Abstract
Myeloperoxidase (MPO) is expressed by myeloid cells for the purpose of catalyzing the formation of hypochlorous acid, from chloride ions and reaction with a hydrogen peroxide-charged heme covalently bound to the enzyme. Most peroxidase enzymes both plant and mammalian are inhibited by benzoic acid hydrazide (BAH)-containing compounds, but the mechanism underlying MPO inhibition by BAH compounds is largely unknown. Recently, we reported MPO inhibition by BAH and 4-(trifluoromethyl)-BAH was due to hydrolysis of the ester bond between MPO heavy chain glutamate 242 (Glu242) residue and the heme pyrrole A ring, freeing the heme linked light chain MPO subunit from the larger remaining heavy chain portion. Here we probed the structure and function relationship behind this ester bond cleavage using a panel of BAH analogs to gain insight into the constraints imposed by the MPO active site and channel leading to the buried protoporphyrin IX ring. In addition, we show evidence that destruction of the heme ring does not occur by tracking the heme prosthetic group and provide evidence that the mechanism of hydrolysis follows a potential attack of the Glu242 carbonyl leading to a rearrangement causing the release of the vinyl-sulfonium linkage between HC-Met243 and the pyrrole A ring.
Keywords: Myeloperoxidase, Inflammation, Benzoic Acid Hydrazide
INTRODUCTION
Myeloperoxidase (MPO) is a heme-dependent peroxidase, but it is the only one capable of consuming hydrogen peroxide (H2O2) to mediate chloride oxidation to hypochlorous acid (HOCl). In its resting state, MPO contains ferric heme (MPO-Fe(III)). Upon reaction with H2O2 the MPO heme is oxidized to a short-lived intermediate termed Compound I (half life ~100 ms; [1]), which contains a ferryl porphyrin π cation radical (MPO-Fe(IV)=O•+π) (Eq. 1) [2, 3]. In the absence of Cl- and in the presence of classical peroxide electron donor (AH2), MPO follows a typical peroxidase catalytic cycle where compound I is reduced back to the ferric state in two sequential one-electron steps (Eq. 2–3). In the first of these, the porphyrin radical is reduced leaving a ferryl heme known as compound II (Eq. 2). A second equivalent of AH2 then reduces compound II to ferric enzyme (Eq. 3). In the process two equivalents of electron donor are oxidized to the corresponding free radical product (AH•) [4, 5].
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
In the presence of Cl−, MPO compound I is uniquely able to oxidize Cl− to HOCl, and in the process compound I is reduced directly to the ferric state (Eq. 4). Neither Compound II (Eq. 3) nor superoxide-inactivated Compound III (Eq. 5) participates in Cl− oxidation. These reactions (Eq. 1–5) occur through octahedral coordination of the active site Fe by the protoporphyrin IX heme and the proximal histidine 336 on the MPO heavy chain (HCHis336).
MPO also auto-catalytically forms three covalent associations with the porphyrin macrocycle the sum of which is an arrangement found nowhere else in nature. An MPO light chain aspartate (LCAsp94) forms an ester with the methyl side chain of pyrrole C. Additionally, a heavy chain glutamate (HCGlu242) forms an ester with the methyl side chain of pyrrole A, and the immediately adjacent methionine (HCMet243) is involved in a vinyl-sulfonium linkage with pyrrole A [6]. Interestingly, these bonds establish, through the prosthetic group itself, a covalent link between MPO’s light and heavy chains and may account for the distinct saddling observed in the MPO heme. The extent of covalent association between mammalian peroxidases and their heme varies. It is completely absent in all non-animal peroxidases including horseradish peroxidase (HRP) [7–9], lignin peroxidase [10], bacterial catalase-peroxidases (KatG) [11, 12], and ascorbate peroxidase [13], indicating that this type of heme modification is not required for classical peroxidase activity. However, mammalian peroxidases like lactoperoxidase (LPO) have two ester linkages analogous to those observed in MPO but lack the vinyl-sulfonium adduct [14, 15]. In LPO, the ester bonds are between the heme b and its single subunit via LPOGlu375 and LPOAsp225 to pyrrole rings A and C, respectively. It is thought that the covalent tethers between mammalian peroxidases and their heme cofactors afford them a certain level of resistance necessary to protect the heme from oxidation by HOCl and HOBr, which they generate [16].
Recently, we reported that incubation of benzoic acid hydrazide (BAH) with MPO in the presence of H2O2 causes a disruption of the linkages that occurred between the heme b and MPO heterodimer subunits [17]. Analysis of H2O2/BAH-treated MPO by SDS-PAGE revealed the co-migration of heme with the light chain, suggesting that cleavage of the HCGlu242 ester and vinyl- HCMet243 sulfonium preceded loss of the LCAsp94 ester bond. Indeed, H2O2/BAH- induced shifts in heme absorption were also consistent with the disruption of its vinyl-sulfonium linkage [17]. The molecular mechanism by which this cleavage takes place and the role of this cleavage in inhibition of MPO remains to be elucidated. There also has been no study to our knowledge that reports correlation between the MPO heme liberation with any other inhibitors that did not involve concomitant Fe loss. A panel of BAH analogs were used here to probe structure and function (i.e. cleavage) relationship to better understand the underlying mechanism by which the disruption occurs. Furthermore, we tracked how a Cy5-hydrazide inhibitor was incorporated into the MPO protein to determine a key event in the reaction mechanism that should parallel the BAH analog mechanism of MPO inhibition. Using peptide mass mapping, we also identified three MPO lysine (Lys) residues (HCLys138, HCLys308, and HCLys463) where benzoic acid radical form adducts following oxidation by compound I. Additionally, we found a number of methionine (Met) residues (LCMet85, LCMet87, HCMet243, HCMet249, HCMet306, and HCMet385) that were differentially oxidized in the presence of BAH with a relatively low concentration of H2O2 compared to the native protein. Oxidation of HCMet243, in particular, may be a direct result of inhibition by BAH but further studies are needed to refine the exact chemical reactions leading to release of the heme from the HC of MPO. Finally, we tested whether BAH could be used to liberate the heme b prosthetic group from the active site of the analogous ester linkages present in LPO. Taken together, these studies provide new insight into the molecular mechanism of BAH inhibition of MPO providing new avenues for future drug discovery efforts to limit the production of peroxidase-derived oxidants in chronic inflammatory diseases [18].
Experimental procedures
Reagent, Proteins and Chemicals
Ultra-pure myeloperoxidase (MPO) purified from human neutrophils was obtained from Lee Biosolutions Inc. (St. Louis, MO) and lactoperoxidase (LPO) of bovine and superoxide dismutase (SOD) were purchased from Worthington Biochemical Corporation (Lakewood, NJ). 4-aminobenzoic acid hydrazide (4-ABAH), benzoic acid hydrazide (BAH), 4-fluorobenzoic acid hydrazide (4-FBAH), 4-nitrobenzoic acid hydrazide (4-NBAH), sodium azide (NaN3), Dimethyl sulphoxide (DMSO) from Alfa Aesar (Ward Hill, MA). Cyanine5 (Cy5) hydrazide was purchased from Lumiprobe Corporation (Hallandale Beach, FL). H2O2, glutathione (GSH), 2-aminobenzoic acid hydrazide (2-ABAH), 4-(trifluoromethyl) benzoic acid hydrazide (4- TFMBAH), 3-(dimethylamino) benzoic acid hydrazide (3-DMABAH) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Human plasma and fetal bovine serum were purchased from Innovative Research Inc. (Novi, MI). For protein staining, Gelcode Blue was purchased from Pierce (Rochford, IL), with tracking of the heme prosthetic group accomplished by use of the chemiluminescent Western Lightning ultra-reagent from PerkinElmer Inc. (Waltham, MA).
Acetate assay buffer, pH 5.6 was prepared by adjusting the pH of sodium acetate buffer with acetic acid. Working solutions of H2O2 were prepared fresh daily by diluting 30% H2O2 (BDH Chemicals, London, UK) according to the extinction coefficient for H2O2 at 240 nm, 39.4 M−1cm−1 [19, 20]. Cy5-hydrazide, 4-ABAH, 2-ABAH, BAH, 4- FBAH, 4-NBAH, 4-TFMBAH, 3-DMABAH, NaN3 and isoniazid were dissolved in DMSO and subsequently diluted into assay buffer. MEBSS buffer (144 mM NaCl, 5.4 mM KCl, 800 µM MgSO4, 800 µM NaH2PO4, 1.2 mM CaCl2, 5.6 mM glucose, 4 mM Hepes, pH 7.4 with 1% fetal bovine serum) was used in the luminescence assay for MPO activity, in accordance with the literature [21].
