that introns are not irreversibly committed to splicing during the early steps of spliceosome assembly. Instead, commitment to splicing increases with the binding of each successive subcomplex and can possibly be regulated by modulating the stabilities of the bound subcomplexes during spliceosome assembly. Indeed, the set of pre-mRNAs that have been tested thus far differ in, among other respects, the sequences of their 5’ and 3’ splice sites, suggesting that differences in the order in which the U1 and U2 snRNPs are observed to co-localize to the different pre-mRNAs may arise from pre-mRNA sequence-dependent differences in the stabilities with which U1 and/or U2 snRNPs bind to each pre-mRNA. Consistent with this possibility, more extensive scrambling of the 5’ splice site results in a loss of detectable U1 snRNP binding in both ensemble [90–92] and single-molecule fluorescence [79] experiments, strongly suggesting that the stability of U1 snRNP binding is highly dependent on the identity of the 5’ splice site.
Despite the success of the single-molecule fluorescence co-localization experiments described in the previous paragraph, additional studies in which the 5’ splice site, the 3’ splice site, and the branch point sequence are systematically mutagenized will be needed to determine how the transient binding of individual subcomplexes during spliceosome assembly across introns is modulated. In addition, the fact that extensive scrambling of the 5’ splice site results in a loss of detectable U1 snRNP binding to the 5’ splice site suggests that the encounter complex between U1 snRNP and the pre-mRNA and, possibly, subsequent U1 snRNP-bound pre-mRNA intermediate states during which U1 snRNP recognizes and selects the 5’ splice site, have yet to be observed. Ultimately, it will be important to extend such experiments so as to investigate the hypothesis that early interactions between U1 snRNP and the C-terminal domain of the large subunit of RNA polymerase II (and potentially chromatin [93]) serve to co-transcriptionally recruit U1 snRNP to nascent pre-mRNA transcripts [94–101]. Likewise, it will ultimately be important to extend these types of experiments to investigate recognition and selection of the 3’ splice site by a subunit of the U2 auxiliary factor (U2AF) [102–104] and the branchsite sequence by SF1 and U2 snRNP [105] during the early steps of spliceosome assembly. Thus, numerous, weakly interacting, highly transient states that are critical for fully understanding splice site selection during spliceosome assembly remain to be identified and characterized, representing an ongoing challenge for the field.
3. Emerging experimental advances for smFRET studies of ligand-binding reactions
3.1 Microscopy developments
As described in Section 1, one of the current limitations in single-molecule fluorescence studies of ligand-binding reactions is the concentration barrier that is created by the relatively high concentrations of fluorophore-labeled ligands that are required to be present in the imaging buffer in order to detect ligand-binding events within the experimental observation time [14]. Because an acceptor will only be efficiently excited via FRET when it is within tens of Å of a donor that is directly excited by an excitation light source (e.g., a laser), smFRET experiments in which a donor-labeled biomolecule of interest is tethered to the surface of the microfluidic, observation flowcell and an acceptor-labeled ligand is supplied in the imaging buffer greatly ameliorate the limitations imposed by the concentration barrier [28,106]. Despite this advantage of smFRET experiments, acceptors can still be directly, albeit inefficiently, excited by the laser that is used to directly excite the donor fluorophore – a low probability event known as excitation crosstalk [10]. At high enough concentrations of acceptor-labeled ligands in the imaging buffer (typically well below the equilibrium dissociation constants of weakly interacting biomolecule-ligand complexes), excitation crosstalk becomes a major source of background fluorescence noise in such experiments, a situation that can ultimately prevent the detection of fluorescence from individual molecules [14].
By preventing the excitation source from penetrating deeper than ~200-300 nm into the imaging buffer in the flowcell, wide-field TIRF microscopy-based smFRET experiments minimize the total amount of noise from excitation crosstalk of acceptor-labeled ligands in the imaging buffer. Unfortunately, the major disadvantage of this approach is that the electron-multiplying charge-coupled device (EMCCD) cameras that are used as detectors in TIRF microscopy-based smFRET experiments operate at time resolutions that are typically too slow to capture the weakest and most transient intermediate states (typically limited to ~30 ms per a full-resolution frame [107]). This tradeoff between lower background noise and faster time resolution is a fundamental limitation of using wide-field smFRET to study encounter complexes. Using a confocal fluorescence microscope with an avalanche photodiode (APD) or a single-photon avalanche diode (SPAD) detector, rather than a TIRF microscope with an EMCCD camera detector, affords significant increases in the sensitivity, signal-to-background ratio, and time resolution (typically ~1 ms per data point) of smFRET experiments [10,108]. Unfortunately, however, this increase in the time resolution comes at the cost of a significant decrease in throughput, as the confocal fluorescence microscopy-based approach can only image one biomolecule at a time, whereas the TIRF microscopy-based approach can simultaneously image hundreds of surface-tethered biomolecules at a time. Such low throughput can be particularly difficult to overcome when attempting to detect weakly interacting, transient biomolecule-ligand complexes and/or conformational states that are rarely-populated. In the ideal setup, one would be able to couple a TIRF microscope with an APD- or SPAD-array detector [107,109]. Such an approach would allow wide-field detection with APDs or SPADs, thereby allowing the high throughput associated with using a conventional TIRF microscope, but with the high time resolution that is associated with using a confocal fluorescence microscope. Towards this goal, smFRET was recently demonstrated using linear arrays of 8 SPADs [110], and so, with further development, commercially available products composed of tens of thousands of SPADs will likely constitute an important future development in the field of single-molecule fluorescence microscopy [109].
An alternative approach to minimizing the excitation crosstalk of acceptor-labeled ligands and improving the signal-to-background ratio of conventional TIRF microscopy-based smFRET experiments without compromising throughput is to remove excess ligand from the excitation volume without changing the local concentration of the ligand in the vicinity of the biomolecule. This can be achieved by locally confining ligands and the biomolecule of interest into spatially resolved regions of the flowcell that are smaller than the excitation volume associated with the detection of single, diffraction-limited spots in the field-of-view. Notable approaches include electrostatic and physical traps [111,112] as well as encapsulation of the biomolecules and ligands in aqueous drops in oil [113]. Perhaps the most promising and physiologically compatible method of local confinement for smFRET experiments is to encapsulate ligands and individual biomolecules in surface-tethered liposomes [114]. These lipid vesicles not only encapsulate ligands with individual biomolecules and confine them to spatially resolved regions on the surface of the microfludic flowcell, but, because the biomolecule is free to diffuse within the surface-tethered liposome, they also serve to further reduce the interactions that a biomolecule of interest might have with the PEG-passivated surface of the mirofluidic flowcell [115]. In addition, since their introduction in 2001 [116], liposomes have been optimized to allow for the free diffusion of small molecules into and out of the liposome [117].
3.2 Fluorophore developments
Although much work has gone into developing robust donor and acceptor pairs with well-behaved photophysics for smFRET studies [118], photobleaching and ‘blinking’ of fluorophores continues to impose limitations on smFRET studies of biomolecular systems [119,120]. The irreversible photobleaching of a fluorophore effectively brings the smFRET experiment, at least for that fluorophore, to an end, thereby limiting the experimental throughput. The reversible blinking of a fluorophore similarly limits the throughput of smFRET experiments and, in addition, can convolute the subsequent analysis of the data (e.g., it is often difficult to distinguish whether changes in EFRET that are observed arise from bone fide changes in the distance between the donor and acceptor or from blinking of the donor or acceptor) [119,120]. The limitations imposed by photobleaching and blinking are particularly challenging for smFRET studies of ligand-binding reactions. Rare events such as the formation of a particularly long-lived complex formed by the generally weak, transient binding of an acceptor-labeled ligand to a surface-tethered, donor-labeled biomolecule of interest, for example, are exceedingly difficult to observe within the limited observation times that are imposed by photobeaching and blinking of the donor fluorophore. The continued development of longer-lived, brighter, and more stable fluorophores, such as Cy3B [121], is therefore an important area of research, as these enhanced fluorophores enable longer and more stable observation times in smFRET experiments [118,122].
