Summary
Several studies have reported reprogramming of fibroblasts to induced cardiomyocytes; however, reprogramming to proliferative induced cardiac progenitor cells (iCPCs) remains to be accomplished. Here we report that a combination of eleven or five cardiac factors along with canonical Wnt and JAK/STAT signaling reprogrammed adult cardiac, lung and tail-tip fibroblasts into iCPCs. The iCPCs were cardiac mesoderm-restricted progenitors, which could be extensively expanded while maintaining multipotency to differentiate into cardiomyocytes, smooth muscle cells and endothelial cells in vitro. Moreover, iCPCs injected into the cardiac crescent of mouse embryos differentiated into cardiomyocytes. iCPCs transplanted into the post-myocardial infarction mouse heart improved survival and differentiated into cardiomyocytes, smooth muscle cells and endothelial cells. Lineage reprogramming of adult somatic cells into iCPCs provides a scalable cell source for drug discovery, disease modeling, and cardiac regenerative therapy.
Introduction
The advent of induced pluripotent stem cells (iPSCs) has revived interest in earlier research showing stable transdifferentiation of somatic cells is possible by forced expression of defined factors (Davis et al., 1987). Previous studies have reported lineage reprogramming into a diverse range of differentiated cells types including neurons (Vierbuchen et al., 2010), hepatocytes (Sekiya and Suzuki, 2011) and cardiomyocytes (CMs) (Ieda et al., 2010; Song et al., 2012). More recently, lineage reprogramming to tissue-specific progenitors has been achieved including neural (Han et al., 2012) and hepatic progenitor cells (Yu et al., 2013). Using transdifferentiation to produce progenitor cells rather than terminally differentiated cell types provides potential advantages for both drug discovery and regenerative medicine applications. Reprogrammed progenitors are proliferative and thus more scalable. Lineage restricted induced progenitor cells may be superior for therapeutic applications due to their ability to proliferate and differentiate into the needed complement of cell types required to fully reconstitute the diseased or damaged tissue. Induced progenitor cells may also provide a more efficient and reproducible platform to obtain tissue-specific terminally differentiated cell types compared to pluripotent stem cells (PSCs).
Cardiac progenitor cells (CPCs) have been identified using various markers in the developing and adult heart. During embryogenesis, CPCs of both first and second heart fields reside in the cardiac crescent. Several studies have isolated CPCs from embryos and embryonic stem cells (ESCs) using transcription factor (TF)-based reporters like Mesp1, Isl1, and Nkx2.5, but a master regulator of the CPC state has not yet been identified (Bondue et al., 2011; Masino et al., 2004; Moretti et al., 2006). Cell surface markers including Cxcr4, Pdgfr-α, Flk1/KDR and SIRPA have been used to identify PSCs-derived CPCs. (Dubois et al., 2011; Kattman et al., 2011). CPCs have also been identified in the adult mammalian heart using markers including Sca1 and cKit which in small animal studies have demonstrated multi-lineage potency following transplantation to the post-MI myocardium (Ellison et al., 2013; Oh et al., 2003). However, in vitro multi-lineage differentiation of adult CPCs has been difficult to demonstrate especially with regard to differentiation to contracting cardiomyocytes (Noseda et al., 2015), and the regenerative capacity of adult c-kit+ CPCs after cardiac injury has been questioned (van Berlo et al., 2014).
Reprogramming to a stem or progenitor cell state requires knowledge of a specific combination of master regulatory factors as well as appropriate culture conditions that can maintain self-renewal and multipotency. Typically the culture conditions for reprogramming mimic those optimized for the in vitro culture of native stem cells based on both empiric optimization and knowledge of developmental signaling pathways. For example, in the case of iPSCs, the distinct culture conditions optimized for mouse and human ESC culture were utilized to generate mouse and human iPSCs, respectively (Takahashi and Yamanaka, 2006; Yu et al., 2007). Likewise, reprogramming to induced neural stem cells employed standard adult neural stem cell medium (Han et al., 2012). In contrast to commonly used neural stem cell medium, variable culture conditions have been used for adult heart-derived CPCs (Ellison et al., 2013; Oh et al., 2003;). It has also proven difficult to generate culture conditions and appropriate signaling to maintain and expand embryonic or PSC-derived CPCs.