SDS-PAGE analysis of (heme b)-LC generation
To determine the effect that BAH analogs and the fluorescent analog Cy5-hydrazide have on the heme catalytic ability of MPO, MPO (1.2 µM) was incubated at room temperature with different BAH analogs following addition of H2O2 (20 µM). These samples of 20 µL were added to non-reducing sample loading buffer (Bio-rad), and loaded without prior heating onto a 4–15% gradient SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Of note, heat treatment was avoided to minimize the autocatalytic cleavage of MPO heavy chain at HCMet243–HCPro244 bond that results in 39 kDa and 20 kDa bands [22]. Protein staining of elaborated gels was performed by GelCode Blue reagent (Pierce) followed by probing of the gel for the movement of the heme b through topical application of Western Lightning reagent (PerkinElmer Inc.). The light production generated as a result of this chemiluminescence reaction was collected using a Fujifilm LAS-1000 luminescence imager. For reactions examining incorporation of Cy5-hydrazide, an additional fluorescence scan was performed with fluorescent-labeled protein bands detected with a FLA-5100 imager with the 670 nM far-red laser and the Cy5 filter set (Fujifilm Inc., Tokyo, Japan). These scans were performed using excitation at λ640 nm and emission at λ670 nm. Total protein staining was done with Gelcode Blue reagent and imaged with Bio-Rad Gel Doc imager using Quantity One version 4.6.9. The luminescence signal of the gel was quantified using ImageJ 64 software (National Institutes of Health, Maryland, USA).
MPO fluorescence assay
MPO peroxidase activity was measured by monitoring the formation of resorufin from the oxidation of ADHP by MPO. This assay was used to determine whether SDS in loading buffer could diminish MPO activity for the gel analysis. Reaction of ADHP (20 µM) and MPO (28 nM) were incubated with or without 2% SDS in assay buffer for 5 mins and initiated by the addition of H2O2 (40 µM) by use of Varioskan Flash plate reader with excitation of λ530nm and emission of λ590nm. SkanIt software 2.4.3 parameters included an interval of 0.1 s for each of 1000 reads.
MPO activity kinetics
To evaluate the inhibitory effect of Cy5-hydrazide on peroxidase activity of MPO, a luminescence assay was performed with luminol as the peroxidase substrate. In reactions of 200 µL total volume, MPO (140 nM) was incubated with the luminol substrate (400 µM) in MEBSS buffer for a series of Cy5-hydrazide concentrations for 5 mins before triggering the reaction by the addition of H2O2 (20 µM). Dispensing of H2O2 was done using the auto-injection function of the Varioskan plate reader controlled by the SkanIt software version 2.4.3. Measurements were taken at 1 s intervals over 2 mins. The inhibitory rate constant for the reaction was not determined due to the inherent difficulty of relating these observed luminescence progress curves to molar product formed, instead apparent IC50 values for Cy5-hydrazide were determined based on the percentage of inhibition of MPO peroxidase activity using Prism software (GraphPad Software, Inc., La Jolla, CA).
Mass spectrometric analysis
To determine the BAH-induced MPO modification, we performed a peptide mass fingerprinting (PMF) experiment using mass spectrometry. The analyzed reactions consisted of MPO alone and MPO with H2O2 in the presence of BAH. These reactions were subjected to tryptic digestion and peptide mapping at Creative Proteomics Inc. (New York, NY). Side-by-side reactions for identical samples of MPO (1.2 µM) with and without H2O2 (40 µM) and BAH (5 mM) in a total of 50 εL and further diluted with 50 εL of 100 mM Tris-HCl (pH 8.5). Then protein disulfide bonds of the diluted sample were reduced for 40 min with 5 mM dithiothreitol at room temperature and alkylated for 40 min with 15 mM iodoacetamide in the dark. The alkylated protein samples were digested overnight at 37°C with sequencing-grade trypsin (Promega Corp., Madison, WI) in a 1:50 enzyme-to-substrate ratio. Following digestion, the peptide mixtures were acidified with trifluoroacetic acid (TFA) to a final concentration of 1% TFA, and desalted by filter-aided sample preparation (FASP) method [23]. Finally, the desalted peptide samples were dried in a vacuum concentrator and these products were analyzed by nanoLC-MS/ MS. For LC-MS/MS analysis of the dried tryptic peptides were dissolved in 10 εL of 0.1% formic acid in water and subjected to Easy-nLC1000 nano-flow UPLC-System (Thermo Fisher Scientific, Dionex Co., CA). The nanoLC separation was accomplished by a four-step gradient from 0.1% formic acid in H2O to 0.1% formic acid in acetonitrile. The Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific, USA) was used to analyze the peptide mixture; raw MS/MS data was analyzed by using MaxQuant version 1.4.1.2 (Max Planck Institute of Biochemistry, Martinsried, Germany). Replicative LC-MS/MS acquisitions and analysis were performed to maximize modification sites identification.
Results
MPO cleavage by H2O2/BAH versus H2O2 alone
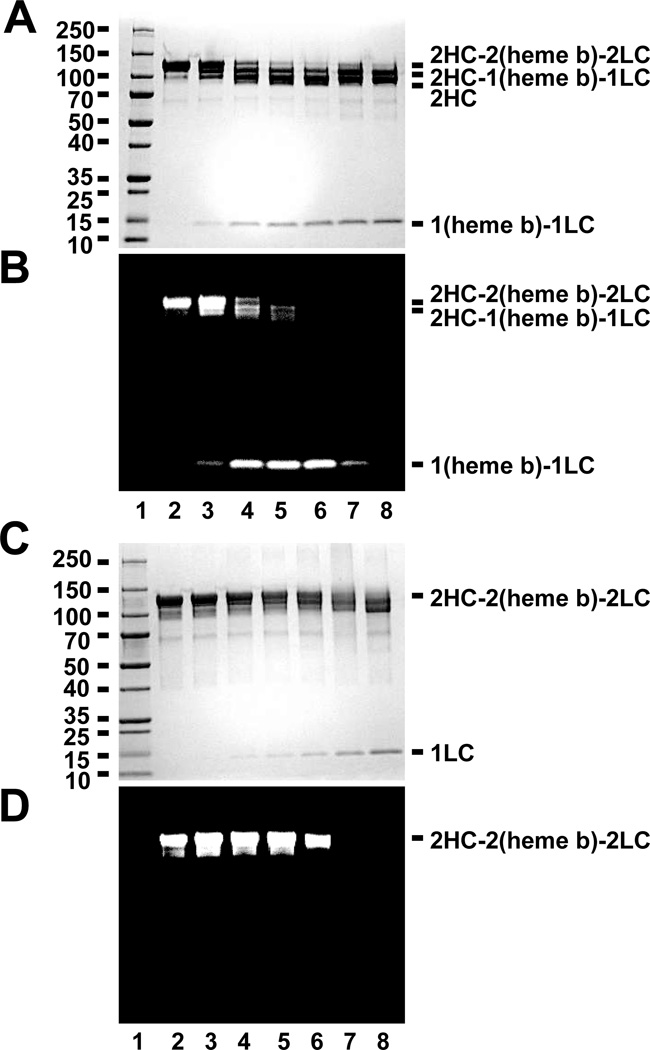
To compare the cleavage products of MPO by BAH, separate reactions were assessed for changes in banding patterns and through tracking the migration of the heme b prosthetic group. Reactions of MPO (1.2 µM) with BAH (2.5 mM, lane 2) in the presence of increasing concentrations of H2O2, namely 8 µM (lane 3), 40 µM (lane 4), 80 µM (lane 5), 160 µM (lane 6), 800 µM (lane 7) and 8 mM (lane 8) for 10 min (Fig. 1A and Fig. 1B) were compared to similar reactions without BAH (Fig. 1C and Fig. 1D). As shown in Figure 1A and 1B, the formation of the (heme b)-LC cleavage product was increased as we increased the concentration of H2O2 from 8 µM to 160 µM. In contrast, treatment of MPO with higher concentrations of H2O2 alone (Fig. 1C and Fig. 1D) showed that the inactivation of MPO by higher concentration of H2O2 (800µM and 8 mM) caused destruction of the heme group as demonstrated by the lack of a functional (heme b)-LC reaction product. Our results agree with the literature observation that incubation of MPO with high relative concentrations of H2O2 would cause lose of the active site Fe molecule [20].
Figure 1. Effect of BAH on MPO compared to the heme destruction generated by high dose H2O2 treatment.
A-B, Total protein-stained gel image (A & C) and luminescence generated images (B & D) for a molecular weight marker (lane 1), MPO alone (1.2 µM, lane 2), and reactions of MPO (1.2 µM) with BAH (5 mM) in the present of increasing concentrations of H2O2, namely 8 µM (lane 3), 40 µM (lane 4), 80 µM (lane 5), 160 µM (lane 6), 800 µM (lane 7) and 8 mM (lane 8) for 10 min. C-D, Similar reactions were set up without BAH for 10 min prior to loading the sample. Experiments conducted as described under “Experimental Procedures.”