Complementing the development of longer-lived, brighter, and more stable fluorophores, considerable work has also gone into optimizing buffer conditions that minimize photobleaching and blinking [119]. Often, photobleaching is mediated via a photochemical reaction between a fluorophore in its electronically excited state and molecular oxygen. As a consequence, oxygen scavenging systems, including mixtures of glucose, gluocose oxidase, and catalase or of protocatechuic acid (PCA) and protocatechuate-3,4-dioxygenase (PCD), are often added to experimental buffer systems to extend fluorophore survival times [123,124]. As early as 1988, β-mercaptoethanol (BME) was being added to buffer systems for single-molecule fluorescence microscopy experiments in order to suppress the blinking of tetramethylrhodamine, presumably by quenching a dark triplet-state of the fluorophore [123]. Since then, triplet-state quenchers, including mixtures of cyclooctatetraene (COT) [23], 4-nitrobenzyl alcohol (NBA) [23], and/or Trolox (a water-soluble analog of vitamin E) [125] have become standard additives in smFRET experiments [126]. Recently, direct, covalent conjugation of triplet-state quenchers such as COT, NBA, or Trolox to the entire class of cyanine dye fluorophores, including Cy3 and Cy5, was shown to significantly enhance the photostability of these fluorophores [127,128]. Although these new fluorophore-quencher conjugates represent a breakthrough in fluorophore development, caution should be exercised in their use, as these conjugates are necessarily larger than the traditional fluorophores on which they are based and are therefore more likely to sterically perturb the biomolecules or ligands to which they are attached.
In addition to the development of fluorophores and/or buffer conditions that resist photobleaching and blinking, the development of fluorophores that enable selective excitation is another promising area of current research [120]. Indeed, one of the major advantages of smFRET is that the acceptor is excited selectively (i.e., only when it is within tens of Å of a donor). Similarly, highly enzyme-specific fluorogenic substrates that fluoresce exclusively upon being enzymatically modified can be used to conduct single-molecule fluorescence microscopy studies of the mechanisms of action of enzymes for which fluorogenic substrates have been developed [12,122]. This strategy, however, is difficult to generalize to all, or even many, enzymes and even more so to non-enzymatic biomolecules. In addition, to our knowledge, fluorogenic strategies have not yet been utilized in smFRET studies. As an alternative to the use of fluorogenic substrates, selective excitation can be achieved through the use of photoswitchable fluorophores [120]. For example, in the PhADE technique, an imaging buffer containing high concentrations of a photoswitchable fluorophore-labeled ligand is activated with a laser pulse and these are allowed to diffuse out of the imaging volume. This effectively removes all of the photoactivated ligands other than those that are bound to the surface-tethered biomolecules from the flowcell, thereby removing much of the background fluorescence that would otherwise arise from photoactivated ligands in solution [129]. Unfortunately, this strategy works only for relatively long-lived, weakly interacting biomolecule-ligand complexes, and, to our knowledge, has not yet been used in smFRET experiments.
Perhaps the most promising recent development in smFRET studies of biomolecular systems is the use of FRET-based quenchers as acceptors for smFRET experiments [130]. By returning from the excited state to the ground state without emitting a photon, quenchers not only free the optical spectrum for the use of additional fluorophores (e.g., for colocalization experiments), but, of particular importance for smFRET studies of ligand-binding reactions, also allow smFRET experiments to be performed under conditions in which very high concentrations of quencher-labeled ligands can be included in the imaging buffer without significantly increasing the fluorescence background. This is because a quencher that is excited by excitation crosstalk will not fluoresce, thereby minimizing the contribution of excitation crosstalk to the fluorescence background. Only a few researchers have thus far used FRET-based quenchers to address biological systems using smFRET [130–134]. Notably, a FRET-based quencher was used to report on the large-scale conformational dynamics of the ribosome during translation elongation [135,136], although this approach was used to free the optical spectrum and expand the number of fluorophores that could be simultaneously detected, rather than to use FRET-based quencher-labeled ligands to overcome the concentration barrier.
3.3 Optical engineering developments
A general strategy for increasing the sensitivity of single-molecule fluorescence experiments is to enhance the fluorescence signal from the molecules of interest. Perhaps the most widely used approach for accomplishing this is plasmon-mediated enhancement, which has traditionally been used to enhance optical spectroscopies (e.g., surface plasmon-enhanced Raman spectroscopy) and, in single-molecule fluorescence applications, can be used to decrease illumination volumes and increase fluorescence signals [137]. Nanofabricated plasmonic bowties [138] and plasmonic nanoantennas [139–141] have both been shown to dramatically increase signal-to-background ratios in single-molecule fluorescence applications. Of particular importance for single-molecule fluorescence studies of ligand-binding reactions, the ‘antenna-in-a-box’ platform has used plasmon-based enhancement to permit the observation of single-molecule fluorescence in the presence of micromolar concentrations of fluorophore in the imaging buffer [142]. Unfortunately, few of these platforms have thus far demonstrated their applicability with smFRET [139] and, while these technologies show promise, the fact that highly specialized facilities, equipment, and technical skills are required to nanofabricate the plasmon-producing structures (e.g., the bowties and antennas) onto the surface of the microfluidic flowcells that are used in single-molecule fluorescence experiments has thus far hindered widespread adoption by the community.
Another general strategy for attaining higher signal-to-background ratios in single-molecule fluorescence studies is to decrease the fluorescence background by reducing the excitation volume. For this purpose, arrays of nanoapertures — nanoscopic wells that have been fabricated into a thin metallic layer that has been deposited onto the surface of a microfluidic flowcell (Fig. 4A) — have proven themselves very useful. Popularized as “zero-mode waveguides” (ZMW), the geometric confinement of light in the nanoaperture leads to a zeptoliter excitation volume at the very bottom of the nanoaperture. This permits the sensitive observation of a single, fluorophore-labeled molecule that has been tethered or otherwise localized to the bottom of the nanoaperture in a background of micromolar concentrations of fluorophore-labeled molecules in the imaging buffer [143]. Nanoaperture arrays are used in Pacific Biosciences’ next-generation, single-molecule real-time (SMRT) DNA sequencing technology (http://www.pacificbiosciences.com). There, a single DNA polymerase is tethered to the bottom of each nanoaperture, and a nucleotide-specific, single-molecule fluorescence signal is observed as the polymerase incorporates each nucleotide into the nascent DNA molecule that is being synthesized [144].
Figure 4.