Recently, mesodermal SSEA1 progenitors have been maintained with robust cardiac differentiation potential (Cao et al., 2013), but to generate and maintain human PSC-derived cardiac-restricted progenitors has required transgenic forced expression of an oncogene; c-Myc (Birket et al., 2015). Thus, the lack of clearly defined culture conditions for the maintenance and expansion of both adult and PSC-derived CPCs has increased the challenge in transdifferentiating cells to CPCs, and likely contributes to the limited success to date in converting fibroblasts to proliferative and multipotent CPCs (Islas et al., 2012).
Here we show that a defined set of cardiac factors complimented by appropriate culture conditions can reprogram adult mouse fibroblasts from three different tissues to iCPCs. iCPCs were stably reprogrammed, cardiac mesoderm-restricted, clonal progenitors that could be extensively passaged, and showed multipotency toward cardiovascular lineages (CMs, SMs, ECs) in vitro and following transplantation to the embryonic cardiac crescent or the adult post-myocardial infarction heart. Cardiac progenitor reprogramming technology holds promise as a scalable cell source for drug discovery, disease modeling, and cardiac regenerative therapy.
RESULTS
Screening for Cardiac Progenitor Cell Inducing Factors
Based on the known expression pattern of genes that are critical during embryonic cardiovascular development, we selected 18 candidate genes to test their reprogramming ability. These included cardiac transcription factors and cardiac chromatin remodeling factors. We also included iPSC factors reasoning that they may facilitate reprogramming. These 22 genes were individually cloned into a doxycycline (dox) inducible lentivirus vector generating an expression library of defined factors (Figure S1A); dox-induced expression of the factors was confirmed at the mRNA and protein level in transduced fibroblasts (Figure S1B-D, extended experimental procedures).
We utilized an Nkx2.5 cardiac reporter mouse model expressing Enhanced Yellow Fluorescent Protein (EYFP) in the developing heart during the initial stages of cardiac development (E7.75 – E 10.5) (Masino et al., 2004). The developmentally restricted Nkx2.5-EYFP reporter identifies early CPCs, but it is inactive during later stages of cardiac development (E11 onwards) and in the adult heart. The Nkx2.5-EYFP mouse was crossed with a transgenic mouse expressing reverse tetracycline transactivator (rtTA) to enable dox-inducible transgene expression. Cardiac fibroblasts were isolated from adult double transgenic (Nkx2.5-EYFP/rtTA) mice by explant culture (Figure 1A and Figure S1E). Immunostaining of the isolated fibroblasts with markers of CPCs (Irx4, Nkx2.5, Gata4) as well as cardiac lineage cells such as CMs (α-actinin, α-MHC), SMs (SM-MHC), and ECs (CD31) revealed no staining (Figure S1F-J).
Figure 1. Screening for iCPC Inducing Factors and Optimal Culture Conditions.