Screening of BAH analogs for generation of the (heme b)-LC reaction product
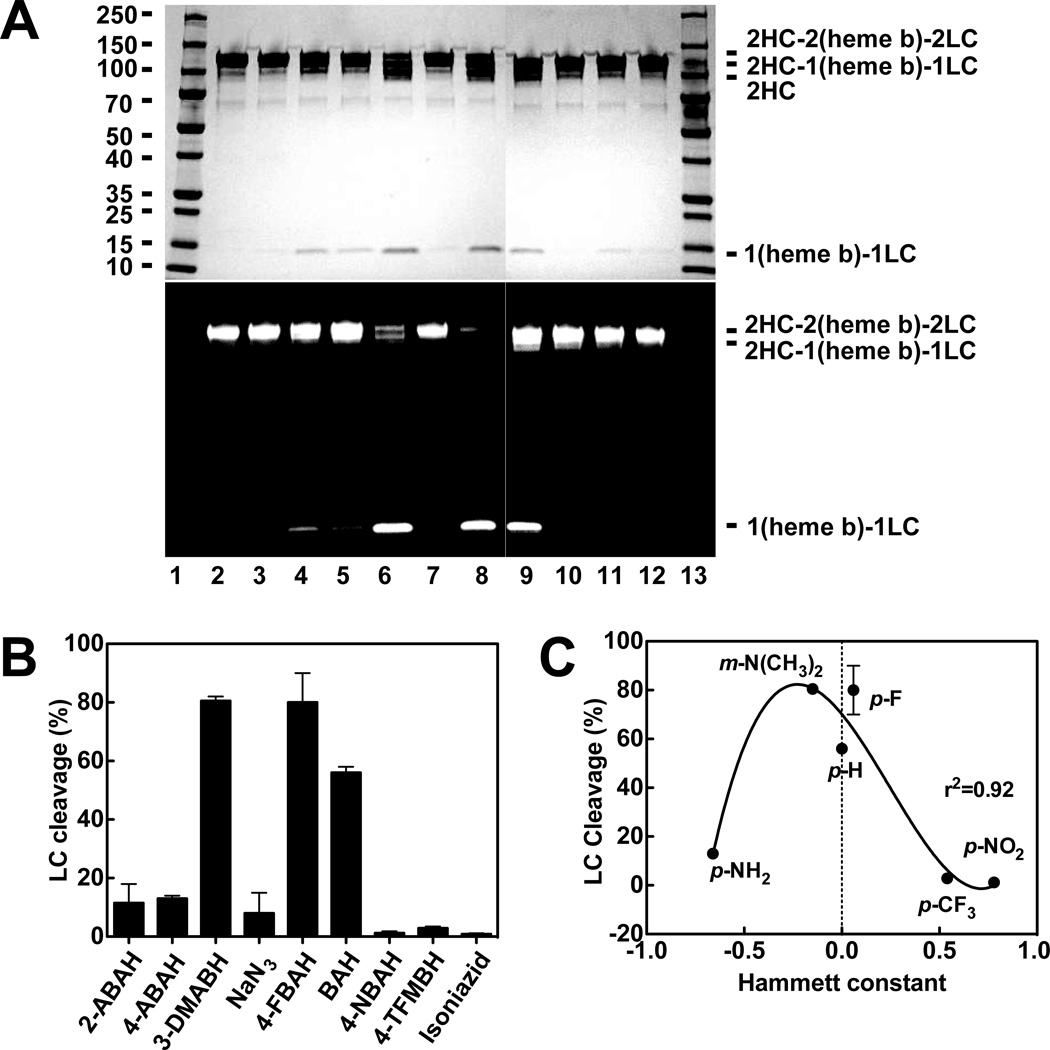
We recently reported the inhibition of MPO by BAH and several of its analogs, determining IC50 values and inhibition constants (Ki) for each one [17]. A table summarizing these data can be found in Supplementary material (Table. S1). In order to explore the impact of hydrazide structure on MPO cleavage, we compared whether BAH analogs can cleave MPO native protein, we investigated the effect of BAH analogs on the MPO protein using SDS-PAGE analysis and our aforementioned ability to track the heme activity. Initially, we assessed the extent of cleavage products generated at concentration equal to 5 folds of the approved IC50 values previously determined [17]. These results indicated that BAH was the most potent modification of this cleavage (data not shown). To determine the extent of cleavage possible, we did the following experiments. MPO was incubated with different BAH analogs at 4mM inhibitor concentration, namely, BAH, NBAH, TFMBAH, isoniazid, 2-ABAH, 4-ABAH, 3-DMABAH, NaN3, and 4-FBAH in the presence of 20 µM H2O2 for 1h. Based on general protein staining and heme-dependent luminescence (Fig. 2A), we found several BAH analogs (i.e. 4-FBAH, BAH and 3-DMABAH) in reactions were cleaved MPO dimer into (heme b)-LC and the HC, but this cleavage product was not present in an appreciable NaN3 and isoniazid. In addition, we quantitated the amount of (heme b)-LC generated by densitometry and expressed these results as LC percentage cleavage shown in Figure 2B. The correlation between the LC cleavage percentages and Hammett constants of BAH analogs were plotted to illustrate the structural and functional cleavage effect of BAH on MPO. We found that three of these compounds, namely, BAH, 4-FBAH and 3-DMBAH incubation mediate the greatest MPO cleavage into the HC and the (heme b)-LC as shown in Figure 2C. These results indicate that the cleavage efficiency of BAH analogs were greatest when the Hammett constant is close to neutral.
Figure 2. Screening of BAH analogs for production of (heme b)-LC cleavage product.
A, A representative protein-stained gel images (upper) and luminescence generated images (lower) for the molecular weight marker (lane 1), MPO alone (1.2 µM, lane 2), 1.2 µM MPO with 20 µM H2O2 (lane 3), 1.2 µM MPO with 20 µM H2O2 incubated for 1 hr with the different BAH analogs each at 4 mM inhibitor concentration, including 2-ABAH (lane 4), 4-ABAH (lane 5), 3-DMABAH (lane 6), NaN3 (lane 7), 4-FBAH (lane 8), BAH (lane 9), 4-NBAH (lane 10), 3-TFMBAH (lane 11) and isoniazid (lane 12). Samples were resolved on a 4–15% SDS gel. B-C, Quantification of the experiment results in Panel A. The cleavage percentage is expressed relative to the protein densitometry for MPO alone. This screen was repeated (n=3) and relative densitometry (%) for the generation of the protein stained (heme b)-LC cleavage product is shown (Panel B) alongside a correlation of (heme b)-LC cleavage product as a function of the Hammett constant (σ) (Panel C), this correlation was analyzed with non-linear regression to a third order polynomial using GraphPad Prism software. Quantification was done from three independent experiments and shown as mean ± S.E.M. Additional methods related to this screen are available under “Experimental Procedures.”
The cleavage of MPO by BAH analogs mediated by formation of MPO compound I
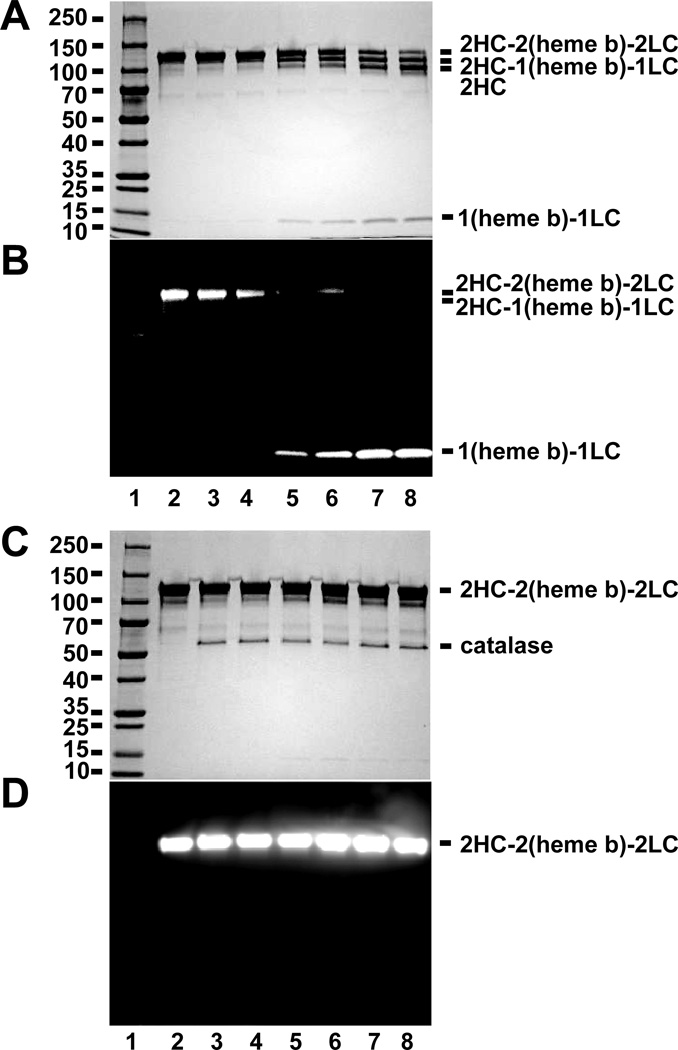
We observed by general protein staining (Fig. 3A) and heme-dependent chemiluminescence (Fig. 3B) the concentration dependent cleavage of MPO by BAH. Notably, no exogenous H2O2 was added to these reactions. To determine whether catalytic amounts of H2O2 were being generated in these reactions, the same reactions were set up with reagents pre-incubated with catalase for 1 hr in prior to mixing. As shown in Figure 3C and 3D, pre-incubation with catalase showed no cleavage with increased BAH concentration. These results indicated that the cleavage of MPO into HC and the (heme b)-LC was dependent on H2O2, implying the participation of MPO compound I in the process. This is consistent with previous reports showing that H2O2 is required to the irreversible inactivation of MPO by 4-ABAH [24].