(A) Schematic of a newly developed nanoaperture array for use in nanoaperture fluorescence microscopy. The gold surfaces in an array of nanoapertures (i.e., nanoscopic wells fabricated onto a gold film that has been deposited on a quartz substrate) is passivated using self-assembled monolayers of thiol-derivatized PEG (PEG-SH SAMs) to protect against non-specific adsorption of acceptor-labeled ligands. Likewise, self-assembled monolayers of a mixture of silane-derivatized PEG and silane-/biotin-derivatized PEG (PEG-Si SAMs) are used to protect against non-specific adsorption of acceptor-labeled ligands and enable tethering of donor-labeled biomolecules using a biotin-streptavidin-biotin bridge, respectively. Because light from the laser excitation source is confined to an exceedingly small volume at the very bottom of the nanoaperture, the excitation crosstalk of acceptor-labeled ligands is significantly minimized relative to the excitation crosstalk produced by the standard TIRF-based microscopy setup depicted in Figure 2. Figure adapted from ref. [106]. (B) Graphical model of the coupled Bayesian HMM used in ebFRET and vbFRET. Two representative signal versus time and state occupancy versus time trajectories in an experiment (left insets). Graphical model showing an HMM for N trajectories with K states. The parameters describing each trajectory (light blue circles) are distributed according to a probability function that depends on a set of hyperparameters (light pink circles). Maximum likelihood methods use a non-Bayesian variant of this HMM, which omits the hyperparameters.
Unfortunately, in the more than ten years since nanoaperture arrays were first introduced for single-molecule fluorescence applications [143], only a handful of biomolecular systems beyond DNA replication have been investigated using this technology [67,136,145–147]. Notably, studies of all but one of these biomolecular systems have been investigated in collaboration with Pacific Biosciences and only one of these studies has used nanoaperture arrays in an smFRET application. As is the case with plasmon-based enhancement technologies, widespread adoption of nanoaperture-array technology has likely been hindered by the limited availability of the resources required for nanofabrication. In addition, the non-specific adsorption of biomolecules to the metallic and glass surfaces of the nanoapertures has likely limited their applications. Non-specific adsorption not only alters working concentrations by sequestering molecules from solution, but it also compromises single-molecule resolution by non-specifically localizing multiple, fluorophore-labeled, molecules to the surfaces near the bottom of the nanoapertures. In the case of SMRT sequencing via DNA replication, a very successful polyvinyl phosphonic acid-based, nanoaperture-surface passivation scheme was developed that minimizes the non-specific adsorption of small, negatively charged nucleotide triphosphates to the surface of aluminum-based nanoapertures [148]. Given the negatively charged nature of the polyvinyl phosphonic acid passivation layer, however, it is likely that this passivation scheme will not be generally applicable to other biomolecules (e.g., positively charged globular proteins).
More recently, an alternative passivation scheme has been reported that uses thiol-based self-assembled monolayers (SAMs) of PEG to robustly passivate gold-based nanoapertures [106]. Given the widespread success of PEG-based passivation schemes in single-molecule fluorescence studies, we anticipate that PEG-passivated nanoaperture arrays will provide a more general solution to the problem of non-specific adsorption of biomolecules to the nanoaperture surfaces and will thereby enable studies of a wide-range of biomolecular systems using nanoaperture arrays (Fig. 4A). Perhaps most excitingly, the use of gold-based nanoapertures such as the PEG-passivated, gold based nanoapertures described by Kinz-Thompson et al. [106] also allows the surface-plasmon of the gold to be used for plasmon-based enhancement of the fluorescence signals at the bottom of the nanoapertures, an approach that has been recently demonstrated [149,150] and that could potentially facilitate the use of nanoaperture arrays for smFRET applications.
4. Emerging computational advances for smFRET studies of ligand-binding reactions
4.1 Review of single-molecule kinetics
In order to discuss recent improvements in the analysis of smFRET data, it is useful to begin with a brief, general review of how kinetic information is obtained from single-molecule experiments and, more specifically, smFRET experiments. Traditional treatments of chemical kinetics are insufficient to describe systems where fluctuations from the average behavior are important, such as systems with small numbers of molecules [151]. Instead, alternative approaches that treat molecules as independent, stochastic entities are used [152]. To interpret the kinetic data yielded by a single molecule from a smFRET experiment, a framework is used which describes the distribution of times that the molecule spends dwelling in a particular state [151–157]. To extract these dwell time distributions from single-molecule EFRET versus time trajectories, dwell-time histograms are built from the results of analyzing the data using methods, such as hidden Markov models (HMM) (see Section 4.2, below), change point analysis, or wavelet analysis [158], that detect the time points at which switching between distinct EFRET states occurs. It is worth noting that a complete theoretical description has been developed that allows high-resolution information about the conformational dynamics of a biomolecular system to be extracted from the joint distribution of EFRET values and fluorescence lifetimes determined from experimentally observed photon bursts [159,160]. Although the joint distribution provides more information regarding the conformational dynamics of the biomolecular system than the distribution of EFRET values does alone, this approach is not generally applicable in traditional, wide-field, TIRF microscopy-based smFRET experiments, as these experiments do not monitor photon arrival times.
4.2 Hidden Markov model developments
One of the most popular approaches to analyzing single-molecule EFRET trajectories has been the application of hidden Markov models (HMMs), which yield the probabilities of transitioning between states as well the ‘idealized’ path between states. HMMs were first used in biology for analyzing conductance time series in single channel ion recordings [161], and first suggested for use with smFRET data in 2003 [162]. Since then, a number of software packages for HMM analysis of smFRET data have been published [163–168]. Although HMM analysis of smFRET data can provide the desired information necessary to develop mechanistic models of biomolecular function, HMM analysis approaches and software packages that use maximum likelihood methods to estimate HMM parameters (e.g., HaMMy [163], QuB [169], and SMART [164]) can result in overfitting of the smFRET data (i.e., overestimating the number of states that can be confidently ascribed to the data), as the value of the likelihood function that is being maximized will always increase with the number of states included in the analysis. This overfitting problem has been recently addressed by HMM analysis approaches and software packages that use Bayesian inference methods to estimate HMMs (e.g., vbFRET [165,166] and ebFRET [167,168]). Despite the success of HMMs for the analysis of smFRET data, however, it is important to note that HMMs are ill-suited for modeling EFRET trajectories containing rapid transitions such as those into and out of energetically unstable, transiently sampled states (e.g., weakly interacting biomoleculeligand complexes with rapid rates of association and dissociation) and are inappropriate for modeling non-Markovian data (e.g., data exhibiting ‘dynamic disorder’ where the probability of a transition changes with time) [165–168].
4.3 Bayesian inference developments
Perhaps the most promising computational approaches for analyzing smFRET data from weakly interacting, transient biomolecule-ligand complexes employ Bayesian inference, and in doing so they are able to ‘learn’ from the data [170]. Bayesian inference approaches have been used in the analysis of smFRET data by removing noise from EFRET trajectories [171] and by analyzing photon bursts [172,173]. Perhaps the most powerful applications, however, are those that use Bayesian inference methods to estimate the HMM parameters that are used to analyze EFRET trajectories [165–168]. The vbFRET software package, for example, uses Bayesian inference on an HMM to select the number of states and rates of transitions between states (i.e., the kinetic model) that best describes each individual EFRET trajectory. This minimizes the overfitting problem faced by approaches and software packages that use maximum likelihood methods to estimate HMMs and subsequently rely on the user or on an ad hoc metric, such as the Bayesian- or Akaike information criteria, to select the kinetic model that best describes the data [165,170]. Moreover, vbFRET shows promise at detecting transiently sampled intermediate states that maximum likelihood approaches for estimating HMMs might otherwise miss, an extremely useful ability when studying weakly interacting, transient biomolecule-ligand complexes using smFRET [165].