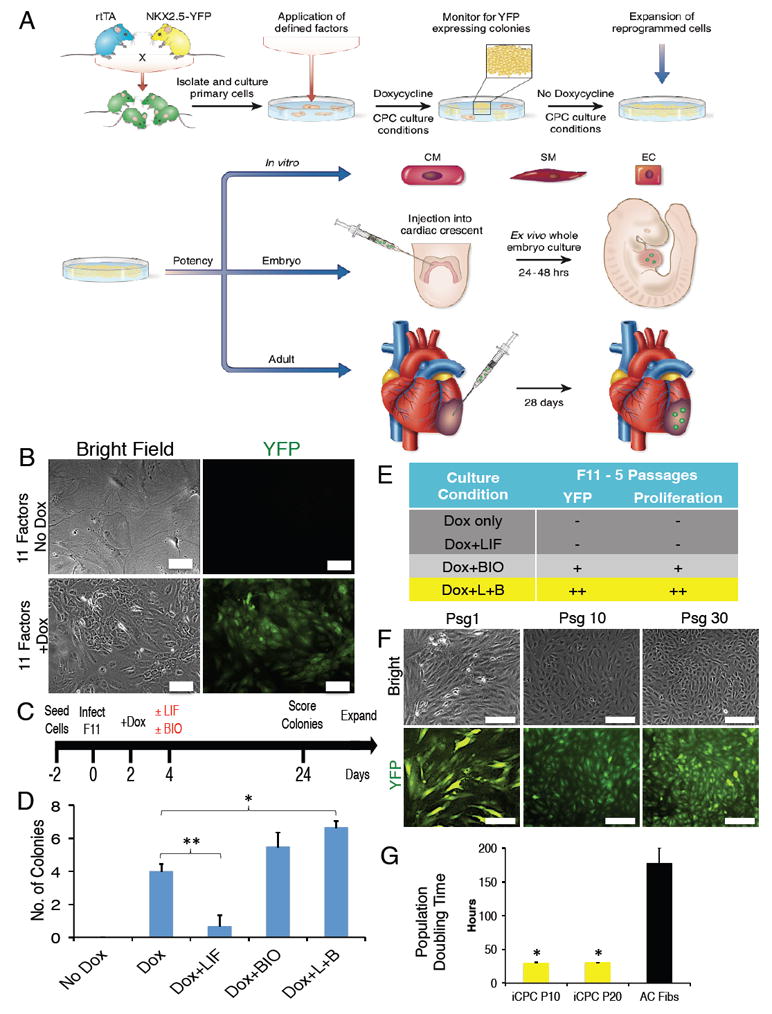
(A) Schematic representation of experimental design depicting direct reprogramming of adult fibroblasts to iCPCs by defined factors and culture conditions, expansion of iCPCs, and in vitro as well as in vivo differentiation of iCPCs into cardiac-lineage cells. (B) Infection with a combination of 11 cardiac factors induced Nkx2.5-EYFP expression in AC Fibs only after dox induction. (C) Strategy to test the impact of culture conditions on F11 reprogramming efficiency as well as the ability of EYFP+ cells to maintain a proliferative state. (D) Number of EYFP+ colonies formed (per 50,000 starting cells) in the respective culture conditions (**p<0.01, *p<0.05). (E) Impact of culture conditions on EYFP+ colonies expanded up to 5 psgs scoring for EYFP+ expression and proliferative ability (Dox only: n=8, Dox+LIF: n=3, Dox+BIO: n=4, Dox+LIF+BIO: n=9). (L=LIF, B=BIO). (F) F11 iCPCs maintained EYFP expression and proliferative ability for at least 30 psgs after dox withdrawal. (G) Population doubling time for psg 10 (P10) and psg 20 (P20) F11 iCPCs as compared to uninfected AC Fibs (n=3). Data presented as mean. Error bars = SEM. Scale bar = 100 μm in B, 200 μm in F. See also Figure S1, S2 and Movie S1.
Uninfected adult cardiac fibroblasts (AC Fibs) did not express EYFP and senesced after 3-4 passages (psgs) (Figure S2A). As a first test of the dox-inducible library for reprogramming, AC Fibs were infected with iPSC factors; dox treatment produced proliferative cells that formed EYFP- iPSC colonies (Figure S2B). Next, we tested infection of AC Fibs with iCM factors (GMT), but even after extended dox induction (6 weeks), we did not observe contracting cells or EYFP+ cells. However, neonatal cardiac fibroblasts infected with iCM factors reprogrammed into spontaneously contracting, EYFP- iCMs after 4 weeks of dox treatment (Figure S2C & Movie S1). These results demonstrate dox-inducible reprogramming with the described vector system. Furthermore, the Nkx2.5-EYFP reporter is not activated during iPSC or iCM reprogramming.