Figure 3. Cleavage of MPO by BAH is dependent on H2O2.
A-B, Protein-stained image (A & C) and luminescence imaging (B & D) for a molecular weight marker (lane 1), MPO alone (1.2 µM, lane 2), and reactions of MPO (1.2 µM) with increasing concentrations of BAH, namely 0.025 mM (lane 3), 0.25 mM (lane 4), 2.5 mM (lane 5), 5 mM (lane 6), 12.5 mM (lane 7) and 25 mM (lane 8) for 10 min. C-D, Similar reactions where BAH and MPO were separately incubated with catalase (20 units) for 1hr prior to mixing. Experiments conducted as described under “Experimental Procedures.”
Inactivation of MPO by Cy5-hydrazide
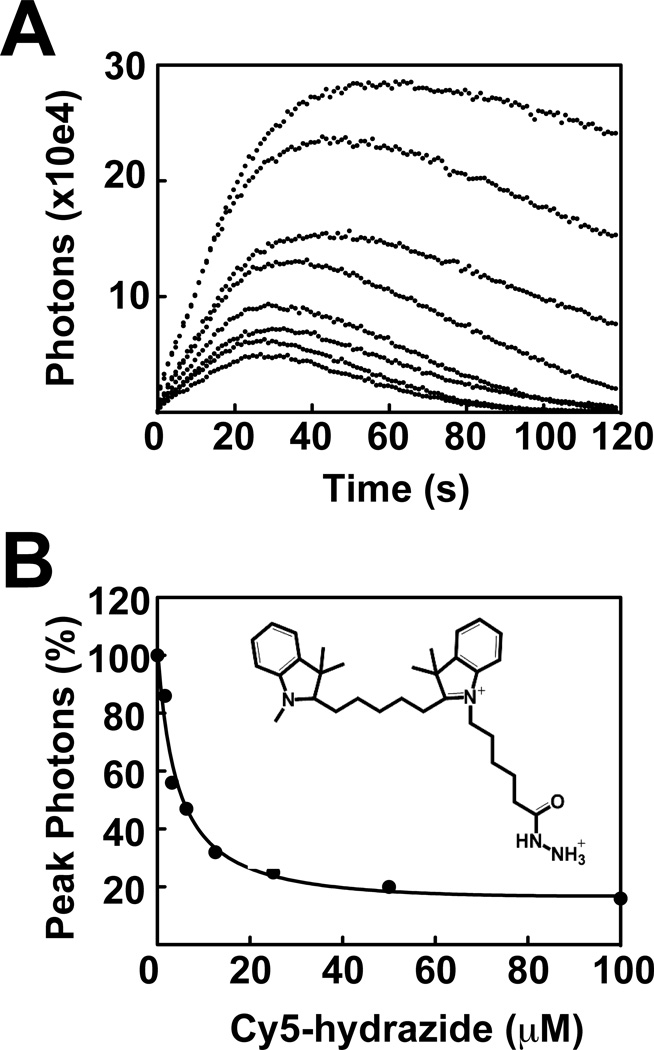
For the tracking of the modification to the MPO subunits, it was necessary inclusion of Cy5-hydrazide produced a concentration-dependent decrease in luminol oxidation catalyzed by MPO/H2O2 (Fig. 4A). Typical reaction traces showed an initial increase in luminescence followed by a steady-state plateau and slow decay of the signal. Increasing concentrations of Cy5-hydrazide resulted in decreased initial rates and extents of luminol oxidation. An IC50 (5.5 µM) for Cy5-hydrazide inhibition of MPO was calculated by plotting the maximum observed luminescence of each kinetic progression curve against Cy5-hydrazide concentration (Fig. 4B). The data were fit to non-linear least squares function in GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA).
Figure 4. Inactivation of MPO by Cy5-hydrazide.
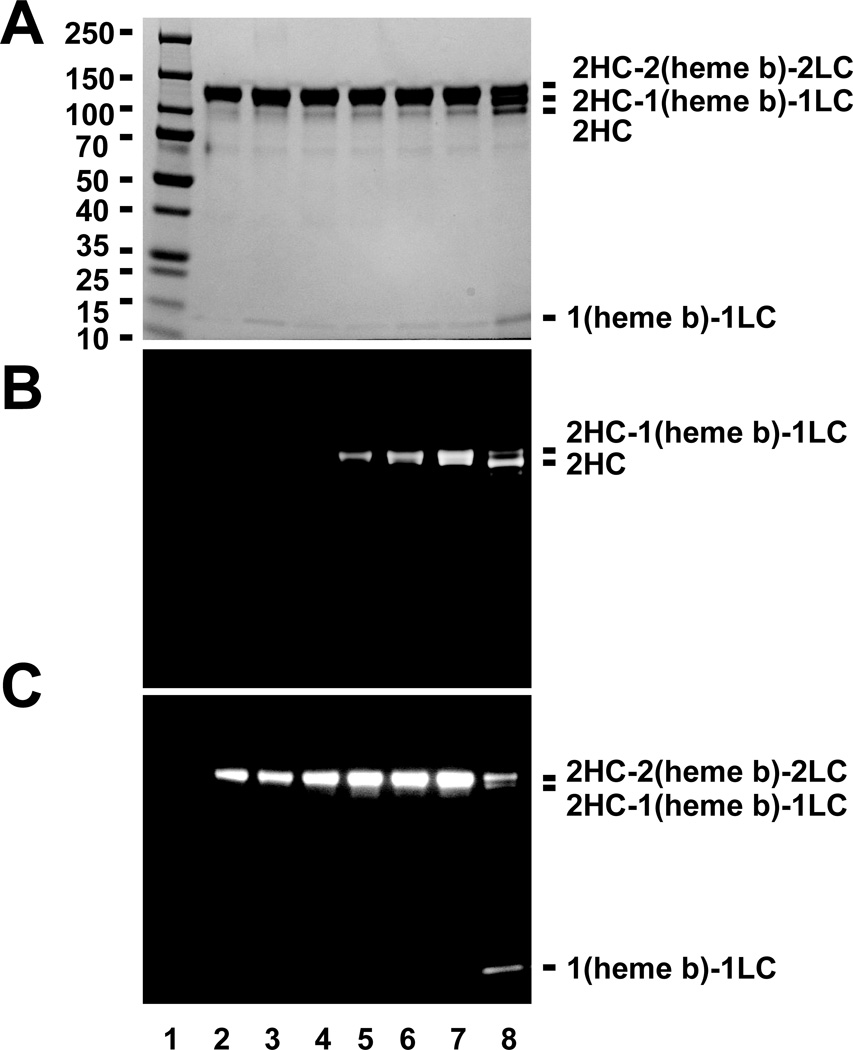
A, Reactions of 400 µM luminol were incubated with MPO (140 nM) in MEBSS buffer in the absence or presence of varied final concentration of Cy5-hydrazide (1.6 µM, 3.125 µM, 6.25 µM, 12.5 µM, 25 µM, 50 µM, 100 µM). The auto-injector function of the Varioskan plate reader was used to initiate the reaction via addition of 20 µM H2O2. B, Apparent IC50 of Cy5-hydrazide (5.5 µM) on MPO was calculated based on the percentage of inhibition of MPO peroxidase activity. Titrations were performed as described in greater detail under “Experimental Procedures.”