One of the major limitations of all HMM-based approaches for smFRET data analysis is that these approaches provide a separate and unique kinetic model for each individual EFRET trajectory. Ultimately, it is left up to the user to determine which single, consensus kinetic model best describes the entire population of kinetic models provided by the HMM-based analysis of the ensemble of EFRET trajectories that are typically obtained from a single smFRET experiment. Recently, van de Meent and coworkers have addressed this problem by expanding on the vbFRET framework and creating ebFRET. ebFRET uses a Bayesian inference method to estimate HMM parameters that is analogous to the one that vbFRET uses, but does so on entire populations of EFRET trajectories rather than on individual EFRET trajectories, thereby using all of the EFRET trajectories to learn the single, consensus kinetic model that best describes all of the available data (Fig. 4B) [167,168]. Because it uses all of the EFRET trajectories, ebFRET has an enhanced and statistically robust ability to detect rarely and transiently sampled states that might appear in some, but not all, of the EFRET trajectories associated with a single smFRET experiment. In addition, ebFRET can distinguish between states that have the same EFRET but differ in their lifetimes. For example, ebFRET has been used to distinguish between two structurally similar states of a biomolecule that yield the same EFRET but that differ in the lifetime of that state due to the presence or absence of a bound ligand [168] – a development that dramatically extends the resolution of smFRET experiments.
One of the major challenges for HMM analyses of data collected from smFRET studies of weakly interacting, transient biomolecule-ligand complexes is that the encounter complexes and many of the intermediate states that are sampled are simply too energetically unstable and too transiently sampled to be reliably modeled with a HMM [165]. This is only likely to become more challenging as ever more sensitive and higher time resolution single-molecule biophysical techniques are developed that increasingly render the most weakly interacting and transient biomolecule-ligand complexes accessible to study. Recently, however, a non-HMM-based Bayesian inference approach called BIASD has been developed that, assuming a two-state model (e.g., the ligand-bound and ligand-free states of a biomolecule), is able to learn both the EFRET values and rates of transitions into and out of transiently sampled states (e.g., the rates of ligand association and dissociation from a biomolecule) in cases where the rates of transitions are on the order of, or even faster than, the experimental time resolution [Kinz-Thompson in preparation]. This method should therefore prove extremely useful in ongoing efforts to study weakly interacting, transient biomolecule-ligand complexes using smFRET.
5. Conclusion
While space constraints necessarily limited the focus of the present article to intermolecular ligand-binding reactions, we note that many of the concepts, experimental techniques, and computational approaches discussed in this article apply equally well to studies of the weakly interacting, transient intermediate states that are sampled during intramolecular biomolecular folding reactions and intramolecular structural rearrangements that are associated with function. Regardless, understanding the origins of specificity in ligand-binding reactions is at the core of many research investigations and, as such, there is a great need to develop technologies that enable such reactions to be investigated at an ever increasing level of detail, a need that single-molecule fluorescence and, specifically, smFRET approaches are uniquely positioned to address.
By initially forming an encounter complex with a potential ligand and subsequently proceeding to form other weakly interacting, transient intermediate states prior to forming a final, stably bound biomolecule-ligand complex, biomolecules can rapidly and efficiently screen potential ligands and control the specificity of binding during ligand-binding reactions. Although space constraints limited us to highlighting only two, aa-tRNA selection by the ribosome and splice site selection during spliceosome assembly, there are now many examples of ligand-binding reactions that are being actively investigated using single-molecule fluorescence colocalization approaches and, at higher resolution, using smFRET approaches [15–17].
Despite the promise and success of smFRET approaches for studies of ligand-binding reactions, there remain several experimental and computational challenges that must be overcome in order to increase the information content of such smFRET studies. In this article, we have reviewed a number of emerging experimental and computational approaches that we consider will be important for overcoming these challenges. We fully anticipate that further development of these approaches, as well as the development of entirely new, currently unforeseen approaches for overcoming these challenges, will enable encounter complexes and other weakly interacting, transient intermediate states to be investigated at an ever increasing level of detail. Such studies promise to reveal the mechanisms that control the selectivity of ligand-binding reactions at unprecedented resolution – knowledge that will allow us not only to understand, but also to modulate, some of the most important processes in all of biology.
Acknowledgements
We would like to thank Professor Aaron Hoskins (University of Wisconsin, Madison) for insightful and helpful discussions as well as a critical reading of the manuscript. This work was supported by a Burroughs Wellcome Fund CABS Award (CABS 1004856), an NSF CAREER Award (MCB 0644262), an NIH-NIGMS grant (R01 GM084288), an American Cancer Society Research Scholar Grant (RSG GMC-117152), and a Camille Dreyfus Teacher-Scholar Award to R.L.G. C.D.K. was supported by the Department of Energy Office of Science Graduate Fellowship Program (DOE SCGF), made possible in part by the American Recovery and Reinvestment Act of 2009, administered by ORISEORAU under contract number DE-AC05-06OR23100.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ubbink M. The courtship of proteins: understanding the encounter complex. FEBS Lett. 2009;583:1060–6. doi: 10.1016/j.febslet.2009.02.046. [DOI] [PubMed] [Google Scholar]
- 2.Schreiber G. Kinetic studies of protein-protein interactions. Curr. Opin. Struct. Biol. 2002;12:41–7. doi: 10.1016/s0959-440x(02)00287-7. [DOI] [PubMed] [Google Scholar]
- 3.Sheinerman FB, Norel R, Honig B. Electrostatic aspects of protein-protein interactions. Curr. Opin. Struct. Biol. 2000;10:153–9. doi: 10.1016/s0959-440x(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 4.Fersht AR. A guide to enzyme catalysis and protein folding. W.H. Freeman and Co.; New York: 1999. Structure and mechanism in protein science. [Google Scholar]
- 5.Hopfield JJ. Kinetic Proofreading: A New Mechanism for Reducing Errors in Biosynthetic Processes Requiring High Specificity. Proc. Natl. Acad. Sci. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57:587–95. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- 7.Volkov AN, Worrall J a R, Holtzmann E, et al. Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR. Proc. Natl. Acad. Sci. 2006;103:18945–50. doi: 10.1073/pnas.0603551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–30. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 9.Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–6. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 10.Tinoco I, Gonzalez RL. Biological mechanisms, one molecule at a time. Genes Dev. 2011;25:1205–31. doi: 10.1101/gad.2050011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solomatin SV, Greenfeld M, Chu S, et al. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463:681–4. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.English BP, Min W, van Oijen AM, et al. Ever-fluctuating single enzyme molecules: Michaelis-Menten equation revisited. Nat. Chem. Biol. 2006;2:87–94. doi: 10.1038/nchembio759. [DOI] [PubMed] [Google Scholar]