Infection of AC Fibs with a mixture of lentiviruses containing all 22 factors resulted in a small number of EYFP+ proliferative colonies only after dox treatment (Figure S2D). Next, we subtracted iPSC factors from the 22-factor library and infected fibroblasts with 18 cardiac factors. Proliferative EYFP+ cells were again observed three weeks after dox treatment (Figure S2D). Reasoning that factors expressed early in cardiac development might have the highest potential to reprogram fibroblasts into iCPCs, we chose 11 early cardiac factors (Mesp1, Mesp2, Gata4, Gata6, Baf60c, SRF, Isl1, Nkx2.5, Irx4, Tbx5, Tbx20) to infect AC Fibs. Infection with 11 factors gave rise to proliferative EYFP+ cells only after dox treatment (Figure 1B).
We analyzed the time course of appearance of EYFP+ cells upon infection with the 11 factors. Single EYFP+ cells were detected as early as day 4 after dox treatment. By 3 weeks these EYFP+ cells developed into two-dimensional, highly proliferative colonies of EYFP+ cells that lost their parental fibroblast morphology and exhibited a high nuclear-cytoplasmic ratio (Figure S2E). Infection with 11 factors reproducibly gave rise to EYFP+ proliferative colonies (4 colonies/50,000 cells; efficiency 0.008%) (Figure1C-D). We manually isolated these EYFP+ colonies and tried to expand them by splitting. However, cells lost EYFP expression and senesced after 3-5 psgs in the ‘dox only’ culture condition (Figure 1E and S2F).
Wnt and JAK/STAT Signaling Promotes Proliferative Reprogrammed Cells
The overexpression of cardiac factors alone, even though sufficient to produce EYFP+ colonies, was insufficient for maintaining EYFP+ cells in a proliferative, reprogrammed state indicating that additional signaling cues might be necessary. Canonical Wnt signaling is critical for proliferation of CPCs (Cao et al., 2013; Qyang et al., 2007) and JAK/STAT signaling is important for normal cardiogenesis (Foshay et al., 2005). Therefore, we tested the effect of supplementing reprogramming medium with BIO (canonical Wnt activator) and/or LIF (JAK/STAT activator) on reprogramming efficiency as well as the ability of EYFP+ cells to maintain a proliferative state (Figure 1C). Surprisingly, addition of LIF alone inhibited the generation of EYFP+ cells and colony formation. Addition of BIO alone resulted in a similar reprogramming efficiency as dox only; however, the EYFP+ cells became spindle-like upon passaging and were not highly proliferative. The LIF+BIO combination produced the brightest EYFP+ cells and the EYFP+ cells were robustly expandable (Figure 1C-E and S2F-H). Hence, both LIF and BIO are included in our reprogramming medium, ‘iCPC induction medium.’ 11-factor infection followed by culture in iCPC induction medium produced 6-9 EYFP+ colonies (per 50,000 starting cells - 0.013% reprogramming efficiency) that could be continuously expanded on splitting (F11 iCPCs). To determine whether LIF+BIO was necessary for initial reprogramming, we tested whether EYFP+ colonies generated by the dox only condition could be expanded by addition of LIF+BIO later during passaging. We observed that addition of LIF+BIO starting at psg1 allowed for robust expansion of dox only EYFP+ cells, indicating that the impact of LIF+BIO was more on the maintenance of the reprogrammed state than on initiation of reprogramming.