Cleavage of MPO by Cy5-hydrazide
A possible consequence of BAH-dependent cleavage of (heme b)-LC from the parent MPO heterodimer is the covalent modification of the protein by the hydrazide. Given the observed inhibition of MPO by Cy5-hydrazide, we sought to use its fluorescent properties to determine whether the process resulted in MPO HC labeling. MPO treated with H2O2 and increasing concentrations of Cy5-hydrazide was evaluated by SDS-PAGE by general protein staining to identify HC-LC cleavage (Fig. 5A), fluorescence to detect Cy5-hydrazide (Fig. 5B), and luminescence to detect protein-linked heme (Fig. 5C). Cy5-hydrazide cleaved MPO dimer into the HC and (heme b)-LC in a manner similar to that seen for BAH, 4-FBAH and DMABAH. Three separated bands of heavy chains (2HC-2(heme b)-2LC dimer, 2HC-1(heme b)-1LC and 2 HC) were produced in reactions containing 5 mM Cy5-hydrazide (Fig. 5A). Further, HC of MPO was labeled with Cy5-hydrazide in a concentration dependent manner (Fig. 5B). Finally, increasing Cy5-hydrazide concentration increased generation of (heme b)-LC (Fig. 5C) consistent with Cy5-hydrazide labeling of the heavy chain. Furthermore, we found the bottom (heme b)-LC band was not modified by direct attachment of the phenyl radical to the heme prosthetic group as the luminescence reaction still where occurring (Fig. 5C). Of note, similar reactions employing fluorescein-hydrazide did not result in labeling of either chain of the MPO protein and did not generate the expected (heme b)-LC fragment (data not shown), presumably due its poor accessibility to the heme active site.
Figure 5. Tracking Cy5 modification following cleavage of the (heme b)-LC from HC of MPO.
Protein-stained bands (A), fluorescence (B), luminescence imaging (C) for a molecular weight marker (lane 1), MPO alone (1.2 µM, lane 2), and reactions of MPO (1.2 µM) with increasing concentrations of Cy5-hydrazide, namely 0.05 mM (lane 3), 0.1 mM (lane 4), 0.5 mM (lane 5), 1 mM (lane 6), 2 mM (lane 7) and 5 mM (lane 8) in the presence of 20 µm H2O2 for 10 min. Additional details related to this experiment can be found in “Experimental Procedures.”
Time-dependent Labeling MPO by Cy5-hydrazide
To determine the relative rates of Cy5-hydrazide labeling and protein cleavage events, we monitored the reaction of MPO with H2O2 and 2.5 mM Cy5-hydrazide at 30 minute intervals for 4 hours. The time course was set up as individual reactions that where stagger started. This allowed us to simultaneously quench the MPO activity by addition of the 2% SDS contained in the gel loading buffer. This experimental design was necessary as we were afraid that the addition of the aliquots from a single reaction into the catalase might not stop these reactions adequately. We were able to validate this quenching protocol by addition of 2 % SDS into fluorescence ADHP assays for reactions of MPO and H2O2. Addition of the SDS completely inactivated the MPO-H2O2 (data not shown). As in Fig. 5, we evaluated SDS-PAGE separated reaction products by general protein staining, fluorescence, and luminescence (Supplementary Fig. 1) Therefore, these results indicated that MPO heavy chain labeled with Cy5-hydrazide in a time dependent manner. Although dependent on maximal intensity parameters used for the fluorescence imaging (Supplementary Fig. 1B), the appearance of the (heme b)-LC product lags behind the Cy5-hydrazide labeling. This indicates that the final release of the (heme b)-LC from the sulfonium linkage maybe a slower process than the initial labeling and ester bond disruption. It is important to note that MPO light chain was still not labeled although light chain is cleaved from the MPO dimer by Cy5-hydrazide as observed in the protein staining gel by Cy5-hydrazide.
Mass spectrometric analysis
To determine which MPO amino acid becomes modified during reaction with hydrazide analogs, we processed identical samples of MPO (1.2 µM) with and without H2O2 (40 µM) and BAH (5 mM). SDS-PAGE analysis of the reactions showed the incubation of with H2O2 and BAH resulted in release of the (heme b)-LC fragment (data not shown). Once confirmed, reaction samples were subjected to tryptic digestion and peptide mapping using LC-MS/MS. To identify the modified sites on MPO, a so-called wqs-104 method was set up, which indicates the modification and reaction of benzoic acid hydrazide (C7H4O) within the MPO protein.. The mass data pooled from the mass spectroscopy analysis was analyzed for accurate mono isotopic mass modifier for C7H5O of 104.03 Da and C7H8N2O of 136.15. The rationale for this approach was based on benzoic acid hydrazide (C7H8N2O) reacted with any amino acid in the MPO protein, hydrazide would be removed thus both products were screened for in both datasets.
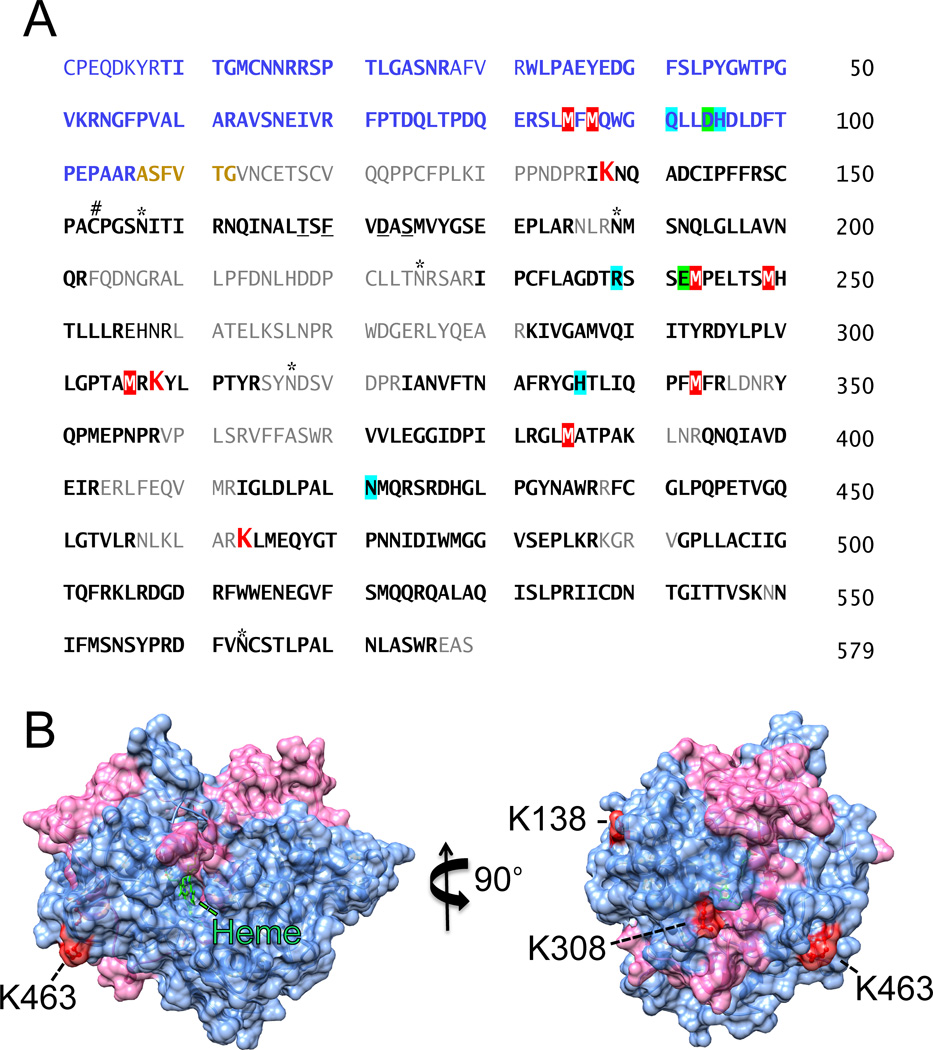
Using the same search parameters for the wqs-104 method, the MPO alone sample lacked the presence of either a 104 or 136 molecular weight adduct. For the inactivated MPO/ H2O2/ BAH sample, three confident wqs-104 modification sites at Lys (K) were identified. As shown in Figure 6A, MS/MS fragmentation of the doubly charged ion (m/z 806.90 Da) gave a sequential b and y ion sequence to match the sequence of one peptide from human MPO (IK138NQADCIPFFR, [M+H]+ 1612.79 Da) with a benzoic acid hydrazide modification. The b and y ions showed that the benzoic acid hydrazide modification site was located at the HCLys138 residue (Table 1). MS/MS fragmentation of the doubly charged ion (m/z 522.78 Da) gave a sequential b and y ion sequence to match the sequence of one peptide from human MPO (K308YLPTYR, [M+H]+ 1044.55 Da) with a benzoic acid radical modified MW. MS/MS fragmentation of the doubly charged ion (m/z 994.16 Da) showed a sequential b and y ion sequence to match the sequence of one peptide from human MPO (K463LMEQYGTPNNIDIWMGGVSEPLKR, [M+H]+ 2980.47 Da) with a benzoic acid hydrazide modification (Supplementary Material Fig. 2). The coverage based on peptides identified by the program represented was 73% and these exact amino acids are indicated by the bold text from the mature MPO protein shown in Figure 6A. To assess the relative proximity of the benzoic acid (added 104 MW) adducts to the central heme and substrate accessibility channel, a representative image was constructed using the coordinates deposited in the Protein Data Bank (NCBI accession code 3F9P). In this representation, the mature MPO protein is a homodimer, each monomer consists of a light chain (106 amino acids, shown as pink) and a heavy chain (467 amino acids, light blue). Modified residues of three Lys residues are depicted in red as shown in Figure 6B.