- 13.Moerner WE, Fromm DP. Methods of single-molecule fluorescence spectroscopy and microscopy. Rev. Sci. Instrum. 2003;74:3597. [Google Scholar]
- 14.Holzmeister P, Acuna GP, Grohmann D, et al. Breaking the concentration limit of optical single-molecule detection. Chem. Soc. Rev. 2013 doi: 10.1039/c3cs60207a. [DOI] [PubMed] [Google Scholar]
- 15.Gambin Y, VanDelinder V, Ferreon ACM, et al. Visualizing a one-way protein encounter complex by ultrafast single-molecule mixing. Nat. Methods. 2011;8:239–41. doi: 10.1038/nmeth.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sivasankar S, Zhang Y, Nelson WJ, et al. Characterizing the initial encounter complex in cadherin adhesion. Structure. 2009;17:1075–81. doi: 10.1016/j.str.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ragunathan K, Liu C, Ha T. RecA filament sliding on DNA facilitates homology search. Elife. 2012;1:e00067. doi: 10.7554/eLife.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–16. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cantor CR, Schimmel PR. Biophysical Chemistry. W.H. Freeman and Co.; New York: 1980. [Google Scholar]
- 20.Förster T. Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann. Phys. 1948;437:55–75. [Google Scholar]
- 21.Stryer L, Haugland RP. Energy Transfer: A Spectroscopic Ruler. Proc. Natl. Acad. Sci. 1967;58:719. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha T, Rasnik I, Cheng W, et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–41. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 23.Blanchard SC, Kim HD, Gonzalez RL, et al. tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. 2004;101:12893–8. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Axelrod D. Cell-substrate contacts illuminated by total internal reflection fluorescence. J. Cell Biol. 1981;89:141–5. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Axelrod D. Chapter 9 Total Internal Reflection Fluorescence Microscopy. Methods Cell Biol. 1989;30:245–270. doi: 10.1016/s0091-679x(08)60982-6. [DOI] [PubMed] [Google Scholar]
- 26.Axelrod D. Chapter 7: Total internal reflection fluorescence microscopy. Methods Cell Biol. 2008;89:169–221. doi: 10.1016/S0091-679X(08)00607-9. [DOI] [PubMed] [Google Scholar]
- 27.Schütz G, Trabesinger W, Schmidt T. Direct observation of ligand colocalization on individual receptor molecules. Biophys. J. 1998;28:2223–2226. doi: 10.1016/S0006-3495(98)77931-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herman B, Krishnan RV, Centonze VE. Microscopic analysis of fluorescence resonance energy transfer (FRET). Methods Mol. Biol. 2004;261:351–70. doi: 10.1385/1-59259-762-9:351. [DOI] [PubMed] [Google Scholar]
- 29.Finka A, Goloubinoff P. Proteomic data from human cell cultures refine mechanisms of chaperone-mediated protein homeostasis. Cell Stress Chaperones. 2013;18:591–605. doi: 10.1007/s12192-013-0413-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewin B. Genes VIII Pearson Education. 2004.
- 31.Voorhees RM, Ramakrishnan V. Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 2013;82:203–36. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 32.Johansson M, Lovmar M, Ehrenberg M. Rate and accuracy of bacterial protein synthesis revisited. Curr. Opin. Microbiol. 2008;11:141–7. doi: 10.1016/j.mib.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 33.Rodnina MV. Quality control of mRNA decoding on the bacterial ribosome. 1st ed. Elsevier Inc.; 2012. [DOI] [PubMed] [Google Scholar]
- 34.Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–62. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frank J, Gonzalez RL. Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu. Rev. Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodenbour JM, Pan T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006;34:6137–46. doi: 10.1093/nar/gkl725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouadloun F, Donner D, Kurland CG. Codon-specific missense errors in vivo. EMBO J. 1983;2:1351–6. doi: 10.1002/j.1460-2075.1983.tb01591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edelmann P, Gallant J. Mistranslation in E. coli. Cell. 1977;10:131–7. doi: 10.1016/0092-8674(77)90147-7. [DOI] [PubMed] [Google Scholar]
- 39.Kramer EB, Farabaugh PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laughrea M, Latulippe J, Filion A-M, et al. Mistranslation in twelve Escherichia coli ribosomal proteins. Cysteine misincorporation at neutral amino acid residues other than tryptophan. Eur. J. Biochem. 1987;169:59–64. doi: 10.1111/j.1432-1033.1987.tb13580.x. [DOI] [PubMed] [Google Scholar]
- 41.Grosjean HJ, de Henau S, Crothers DM. On the physical basis for ambiguity in genetic coding interactions. Proc. Natl. Acad. Sci. 1978;75:610–4. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kierzek R, Burkard ME, Turner DH. Thermodynamics of single mismatches in RNA duplexes. Biochemistry. 1999;38:14214–23. doi: 10.1021/bi991186l. [DOI] [PubMed] [Google Scholar]
- 43.Rodnina MV, Wintermeyer W. Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu. Rev. Biochem. 2001;70:415–35. doi: 10.1146/annurev.biochem.70.1.415. [DOI] [PubMed] [Google Scholar]
- 44.Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodnina MV, Pape T, Fricke R, et al. Initial Binding of the Elongation Factor Tu·GTP·Aminoacyl-tRNA Complex Preceding Codon Recognition on the Ribosome. J. Biol. Chem. 1996;271:646–652. doi: 10.1074/jbc.271.2.646. [DOI] [PubMed] [Google Scholar]
- 46.Thompson RC, Stone PJ. Proofreading of the codon-anticodon interaction on ribosomes. Proc. Natl. Acad. Sci. 1977;74:198–202. doi: 10.1073/pnas.74.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruusala T, Ehrenberg M, Kurland CG. Is there proofreading during polypeptide synthesis? EMBO J. 1982;1:741–5. doi: 10.1002/j.1460-2075.1982.tb01240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999;18:3800–7. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koshland DE. Application of a Theory of Enzyme Specificity to Protein Synthesis. Proc. Natl. Acad. Sci. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blanchard SC, Gonzalez RL, Kim HD, et al. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 2004;11:1008–14. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 51.Gonzalez RL, Chu S, Puglisi JD. Thiostrepton inhibition of tRNA delivery to the ribosome. RNA. 2007;13:2091–7. doi: 10.1261/rna.499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, Qin H, Kudaravalli RD, et al. Single-molecule structural dynamics of EF-G--ribosome interaction during translocation. Biochemistry. 2007;46:10767–75. doi: 10.1021/bi700657d. [DOI] [PubMed] [Google Scholar]
- 53.Lee T-H, Blanchard SC, Kim HD, et al. The role of fluctuations in tRNA selection by the ribosome. Proc. Natl. Acad. Sci. 2007;104:13661–5. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Effraim PR, Wang J, Englander MT, et al. Natural amino acids do not require their native tRNAs for efficient selection by the ribosome. Nat. Chem. Biol. 2009;5:947–53. doi: 10.1038/nchembio.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geggier P, Dave R, Feldman MB, et al. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J. Mol. Biol. 2010;399:576–95. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fei J, Wang J, Sternberg SH, et al. A highly purified, fluorescently labeled in vitro translation system for single-molecule studies of protein synthesis. 1st ed. Elsevier Inc.; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pestka S. Inhibitors of ribosome functions. Annu. Rev. Microbiol. 1971;25:487–562. doi: 10.1146/annurev.mi.25.100171.002415. [DOI] [PubMed] [Google Scholar]