To determine whether continued forced expression of cardiac factors was required to maintain the iCPC state, we withdrew dox from the iCPC induction medium (iCPC maintenance medium) after 2 psgs and assessed whether EYFP+ cells remained proliferative. Cells maintained EYFP expression as well as their proliferative ability for over 30 psgs (Figure 1F). The EYFP+ cells continued to reduce in size during the initial psgs until psg 2-3, when they reached a steady state after which their morphology remained unchanged during further passaging. Next, we determined the population doubling time of iCPCs that had been passaged in iCPC maintenance medium for 10 and 20 psgs. Both psg 10 and 20 iCPCs had similar population doubling time of about 30 hours, which was significantly less than AC Fibs (Figure 1G). These results suggest that iCPCs were stably reprogrammed and maintained their proliferative state in the presence of LIF+BIO and without exogenous induction of cardiac factors by dox.
5-Factors are Sufficient to Reprogram Adult Cardiac Fibroblasts to iCPCs
We wanted to determine whether iCPCs could be reprogrammed using a subset of the 11 factors. Initially, we tested a core combination of three factors (within the 11-factor combination) that are expressed earliest in cardiac development: Mesp1, Tbx5 and Gata4 (MTG). We infected AC Fibs with MTG and cultured them in iCPC induction medium, but we did not observe emergence of any EYFP+ colonies for up to 4 weeks. Therefore, we tested if addition of remaining factors within the 11-factor pool to MTG can induce colony formation. Addition of Baf60c, Isl1 and Nkx2.5 to MTG reproducibly generated EYFP+ colonies after 3 weeks (Figure 2A, Figure S2I). Although the 4-factor combinations produced EYFP+ cells, their proliferative ability as well as EYFP expression progressively declined with subsequent passaging in iCPC maintenance medium. This suggested that 4-factor combinations induced partial reprogramming and were insufficient to epigenetically stabilize cells in the iCPC state. Since MTG+Nkx2.5 (MTGN) produced the most colonies, we tested if addition of the other factors that produced colonies in the 4 factor combination, Baf60c or Isl1, to MTGN could facilitate stable reprogramming to iCPCs (Figure 2A, Figure S2J). Addition of both Baf60c and Isl1 to MTGN produced proliferative colonies, but Baf60c+MTGN (MTGNB) produced the most expandable colonies post dox withdrawal and hence was selected for further characterization. Infection with MTGNB (5F) reproducibly gave rise to expandable F5 iCPCs (~7.25 colonies/50,000 cells) (Figure 2B), which could be stably expanded in iCPC maintenance medium (without dox) for at least 20 psgs (Figure S3A). Three 5F iCPC lines underwent karyotype analysis, which demonstrated that 2 of 3 lines exhibited normal karyotypes while the third line had numerical and structural abnormalities (Figure S3B). These results suggest that 5 Factors stably reprogrammed AC Fibs into proliferative iCPCs.