Figure 6. Identification of modified sites on myeloperoxidase after incubation with BAH and H2O2.
A, LC-MS/ MS analysis rectified 73% coverage of the mature MPO protein. The amino acid sequence of the heterodimer MPO is shown with the light chain designated by the blue text and the heavy chain by the black text. Peptides coverage generated from the tryptic digestion of MPO followed by nanoLC-MS/MS analysis is shown in bold, oxidized methionine residues are highlighted in red with white text, and each modified lysine residues (HCLys138, HCLys308, and HCLys463) are shown in larger red text. Orange text indicates a hexa-peptide excised from the mature MPO protein, thus not factored into the coverage. Highlighted in green are LCAsp94 and HCGlu242 that form ester bond linkages with the heme b. Other important residues are marked including catalytic residues (turquoise), glycosylation sites (*), the bridging cysteine (#) and calcium-binding residues (underlined). (B) The 3D structure of the MPO monomer (PDB #3F9P) is shown with HCLys138, HCLys308, and HCLys463 indicated in red. The mature MPO is a homodimer, each monomer consists of a light chain (106 amino acids, pink) and a heavy chain (467 amino acids, light blue). The structure was oriented with a central heme is in the center and then rotated to the right. The image was created using Chimera (www.cgl.ucsf.edu/chimera).
Table 1.
Mass spectrometry analysis of tryptic MPO peptides indicated three peptides had an increased mass corresponding to the formation of the benzoic acid adducts. Comparison was made between peptides observed in the MPO protein alone and a reaction of MPO (1.2 µM) with BAH (5 mM) in the presence of H2O2 (40 µM) in 50mM sodium acetate buffer at pH 5.7. The statistical probabilities for the correct identification peptides are shown by the peptide–spectrum matches with the Sequest score (XCorr) and the posterior error probability (PEP) values. The smaller the PEP value, the more certainty there is the identification of a correctly matched peptide.
| MPO amino acid |
Identified modified peptide | Oxidized residue |
MH+ [Da] | XCorr | PEP |
|---|---|---|---|---|---|
| HCLys138 | IkNQADcIPFFR | M2 | 1612.79 | 2.25 | 7.86E-3 |
| HCLys308 | kYLPTYR | M1 | 1044.55 | 2.18 | 5.9E-2 |
| HCLys463 | kLMEQYGTPNNIDIWMGGVSEPLKR | M1 | 2980.47 | 4.13 | 6.4E-5 |
In addition, mass spectrometry analysis of tryptic MPO peptides indicated eight peptides had an increased mass corresponding to oxidization of methionine (Met) residues. Comparison with the MPO protein alone, there are 7 methionine (Met) residues (LCMet85, LCMet87, HCMet243, HCMet249, HCMet306, HCMet343 and HCMet385) were differentially oxidized for MPO in a reaction of MPO (1.2 µM) with BAH (5 mM) in the presence of sub-inhibitory concentrations of H2O2 (40 µM) in 50mM sodium acetate buffer as shown in Table 2. Comparison of the oxidized Met residues reported by Paumann-Page et al. for inhibitory concentrations of H2O2 (1.5 mM) eliminated additional oxidation seen in HCMet287, HCMet411, HCMet422, HCMet465 and HCMet478 [19]. Interestingly, the HCMet411 and HCMet465 residues on MPO can be eliminated from consideration also because they were oxidized in the reaction of MPO without H2O2 and may be indicate that some particular Met residues are oxidized at a basal level in MPO.
Table 2.
Mass spectrometry analysis of tryptic MPO peptides indicated eight peptides had an increased mass corresponding to oxidization of methionine (Met) residues. Comparison was made between peptides observed in the MPO protein alone and a reaction of MPO (1.2 µM) with BAH (5 mM) in the presence of H2O2 (40 µM) in 50mM sodium acetate buffer at pH 5.7. The HCMet243 and HCMet249 peptide was identified as the sole peptide not previously identified as a sensitive to oxidization by inhibitory concentrations (1.5 mM) of H2O2 as previous reported by Paumann-Page et al. [19]. The statistical probabilities for the correct identification peptides are shown by the peptide–spectrum matches with the Sequest score (XCorr) and the posterior error probability (PEP) values. The smaller the PEP value, the more certainty there is the identification of a correctly matched peptide.
| MPO Amino Acid | Identified Modified Peptide | Oxidized Residue |
MH+ [Da] | XCorr | PEP |
|---|---|---|---|---|---|
| LCMet85, LCMet87 | SLmFmQWGQLLDHDLDFTPEPAAR | M3; M5 | 2850.32 | 2.95 | 1.71E-2 |
| LCMet87 | SLMFmQWGQLLDHDLDFTPEPAAR | M5 | 2834.32 | 4.41 | 6.42E-8 |
| HCMet243, HCMet249 | SSEmPELTSmHTLLLR | M4; M10 | 1876.91 | 2.89 | 9.39E-6 |
| HCMet306 | KIVGAMVQIITYRDYLPLVLGPTAmR | M25 | 2934.63 | 2.91 | 4.43E-2 |
| HCMet343 | IANVFTNAFRYGHTLIQPFmFR | M20 | 2659.36 | 4.07 | 4.7E-5 |
| HCMet385 | GLmATPAK | M3 | 804.42 | 1.69 | 2.4E-2 |
The cleavage specificity of MPO by BAH
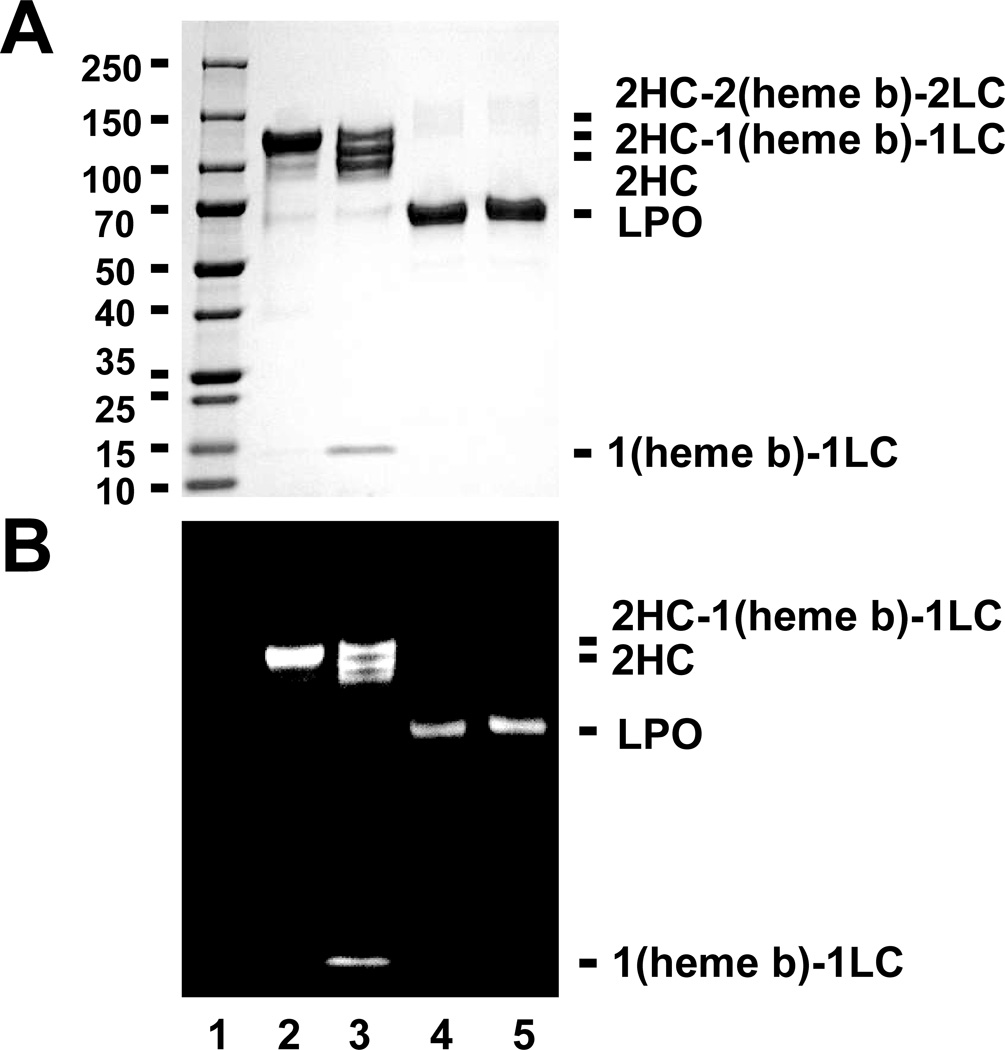
To evaluate whether this unique mechanism of ester bond hydrolysis and heme release is specific to MPO or broadly to all such linked peroxidases, we analyzed identical conditions using either MPO or LPO. As previously mentioned, LPO is also a mammalian peroxidase and, similar to MPO, it possesses 2 ester bonds between the heme pyrrole rings A and C. BAH mediated cleavage was seen for MPO and not LPO. Figure 7 is the results of obtained for general protein staining (Fig. 7A) and heme-dependent chemiluminescence (Fig. 7B). Treatment of LPO with BAH and H2O2 had no effect on the protein as monitored by either method. The LPO protein remained intact (Fig. 7A), and the same quantity active heme remained associated with the LPO protein (Fig. 7B). We found no evidence for the hydrolysis of the ester bonds in the case of LPO incubation with BAH as determined by the lack of degradation or cleavage bands or loss of catalytic potential due to heme liberation for the parent protein (Fig. 7). It is possible that lack of cleavage of the heme in the case of LPO is governed by other structural constraints that the BAH does not match well with and that another hydrazide could fit the LPO lock more efficiently and allow for release the heme b for this peroxidase. These results indicate that BAH-mediated peroxidase cleavage is specific to MPO and suggest that fluorescent hydrazides could be applied for the specific labeling of MPO.