- 58.Vázquez D. Inhibitors of Protein Biosynthesis. Springer-Verlag; Berlin: 1979. [DOI] [PubMed] [Google Scholar]
- 59.Blanchard SC, Cooperman BS, Wilson DN. Probing translation with small-molecule inhibitors. Chem. Biol. 2010;17:633–45. doi: 10.1016/j.chembiol.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carbon J, David H. Studies on the thionucleotides in transfer ribonucleic acid. Addition of N-ethylmaleimide and formation of mixed disulfides with thiol compounds. Biochemistry. 1968;7:3851–8. doi: 10.1021/bi00851a010. [DOI] [PubMed] [Google Scholar]
- 61.Seo H-S, Abedin S, Kamp D, et al. EF-G-dependent GTPase on the ribosome. conformational change and fusidic acid inhibition. Biochemistry. 2006;45:2504–14. doi: 10.1021/bi0516677. [DOI] [PubMed] [Google Scholar]
- 62.Stark H, Rodnina MV, Wieden H-J, et al. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Struct. Biol. 2002;9:849–54. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- 63.Valle M, Zavialov A, Li W, et al. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- 64.Skogerson L, Moldave K. Evidence for aminoacyl-tRNA binding, peptide bond synthesis, and translocase activities in the aminoacyl transfer reaction. Arch. Biochem. Biophys. 1968;125:497–505. doi: 10.1016/0003-9861(68)90607-3. [DOI] [PubMed] [Google Scholar]
- 65.Cool RH, Parmeggiani a. Substitution of histidine-84 and the GTPase mechanism of elongation factor Tu. Biochemistry. 1991;30:362–6. doi: 10.1021/bi00216a008. [DOI] [PubMed] [Google Scholar]
- 66.Wolf H, Chinali G, Parmeggiani a. Mechanism of the inhibition of protein synthesis by kirromycin. Role of elongation factor Tu and ribosomes. Eur. J. Biochem. 1977;75:67–75. doi: 10.1111/j.1432-1033.1977.tb11504.x. [DOI] [PubMed] [Google Scholar]
- 67.Uemura S, Aitken CE, Korlach J, et al. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–7. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larson JD, Rodgers ML, Hoskins A a. Visualizing cellular machines with colocalization single molecule microscopy. Chem. Soc. Rev. 2014;43:1189–200. doi: 10.1039/c3cs60208g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoskins A a, Gelles J, Moore MJ. New insights into the spliceosome by single molecule fluorescence microscopy. Curr. Opin. Chem. Biol. 2011;15:864–70. doi: 10.1016/j.cbpa.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fabrizio P, Dannenberg J, Dube P, et al. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol. Cell. 2009;36:593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 71.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 72.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 73.Semlow DR, Staley JP. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem. Sci. 2012;37:263–73. doi: 10.1016/j.tibs.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reed R. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 2000;12:340–5. doi: 10.1016/s0955-0674(00)00097-1. [DOI] [PubMed] [Google Scholar]
- 75.Egecioglu DE, Chanfreau G. Proofreading and spellchecking: a two-tier strategy for pre-mRNA splicing quality control. RNA. 2011;17:383–9. doi: 10.1261/rna.2454711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pickrell JK, Marioni JC, Pai A a, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–72. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fox-Walsh KL, Hertel KJ. Splice-site pairing is an intrinsically high fidelity process. Proc. Natl. Acad. Sci. 2009;106:1766–71. doi: 10.1073/pnas.0813128106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mitrovich QM, Tuch BB, De La Vega FM, et al. Evolution of yeast noncoding RNAs reveals an alternative mechanism for widespread intron loss. Science. 2010;330:838–41. doi: 10.1126/science.1194554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoskins A a, Friedman LJ, Gallagher SS, et al. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–95. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Albert BJ, McPherson P a, O'Brien K, et al. Meayamycin inhibits pre-messenger RNA splicing and exhibits picomolar activity against multidrug-resistant cells. Mol. Cancer Ther. 2009;8:2308–18. doi: 10.1158/1535-7163.MCT-09-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Corrionero A, Miñana B, Valcárcel J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 2011;25:445–59. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Newman AJ, Nagai K. Structural studies of the spliceosome: blind men and an elephant. Curr. Opin. Struct. Biol. 2010;20:82–9. doi: 10.1016/j.sbi.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 83.Abelson J, Blanco M, Ditzler M a, et al. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nat. Struct. Mol. Biol. 2010;17:504–12. doi: 10.1038/nsmb.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crawford DJ, Hoskins A a, Friedman LJ, et al. Single-molecule colocalization FRET evidence that spliceosome activation precedes stable approach of 5’ splice site and branch site. Proc. Natl. Acad. Sci. 2013;110:6783–8. doi: 10.1073/pnas.1219305110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo Z, Karunatilaka KS, Rueda D. Single-molecule analysis of protein-free U2-U6 snRNAs. Nat. Struct. Mol. Biol. 2009;16:1154–9. doi: 10.1038/nsmb.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Karunatilaka KS, Rueda D. Post-transcriptional modifications modulate conformational dynamics in human U2-U6 snRNA complex. RNA. 2014;20:16–23. doi: 10.1261/rna.041806.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miller LW, Cai Y, Sheetz MP, et al. In vivo protein labeling with trimethoprim conjugates: a flexible chemical tag. Nat. Methods. 2005;2:255–7. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- 88.Juillerat A, Gronemeyer T, Keppler A, et al. Directed Evolution of O6-Alkylguanine-DNA Alkyltransferase for Efficient Labeling of Fusion Proteins with Small Molecules In Vivo. Chem. Biol. 2003;10:313–317. doi: 10.1016/s1074-5521(03)00068-1. [DOI] [PubMed] [Google Scholar]
- 89.Friedman LJ, Chung J, Gelles J. Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys. J. 2006;91:1023–31. doi: 10.1529/biophysj.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ruby S, Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988;242:1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- 91.Seraphin B, Kretzner L, Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5’ cleavage site. EMBO J. 1988;7:2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ruby SW. Dynamics of the U1 Small Nuclear Ribonucleoprotein during Yeast Spliceosome Assembly. J. Biol. Chem. 1997;272:17333–17341. doi: 10.1074/jbc.272.28.17333. [DOI] [PubMed] [Google Scholar]