Figure 2. Cardiac Factors Stably Reprogram Adult Cardiac Fibroblasts into Proliferative iCPCs.
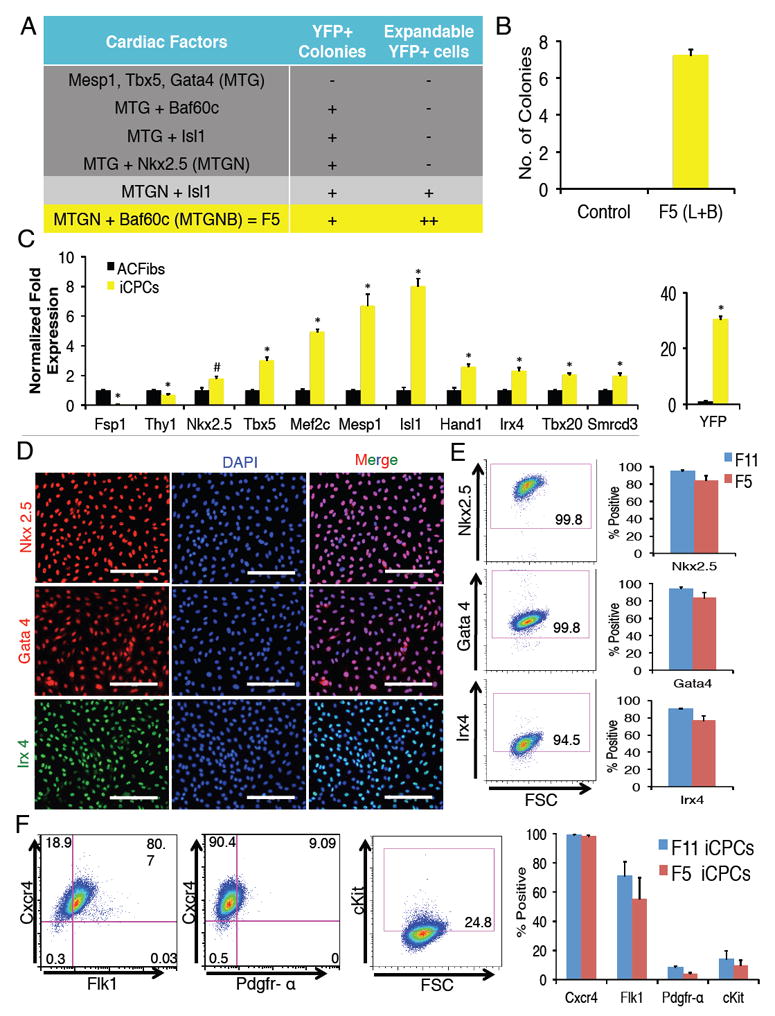
(A) Factor combinations tested both for ability to produce Nkx2.5-EYFP+ colonies and to expand them for at least 5 psgs without dox. (n=3-5) (B) Number of EYFP+ colonies produced after infection with 5 factors (MTGNB) and culture in iCPC induction medium for 3 weeks (per 50,000 seeded cells, n=8). (L=LIF, B=BIO). (C) qPCR analysis of F11 iCPCs showed upregulation of CPC markers and downregulation of fibroblast markers. Data represent normalized fold expression relative to uninfected AC Fibs (*p<0.01, #p<0.05). (D) Immunofluorescence labeling of F11 iCPCs showed nuclear localization of cardiac TFs Nkx2.5, Gata4, and Irx4. (E) Flow cytometry analysis revealed that majority of F11 iCPCs expressed cardiac TFs. F11 & F5 iCPCs showed comparable expression of cardiac TFs. (F) Flow cytometry analyses showed that F11 iCPCs expressed cell surface makers such as Cxcr4, Flk1, Pdgfr-α, cKit that are associated with CPCs (n=3). F11 & F5 iCPCs showed comparable expression of CPC associated cell surface markers. Error bars = SEM. Scale bars = 200 μm. See also Figure S1, S2 and S3 (for F5 iCPCs).
Cardiac Factors Stably Reprogram Adult Cardiac Fibroblasts into Cardiac Mesoderm-restricted iCPCs
To analyze iCPC-gene expression we performed qPCR analysis on psg 1 iCPCs that revealed upregulation of key CPC transcription factors including Nkx2.5, Tbx5, Mef2c, Mesp1, Tbx20, and Irx4 accompanied by down regulation of the fibroblast-specific gene Fsp1 (Figure 2C). These results indicate that iCPCs initiated the cardiac program at the expense of the fibroblast program, a hallmark of lineage reprogramming. Next, we performed immunostaining for CPC-related TFs. In contrast to AC Fibs, which did not immunolabel for Nkx2.5, Gata4 or Irx4 (Figure S1F), iCPCs exhibited nuclear localization of these TFs that remained constant across psgs 5-25 (Figure 2D, S3C S4A-C). Flow cytometry demonstrated that greater than 80% of the iCPC expressed Nkx2.5, Gata4, and Irx4 (Figure 2E). Further, we assessed whether iCPCs expressed cell surface markers associated with CPCs (Kattman et al., 2011; Nelson et al., 2008). Flow cytometry analysis revealed that iCPCs homogenously expressed Cxcr4; however, only a fraction of iCPCs expressed Flk1, Pdgfr-α or cKit (Figure 2F, S3D). We found no protein expression for pluripotency (Oct4) or cardiac lineage differentiation markers (α-actinin, SM-MHC, CD31) even after extensive passaging (Figure S4F). Comparable results were observed for these experiments whether 11F or 5F iCPCs were used.