Figure 7. BAH mediated release of heme b is specific to MPO.
Total protein-stained gel images (Panel A) and luminescence generated images (Panel B) for a molecular weight marker (lane 1), MPO alone (1.2 µM, lane 2), and reactions of MPO (1.2 µM) with 20 mM BAH in the presence of 20 µM H2O2 (lane 3), LPO (1.5 µM) alone (lane 4), and LPO (1.5 µM) with 20 mM BAH in the presence of 20 µM H2O2 (lane 5) at room temperature for 30 min. Experiments conducted as described under “Experimental Procedures.”
DISCUSSION
This study had 2 broad goals (1) to probe the structure and function relationship between BAH mediated cleavage of ester linkages from the heme b prosthetic group to the parent MPO heterodimer subunits and (2) to gain insight into the underlying chemistry that allows this cleavage to occur. The concentration dependent relationship for reactions in the presence and absence of BAH in mixtures of increasing H2O2 concentration presented in Figure 1 clearly demonstrates that the underlying mechanism of inactivation of MPO by BAH is fundamentally different from the heme destruction caused by high levels of H2O2 [20]. Most obviously, the latter results in the disruption of the porphyrin macrocycle and release of iron (Fe) from the cofactor, which is evident in lanes 7 and 8 in both cases. In contrast, the hydrazide-dependent mechanism leaves the heme intact over a significant range of concentration and this activity with respect to oxidation to produce chemiluminescent signal remains associated with the LC-heme product (Fig. 1).
Hydrazide-containing compounds are well-documented inhibitors of MPO and others peroxidases, including studies of HRP by phenylhydrazide [25, 26] and microperoxidase-11 by 4-ABAH [27]. The inhibition of various peroxidases have been studied for a number of these enzymes but most demonstrate loss of the Fe coordination by the modification of the heme b ring as is the case for the hydrazide-induced carbene Fe-porphyrin complex described for cytochrome P450 [28] and or methionine-oxidation of MPO by incubation with excess H2O2 [20]. In both of these cases, the Fe is lost due to rearrangement of electrons in the architecture of the heme. However, hydrazide-mediated ester bond disruption is a new observation in the field of MPO inactivation [17]. Here we probed the ability of different BAH analogs to fit into the active site channel of MPO and how modifications to the inhibitor might impact their relative access to the HC ester bond position between the pyrrole ring C and a distal HCGlu242. To do this we again tracked the heme b via chemiluminescence signal (Fig. 2A-B) and correlated the extent of (heme b)-LC protein band development with their respective Hammett coefficients (σ) for the meta- and para-substituted compounds (Fig. 2C). In the Hammett relationship analysis, positive σ indicates an electron-withdrawing group and a negative value for σ is an electron-donating group, so –CF3 has an overall + 0.54 electron-withdrawing potential and the –NH2 substituent carries a σ value of −0.66. Interestingly, maximal generation of (heme b)-LC product occurs at near neutral electron potential indicating that the cleavage is not dependent on the electron potential associated with the aromatic ring and may instead be dependent on accessibility to the heme b ring in the MPO active site (Fig. 2C). Once accessibility was a potential concern of the inhibitors to the heme b prosthetic group, we want to confirm the compound I was the oxidizing species and that H2O2 was expressly required (Fig. 3).
To probe the mechanism by which BAH analogs mediate the disruption of HCGlu242 ester bond linkage, we characterized the inhibitory effect of a fluorescence hydrazide analog as a means to track the underlying chemistry. Luminescence-based kinetic results showed that indeed addition of Cy5-hydrazide and H2O2 mediated inhibition of MPO (Fig. 4) and follow-up SDS gel analysis of the MPO/Cy5-hydrazide/H2O2 system confirmed disruption of the ester bond at HCGlu242 (Fig. 4) and vinyl-sulfonium linkage. The loss of vinyl-sulfonium bond must occur either directly through electron rearrangements triggered by the initial disruption of the ester bond linkage or by a separate nucleophilic attack event. Of note, access of the inhibitor-tethered probes to the catalytic site directly affected the cleavage and resulting labeling. For instance, both cleavage and labeling were not seen for other fluorescein reporters hydrazides, presumably due lack of the aliphatic hydrocarbon chains that are present in the Cy5-hydrazide (see Fig. 4B inset for structure of the Cy5-hydrazide used here). It appears from our data (Fig. 5) that loss of the vinyl-sulfonium bond maybe an overall slower process evident by the delay or lag seen in the appearance of the (heme b)-LC band relative to the Cy5-hydrazide labeling of the HC (Fig. 5B & Supplementary Fig. 1B). The labeling did not appear non-specific in nature under our conditions, as the parent MPO bands were not modified by the Cy5-hydrazide during either the concentration dependence (Fig. 5) or the time course shown (Supplementary Fig. 1), thus indicating modification of MPO by the Cy5-hydrazide only followed the cleavage event.
To better understand the sites of modification of MPO caused by BAH inhibition, a peptide mass fingerprinting (PMF) study was performed comparing MPO alone and MPO treated with a relatively low concentrations H2O2 (40 µM) in the presence of BAH. Our goal was to determine sites that may help to explain the Cy5-hydrazide inhibition and labeling experiments. Coverage of identified proteins was 73% of the mature MPO protein (Fig. 5) and scanning for the peptides modifications of either 136 or 104 Da showed three Lys (HCLys138, HCLys308, and HCLys463) where benzoic acid adducts (104 Da) had formed following oxidation of the BAH by compound I causing the release of the NH2-NH2. The literature does report that formation of phenyl radicals follows incubation of phenylhydrazide with HRP in the presence of H2O2, whereby the N2 is lost upon radical formation and the phenyl radical adds directly to the heme b ring, which presumable causes loss of the centrally bonded Fe [26]. A similar radical formation would explain the addition of the 104 Da adduct to each of the Lys residues in lieu of the possible full BAH adduct (136 Da), but unlike the case with HRP we have no evidence for the any these radicals being added to the covalently tethered heme ring of MPO. Unfortunately, these Lys modifications once mapped on the MPO crystal structure (Fig. 5B) demonstrated the modification were peripheral to the heme and important active site residues, thus implying these alteration do not explain the inhibition of BAH by MPO. In fact, these benzoic acid radicals may modify the Lys residues on the back of the neighboring juxtaposed MPO molecule in the dimer.
One draw back of this PMF method is that to identify the modification you must have prior knowledge of the underlying chemistry and deviation from the predictions are not readily apparent. It is interesting that we can observe oxidation of the HCMet243, presumably due to the loss of the vinyl-sulfonium bond consistent with our previous results [17]. Optimally, application of the fluorescence probe tracker (i.e. Cy5-hydrazide) could be used to address this fundamental question as to where the R-group of a R-NH2-NH2 compound (i.e. R-group is Cy5) would ultimately reside. Solubility issues of MPO labeled with Cy5-hydrazide hampered initial mass spectrometer studies and thus we had to revert back to aforementioned BAH-based studies. Use of Cy5-hydrazide demonstrated that the dye did not co-migrate with the (heme b)-LC band on the SDS-gel (see Fig. 4 & Supplementary Fig. 1). Collectively, our data only supports the scenario, whereby the Cy5 labels the HC of MPO. It is possible would apply to any hydrazide analog capable of mediating the hydrolysis of the ester bond at HCGlu242 including BAH, 4-FBAH, and DMABAH. It is possible that in order to mediate the cleavage of the ester bond and specific labeling of the HC is transferred as the cleavage occurs potentially through a nucleophilic substitution (i.e. SN2) reaction with the terminal NH2- group of the Cy5-hydrazide and the carbonyl of the HCGlu242 directly bonded to the ester linkage. To be clear, this potential mechanism would allow for bond formation between the amino group and carbonyl, as the ester bond is lost. We determined that BAH could not mediate the necessary cleavage in the absence of the H2O2 (Fig. 6C-D) indicating a role for MPO driven catalysis in the mechanism of disruption of ester bond at HCGlu242. It is apparent that a further chemical rearrangement leads to oxidation of the HCMet243 as indicated by the PMF results but the exact chemical nature of transient species remains illusive and future studies needed to determine to characterize this further.