- 93.Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5:e1000682. doi: 10.1371/journal.pgen.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kotovic KM, Lockshon D, Boric L, et al. Cotranscriptional Recruitment of the U1 snRNP to Intron-Containing Genes in Yeast. Mol. Cell. Biol. 2003;23:5768–5779. doi: 10.1128/MCB.23.16.5768-5779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lacadie S a, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5’ss base pairing in yeast. Mol. Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 96.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 2006;13:815–22. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 97.Han J, Xiong J, Wang D, et al. Pre-mRNA splicing: where and when in the nucleus. Trends Cell Biol. 2011;21:336–43. doi: 10.1016/j.tcb.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Darnell RB. RNA protein interaction in neurons. Annu. Rev. Neurosci. 2013;36:243–70. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reed R. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 100.Pandit S, Wang D, Fu X-D. Functional integration of transcriptional and RNA processing machineries. Curr. Opin. Cell Biol. 2008;20:260–5. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Muñoz MJ, de la Mata M, Kornblihtt AR. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends Biochem. Sci. 2010;35:497–504. doi: 10.1016/j.tibs.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 102.Merendino L, Guth S, Bilbao D, et al. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3’ splice site AG. Nature. 1999;402:838–41. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- 103.Wu S, Romfo CM, Nilsen TW, et al. Functional recognition of the 3’ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–5. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 104.Zorio DA, Blumenthal T. Both subunits of U2AF recognize the 3’ splice site in Caenorhabditis elegans. Nature. 1999;402:835–8. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]
- 105.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–12. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 106.Kinz-Thompson CD, Palma M, Pulukkunat DK, et al. Robustly Passivated, Gold Nanoaperture Arrays for Single-Molecule Fluorescence Microscopy. ACS Nano. 2013;7:8159–8166. doi: 10.1021/nn403447s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Michalet X, Siegmund OHW, Vallerga JV, et al. Detectors for single-molecule fluorescence imaging and spectroscopy. J. Mod. Opt. 2007;54:239. doi: 10.1080/09500340600769067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tan Y, Hanson JA, Chu J, et al. Confocal Single-Molecule FRET for Protein Conformational Dynamics. In: Livesay DR, editor. Protein Dynamics: Methods and Protocols. Humana Press; New York: 2014. pp. 51–62. [DOI] [PubMed] [Google Scholar]
- 109.Michalet X, Colyer RA, Scalia G, et al. Development of new photon-counting detectors for single-molecule fluorescence microscopy. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2013;368:20120035. doi: 10.1098/rstb.2012.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ingargiola A, Panzeri F, Sarkosh N, et al. 8-spot smFRET analysis using two 8-pixel SPAD arrays. Proc. SPIE. 2013;8590:85900E–85900E–11. doi: 10.1117/12.2003704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cohen AE, Moerner WE. Method for trapping and manipulating nanoscale objects in solution. Appl. Phys. Lett. 2005;86:093109. [Google Scholar]
- 112.Shon MJ, Cohen AE. Mass action at the single-molecule level. J. Am. Chem. Soc. 2012;134:14618–23. doi: 10.1021/ja3062425. [DOI] [PubMed] [Google Scholar]
- 113.Reiner JE, Crawford a. M, Kishore RB, et al. Optically trapped aqueous droplets for single molecule studies. Appl. Phys. Lett. 2006;89:013904. [Google Scholar]
- 114.Joo C, Balci H, Ishitsuka Y, et al. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 115.Rasnik I, McKinney S a, Ha T. Surfaces and orientations: much to FRET about? Acc. Chem. Res. 2005;38:542–8. doi: 10.1021/ar040138c. [DOI] [PubMed] [Google Scholar]
- 116.Boukobza E, Sonnenfeld A, Haran G. Immobilization in Surface-Tethered Lipid Vesicles as a New Tool for Single Biomolecule Spectroscopy. J. Phys. Chem. B. 2001;105:12165–12170. [Google Scholar]
- 117.Cisse I, Okumus B, Joo C, et al. Fueling protein DNA interactions inside porous nanocontainers. Proc. Natl. Acad. Sci. 2007;104:12646–50. doi: 10.1073/pnas.0610673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zheng Q, Juette MF, Jockusch S, et al. Ultra-stable organic fluorophores for single-molecule research. Chem. Soc. Rev. 2014;43:1044–56. doi: 10.1039/c3cs60237k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ha T, Tinnefeld P. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu. Rev. Phys. Chem. 2012;63:595–617. doi: 10.1146/annurev-physchem-032210-103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stennett EMS, Ciuba M a, Levitus M. Photophysical processes in single molecule organic fluorescent probes. Chem. Soc. Rev. 2014;43:1057–75. doi: 10.1039/c3cs60211g. [DOI] [PubMed] [Google Scholar]
- 121.Cooper M, Ebner A, Briggs M, et al. Cy3B: improving the performance of cyanine dyes. J. Fluoresc. 2004;14:145–50. doi: 10.1023/b:jofl.0000016286.62641.59. [DOI] [PubMed] [Google Scholar]
- 122.Wysocki LM, Lavis LD. Advances in the chemistry of small molecule fluorescent probes. Curr. Opin. Chem. Biol. 2011;15:752–9. doi: 10.1016/j.cbpa.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 123.Kishino A, Yanagida T. Force measurements by micromanipulation of a single actin filament by glass needles. Nature. 1988;334:74–6. doi: 10.1038/334074a0. [DOI] [PubMed] [Google Scholar]
- 124.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–35. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods. 2006;3:891–3. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 126.Dave R, Terry DS, Munro JB, et al. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys. J. 2009;96:2371–81. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Altman RB, Terry DS, Zhou Z, et al. Cyanine fluorophore derivatives with enhanced photostability. Nat. Methods. 2012;9:68–71. doi: 10.1038/nmeth.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Altman RB, Zheng Q, Zhou Z, et al. Enhanced photostability of cyanine fluorophores across the visible spectrum. Nat. Methods. 2012;9:428–9. doi: 10.1038/nmeth.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Loveland AB, Habuchi S, Walter JC, et al. A general approach to break the concentration barrier in single-molecule imaging. Nat. Methods. 2012;9:987–92. doi: 10.1038/nmeth.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schwartz JJ, Quake SR. Single molecule measurement of the “speed limit” of DNA polymerase. Proc. Natl. Acad. Sci. 2009;106:20294–9. doi: 10.1073/pnas.0907404106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Le Reste L, Hohlbein J, Gryte K, et al. Characterization of dark quencher chromophores as nonfluorescent acceptors for single-molecule FRET. Biophys. J. 2012;102:2658–68. doi: 10.1016/j.bpj.2012.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Takada T, Takeda Y, Fujitsuka M, et al. “Signal-on” detection of DNA hole transfer at the single molecule level. J. Am. Chem. Soc. 2009;131:6656–7. doi: 10.1021/ja9009919. [DOI] [PubMed] [Google Scholar]
- 133.Wang T-H, Peng Y, Zhang C, et al. Single-molecule tracing on a fluidic microchip for quantitative detection of low-abundance nucleic acids. J. Am. Chem. Soc. 2005;127:5354–9. doi: 10.1021/ja042642i. [DOI] [PubMed] [Google Scholar]
- 134.Orte A, Clarke RW, Klenerman D. Fluorescence coincidence spectroscopy for single-molecule fluorescence resonance energy-transfer measurements. Anal. Chem. 2008;80:8389–97. doi: 10.1021/ac8009092. [DOI] [PubMed] [Google Scholar]
- 135.Chen J, Tsai A, Petrov A, et al. Nonfluorescent quenchers to correlate single-molecule conformational and compositional dynamics. J. Am. Chem. Soc. 2012;134:5734–7. doi: 10.1021/ja2119964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen J, Petrov A, Tsai A, et al. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat. Struct. Mol. Biol. 2013;20:718–27. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Novotny L, van Hulst N. Antennas for light. Nat. Photonics. 2011;5:83–90. [Google Scholar]
- 138.Kinkhabwala A, Yu Z, Fan S, et al. Large single-molecule fluorescence enhancements produced by a bowtie nanoantenna. Nat. Photonics. 2009;3:654–657. [Google Scholar]
- 139.Acuna GP, Möller FM, Holzmeister P, et al. Fluorescence enhancement at docking sites of DNA-directed self-assembled nanoantennas. Science. 2012;338:506–10. doi: 10.1126/science.1228638. [DOI] [PubMed] [Google Scholar]