To characterize the transcriptome of iCPCs, we performed RNA-seq analysis on early psg (1-3) as well as late psg (8-10) iCPCs. As a positive control we utilized gene expression values for a mESC-derived CPC population previously described by (Wamstad et al., 2012), and the gene expression was compared with uninfected AC Fibs. We found that genes involved in cardiovascular development including TFs (Tbx3, Hes1, Prrx1, Foxa2, Gata4/6, Meis1, Gli2), signaling molecules (LIF, Vegfc, Grem1, Fgf2), cell surface markers (cKit, Pdgfr- α, Notch1, Gpc3) and chromatin remodeling genes (Smarcd3, Hdac 2/5/7/10, Jarid2) were increasingly upregulated as iCPCs were passaged (Figure 3A). In contrast, fibroblast-specific genes (Postn, Twist2, Thy1) were increasingly downregulated with passaging (Figure 3B). Furthermore, CM differentiation markers (Actc1, Myh6, Myl2, Myl7) were not expressed in iCPCs. Interestingly, genes associated with SM (Cnn1, Myh11) and EC (Pecam1) were upregulated in one early psg replicate. However, these genes were downregulated in late psg iCPCs. Primitive streak genes (Gsc, Mixl1, T) were not detected (Figure S4D). Likewise, progenitor genes for endoderm, ectoderm and non-cardiac mesoderm were not expressed by iCPCs (Figure S4E). mESC-CPCs used by (Wamstad et al., 2012) were derived from the whole population of cells after 5 days of differentiation. This population was highly enriched for CPCs but potentially contained a minority of cells from non-cardiac lineages. Hence, mESC-CPC samples had higher expression for some endoderm, ectoderm and non-cardiac mesoderm markers. Additionally, Bmp (4/6/7) genes that induce cardiac differentiation were also downregulated. Importantly, iCPCs did not express markers of pluripotent stem cells (Pou5f1, Esrrb, Dppa2/3, Lin28a, Sox2) (Figure S4D); however, we did observe upregulation of Nanog (see discussion). A pairwise Pearson’s correlation analysis of all expressed genes revealed that transcription profiles of both low and high psg iCPCs have higher correlation with mESC-CPCs as compared to mESC-CMs (Wamstad et al., 2012) (Figure 3C). Gene Ontology (GO) terms associated with upregulated genes in iCPCs include categories such as “positive regulation of cell proliferation,” “negative regulation of cell differentiation,” and “cardiovascular system development,” whereas terms associated with the downregulated genes include categories such as “cell adhesion,” “cell differentiation,” and “apoptosis” (Figure 3D). In aggregate, these data suggest that iCPCs are cardiac mesoderm-restricted precursors.
Figure 3. iCPCs Exhibit Cardiac-Mesoderm Restricted Gene Expression, are Multipotent and Differentiate into Cardiomyocytes, Smooth Muscle Cells and Endothelial Cells in vitro.