Taken together, our results show that BAH and other benzyl hydrazide can mediate this unique ester bond hydrolysis that is different from the inhibition recently described for H2O2 [19]. We show that BAH must be oxidized by compound I to mediate the cleavage of the heme anchoring ester linkages. We showed that certain fluorescent reporter hydrazide compounds mediated a similar cleavage event as seen for BAH of the MPO protein releasing the still active (heme b)-LC subunit from the parent HC of MPO. This labeling is predicted to be possible through a nucleophilic attach at the carbonyl at HCGlu242 leading to chemical rearrangements resulting in the loss of the sulfonium ion linkage between HCMet243 and the pyrrole A ring. Although unable to provide this by MS analysis, we did observe oxidation of the HCMet243 to a sulfoxide, which resulted in the loss of the vinyl-sulfonium bond. We also presented evidence for modification to the MPO probe based on BAH reaction in the presence of H2O2 causes other important changes to the MPO protein including benzoic acid radical adduct formation of three Lys (HCLys138, HCLys308, and HCLys463) residues and oxidation of the HCMet243 and HCMet249 residues can not be ascribed to simple incubation with H2O2. Finally, we showed that this unique cleavage is specific to MPO and does not occur to an appreciable level in the analogous mammalian peroxidase LPO (Figure 7). Our findings presented here may provide new avenues to fuel drug discovery efforts to designing new MPO inhibitors that would limit inflammation.
Supplementary Material
The abbreviations used are
- MPO
myeloperoxidase
- LC
light chain of myeloperoxidase
- HC
heavy chain of myeloperoxidase
- Cy5-hydrazide
cyanine5 hydrazide
- ABAH
aminobenzoic acid hydrazide
- 2-ABAH
2-aminobenzoic acid hydrazide
- 4-ABAH
4-aminobenzoic acid hydrazide
- BAH
benzoic acid hydrazide
- 4-FBAH
4-fluorobenzoic acid hydrazide
- 4-NBAH
4-nitrobenzoic acid hydrazide
- 4-TFMBAH
4-(trifluoromethyl) benzoic acid hydrazide
- 3-DMABAH
3-(dimethylamino) benzoic acid hydrazide
- HOCl
hypochlorous acid
- H2O2
hydrogen peroxide
- GSH
glutathione
- HRP
horseradish peroxidase
- LPO
lactoperoxidase
- SOD
superoxide dismutase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supported by the National Institutes of Health through grants provided by the National Heart, Lung, and Blood Institute, R00HL094533 (to P.P.) & R01HL114477 (to P.P.), the National Institute of Allergy and Infectious Diseases grant 2R44AI085840-02 (to P.P.) and Auburn University Research Initiative Cancer Graduate Fellowship (to J.H.).
References
- 1.Harrison JE, Araiso T, Palcic MM, Dunford HB. Biochemical and biophysical research communications. 1980;94:34–40. doi: 10.1016/s0006-291x(80)80183-5. [DOI] [PubMed] [Google Scholar]
- 2.Everse J. Free radical biology & medicine. 1998;24:1338–1346. doi: 10.1016/s0891-5849(97)00451-6. [DOI] [PubMed] [Google Scholar]
- 3.Kettle AJ, van Dalen CJ, Winterbourn CC. Redox report: communications in free radical research. 1997;3:257–258. doi: 10.1080/13510002.1997.11747120. [DOI] [PubMed] [Google Scholar]
- 4.Andrews PC, Krinsky NI. Analytical biochemistry. 1982;127:346–350. doi: 10.1016/0003-2697(82)90185-3. [DOI] [PubMed] [Google Scholar]
- 5.Andrews PC, Krinsky NI. The Journal of biological chemistry. 1982;257:13240–13245. [PubMed] [Google Scholar]
- 6.Furtmuller PG, Zederbauer M, Jantschko W, Helm J, Bogner M, Jakopitsch C, Obinger C. Archives of biochemistry and biophysics. 2006;445:199–213. doi: 10.1016/j.abb.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 7.Huang L, Wojciechowski G, de Montellano PRO. Journal of Biological Chemistry. 2006;281:18983–18988. doi: 10.1074/jbc.M602307200. [DOI] [PubMed] [Google Scholar]
- 8.Wojciechowski G, Huang L, Ortiz de Montellano PR. Journal of the American Chemical Society. 2005;127:15871–15879. doi: 10.1021/ja054084t. [DOI] [PubMed] [Google Scholar]
- 9.Colas C, Kuo JM, Ortiz de Montellano PR. The Journal of biological chemistry. 2002;277:7191–7200. doi: 10.1074/jbc.M109523200. [DOI] [PubMed] [Google Scholar]
- 10.Poulos TL, Edwards SL, Wariishi H, Gold MH. The Journal of biological chemistry. 1993;268:4429–4440. doi: 10.2210/pdb1lga/pdb. [DOI] [PubMed] [Google Scholar]
- 11.Yamada Y, Fujiwara T, Sato T, Igarashi N, Tanaka N. Nature structural biology. 2002;9:691–695. doi: 10.1038/nsb834. [DOI] [PubMed] [Google Scholar]
- 12.Baker RD, Cook CO, Goodwin DC. Biochemistry. 2006;45:7113–7121. doi: 10.1021/bi052392y. [DOI] [PubMed] [Google Scholar]
- 13.Patterson WR, Poulos TL. Biochemistry. 1995;34:4331–4341. doi: 10.1021/bi00013a023. [DOI] [PubMed] [Google Scholar]
- 14.Colas C, de Montellano PRO. Journal of Biological Chemistry. 2004;279:24131–24140. doi: 10.1074/jbc.M401687200. [DOI] [PubMed] [Google Scholar]
- 15.Colas C, Ortiz de Montellano PR. Chemical reviews. 2003;103:2305–2332. doi: 10.1021/cr0204303. [DOI] [PubMed] [Google Scholar]
- 16.Huang L, Ortiz de Montellano PR. Archives of biochemistry and biophysics. 2006;446:77–83. doi: 10.1016/j.abb.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Huang J, Smith F, Panizzi P. Archives of biochemistry and biophysics. 2014;548:74–85. doi: 10.1016/j.abb.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forman HJ, Augusto O, Brigelius-Flohe R, Dennery PA, Kalyanaraman B, Ischiropoulos H, Mann GE, Radi R, Roberts LJ, 2nd, Vina J, Davies KJ. Free radical biology & medicine. 2015;78:233–235. doi: 10.1016/j.freeradbiomed.2014.10.504. [DOI] [PubMed] [Google Scholar]
- 19.Beers RF, Jr, Sizer IW. The Journal of biological chemistry. 1952;195:133–140. [PubMed] [Google Scholar]
- 20.Paumann-Page M, Furtmuller PG, Hofbauer S, Paton LN, Obinger C, Kettle AJ. Archives of biochemistry and biophysics. 2013;539:51–62. doi: 10.1016/j.abb.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross S, Gammon ST, Moss BL, Rauch D, Harding J, Heinecke JW, Ratner L, Piwnica-Worms D. Nature medicine. 2009;15:455–461. doi: 10.1038/nm.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor KL, Pohl J, Kinkade JM., Jr The Journal of biological chemistry. 1992;267:25282–25288. [PubMed] [Google Scholar]
- 23.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Nature methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 24.Kettle AJ, Gedye CA, Winterbourn CC. The Biochemical journal. 1997;321(Pt 2):503–508. doi: 10.1042/bj3210503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ator MA, David SK, Ortiz de Montellano PR. The Journal of biological chemistry. 1987;262:14954–14960. [PubMed] [Google Scholar]
- 26.Ator MA, Ortiz de Montellano PR. The Journal of biological chemistry. 1987;262:1542–1551. [PubMed] [Google Scholar]
- 27.Arvadia P, Narwaley M, Whittal RM, Siraki AG. Archives of biochemistry and biophysics. 2011;515:120–126. doi: 10.1016/j.abb.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 28.Lange M, Mansuy D. Tetrahedron Lett. 1981;22:2561–2564. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.