- 140.Yuan H, Khatua S, Zijlstra P, et al. Thousand-fold enhancement of single-molecule fluorescence near a single gold nanorod. Angew. Chem. Int. Ed. Engl. 2013;52:1217–21. doi: 10.1002/anie.201208125. [DOI] [PubMed] [Google Scholar]
- 141.Estrada LC, Aramendía PF, Martínez OE. 10000 Times Volume Reduction for Fluorescence Correlation Spectroscopy Using Nano-Antennas. Opt. Express. 2008;16:20597–602. doi: 10.1364/oe.16.020597. [DOI] [PubMed] [Google Scholar]
- 142.Punj D, Mivelle M, Moparthi SB, et al. A plasmonic “antenna-in-box” platform for enhanced single-molecule analysis at micromolar concentrations. Nat. Nanotechnol. 2013;8:512–6. doi: 10.1038/nnano.2013.98. [DOI] [PubMed] [Google Scholar]
- 143.Levene MJ, Korlach J, Turner SW, et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–6. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 144.Eid J, Fehr A, Gray J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–8. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 145.Tsai A, Petrov A, Marshall RA, et al. Heterogeneous pathways and timing of factor departure during translation initiation. Nature. 2012;487:390–3. doi: 10.1038/nature11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tsai A, Uemura S, Johansson M, et al. The Impact of Aminoglycosides on the Dynamics of Translation Elongation. Cell Rep. 2013;3:497–508. doi: 10.1016/j.celrep.2013.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sameshima T, Iizuka R, Ueno T, et al. Single-molecule study on the decay process of the football-shaped GroEL-GroES complex using zero-mode waveguides. J. Biol. Chem. 2010;285:23159–64. doi: 10.1074/jbc.M110.122101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Korlach J, Marks PJ, Cicero RL, et al. Selective aluminum passivation for targeted immobilization of single DNA polymerase molecules in zero-mode waveguide nanostructures. Proc. Natl. Acad. Sci. 2008;105:1176–81. doi: 10.1073/pnas.0710982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wenger J, Gérard D, Dintinger J, et al. Emission and excitation contributions to enhanced single molecule fluorescence by gold nanometric apertures. Opt. Express. 2008;16:3008. doi: 10.1364/oe.16.003008. [DOI] [PubMed] [Google Scholar]
- 150.Gérard D, Wenger J, Bonod N, et al. Nanoaperture-enhanced fluorescence: Towards higher detection rates with plasmonic metals. Phys. Rev. B. 2008;77:045413. [Google Scholar]
- 151.McQuarrie DA. Stochastic Approach to Chemical Kinetics. J. Appl. Probab. 1967;4:413–478. [Google Scholar]
- 152.Bartholomay AF. Enzymatic Reaction-Rate Theory: A Stochastic Approach. Ann. N. Y. Acad. Sci. 1962;96:897–912. doi: 10.1111/j.1749-6632.1962.tb54110.x. [DOI] [PubMed] [Google Scholar]
- 153.Schnitzer MJ, Block SM. Statistical kinetics of processive enzymes. Cold Spring Harb. Symp. Quant. Biol. 1995;60:793–802. doi: 10.1101/sqb.1995.060.01.085. [DOI] [PubMed] [Google Scholar]
- 154.Kou SC, Cherayil BJ, Min W, et al. Single-molecule Michaelis-Menten equations. J. Phys. Chem. B. 2005;109:19068–81. doi: 10.1021/jp051490q. [DOI] [PubMed] [Google Scholar]
- 155.Shaevitz JW, Block SM, Schnitzer MJ. Statistical kinetics of macromolecular dynamics. Biophys. J. 2005;89:2277–85. doi: 10.1529/biophysj.105.064295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Moffitt JR, Chemla YR, Bustamante C. Methods in statistical kinetics. 1st ed. Elsevier Inc.; 2010. [DOI] [PubMed] [Google Scholar]
- 157.Barkai E, Jung Y, Silbey R. Theory of single-molecule spectroscopy: beyond the ensemble average. Annu. Rev. Phys. Chem. 2004;55:457–507. doi: 10.1146/annurev.physchem.55.111803.143246. [DOI] [PubMed] [Google Scholar]
- 158.Yang H. Change-Point Localization and Wavelet Spectral Analysis of Single-Molecule Time Series. In: Komatsuzaki T, Kawakami M, Takahashi S, Yang H, et al., editors. Single-Molecule Biophysics: Experiment and Theory. Vol. 146. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2011. pp. 219–243. [Google Scholar]
- 159.Gopich IV, Szabo A. Theory of Single-Molecule FRET Efficiency Histograms. In: Komatsuzaki T, Kawakami M, Takahashi S, Yang H, et al., editors. Single-Molecule Biophysics: Experiment and Theory. Vol. 146. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2011. pp. 245–297. [Google Scholar]
- 160.Gopich IV, Szabo A. Theory of the energy transfer efficiency and fluorescence lifetime distribution in single-molecule FRET. Proc. Natl. Acad. Sci. 2012;109:7747–52. doi: 10.1073/pnas.1205120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chung SH, Moore JB, Xia LG, et al. Characterization of single channel currents using digital signal processing techniques based on Hidden Markov Models. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1990;329:265–85. doi: 10.1098/rstb.1990.0170. [DOI] [PubMed] [Google Scholar]
- 162.Andrec M, Levy RM, Talaga DS. Direct Determination of Kinetic Rates from Single-Molecule Photon Arrival Trajectories Using Hidden Markov Models. J. Phys. Chem. A. 2003;107:7454–7464. doi: 10.1021/jp035514+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.McKinney S a, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 2006;91:1941–51. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Greenfeld M, Pavlichin DS, Mabuchi H, et al. Single Molecule Analysis Research Tool (SMART): an integrated approach for analyzing single molecule data. PLoS One. 2012;7:e30024. doi: 10.1371/journal.pone.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bronson JE, Fei J, Hofman JM, et al. Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys. J. 2009;97:3196–205. doi: 10.1016/j.bpj.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bronson JE, Hofman JM, Fei J, et al. Graphical models for inferring single molecule dynamics. BMC Bioinformatics. 2010;11(Suppl 8):S2. doi: 10.1186/1471-2105-11-S8-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Van De Meent J-W, Bronson JE, Wood F, et al. Hierarchically-coupled hidden Markov models for learning kinetic rates from single-molecule data.. Proc. 30th Int. Conf. Mach. Learn.; 2013. [PMC free article] [PubMed] [Google Scholar]
- 168.Van de Meent J-W, Bronson JE, Wiggins CH, et al. Empirical Bayes Methods Enable Advanced Population-Level Analyses of Single-Molecule FRET Experiments. Biophys. J. 2014;106:1327–1337. doi: 10.1016/j.bpj.2013.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Qin F, Auerbach a, Sachs F. Maximum likelihood estimation of aggregated Markov processes. Proc. Biol. Sci. 1997;264:375–83. doi: 10.1098/rspb.1997.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bishop CM. Pattern Recognition and Machine Learning. Springer; New York: 2006. [Google Scholar]
- 171.Taylor JN, Makarov DE, Landes CF. Denoising single-molecule FRET trajectories with wavelets and Bayesian inference. Biophys. J. 2010;98:164–73. doi: 10.1016/j.bpj.2009.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Devore MS, Gull SF, Johnson CK. Reconstruction of Calmodulin Single-Molecule FRET States, Dye-Interactions, and CaMKII Peptide Binding by MultiNest and Classic Maximum Entropy. Chem. Phys. 2013;422:238–245. doi: 10.1016/j.chemphys.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Okamoto K, Sako Y. Variational Bayes analysis of a photon-based hidden Markov model for single-molecule FRET trajectories. Biophys. J. 2012;103:1315–24. doi: 10.1016/j.bpj.2012.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Perez CE, Gonzalez RL. In vitro and in vivo single-molecule fluorescence imaging of ribosome-catalyzed protein synthesis. Curr. Opin. Chem. Biol. 2011;15:853–63. doi: 10.1016/j.cbpa.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Shcherbakova I, Hoskins A a, Friedman LJ, et al. Alternative spliceosome assembly pathways revealed by single-molecule fluorescence microscopy. Cell Rep. 2013;5:151–65. doi: 10.1016/j.celrep.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]