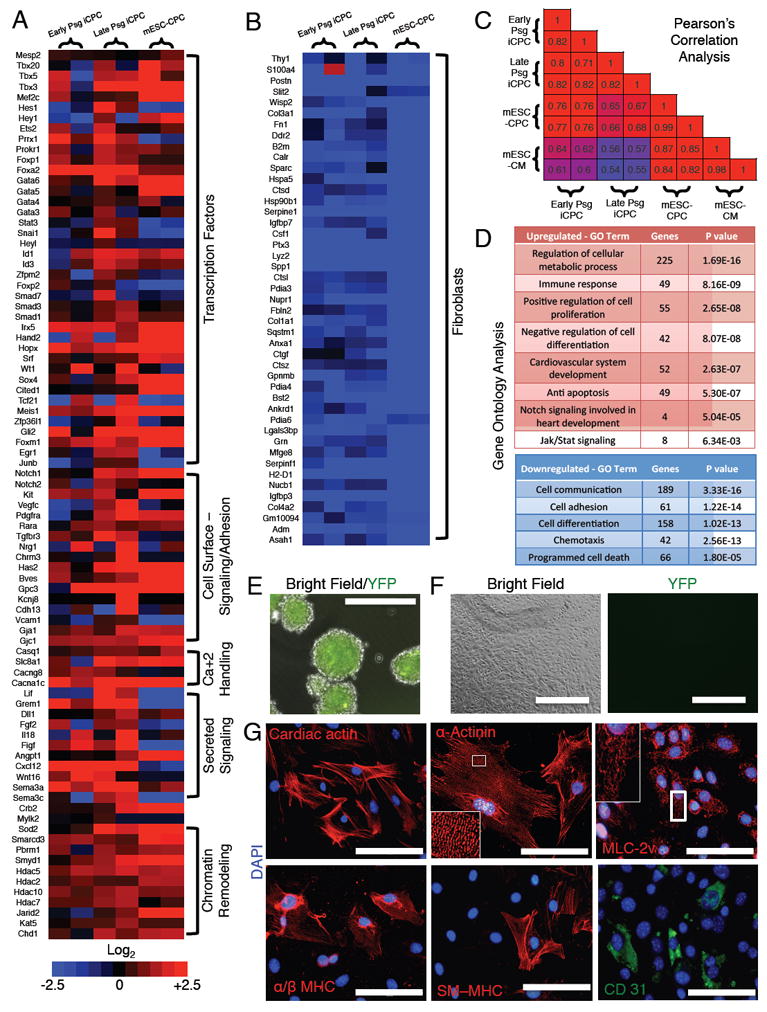
(A-B) Heat maps of RNA Seq data illustrating differentially expressed genes in early psg (1-3), late psg (8-10) iCPCs and mESC-CPCs (Wamstad et al., 2012) as compared to AC Fibs. (n=2, biological replicates in each group). (A) Genes involved in embryonic cardiovascular development were increasingly upregulated in iCPC with psgs. (B) Fibroblasts genes were strongly downregulated. (C) Pearson’s correlation analysis of all expressed genes among low psg iCPCs, high psg iCPCs, mESC-CPCs and mESC-CMs (Wamstad et al., 2012). Numbers indicate Pearson’s R values. (D) Gene ontology analysis performed for upregulated and downregulated genes in late psg iCPCs as compared to AC Fibs. (E) F11 iCPCs aggregated in cardiac differentiation medium were EYFP+ at day 2. (F) F11 iCPC aggregates were plated and cultured in low serum conditions and lost Nkx2.5-EYFP expression by day 20. (G) Immunocytochemistry on plated cells revealed expression of CM markers such as cardiac actin, α-actinin (note highly organized sarcomere staining), MLC-2v, α/β MHC, a SM marker SM-MHC and EC marker CD31. Scale bars = 400 μm in E & F, 100 μm in G. See also Figure S3 and S5
iCPCs Differentiate into Cardiomyocytes, Smooth Muscle Cells and Endothelial Cells
To determine whether iCPCs were capable of differentiation into cardiovascular lineages, iCPCs were aggregated in cardiac differentiation medium. iCPCs maintained EYFP expression in aggregates. However, 20 days after plating, cells lost EYFP expression, suggesting that the iCPCs exited the progenitor state and differentiated (Figure 3E-F). Immunocytochemistry revealed differentiated cells expressing CM (cardiac actin, α-actinin, MLC-2a, MLC-2v, α/β MHC), SM (SM-MHC) or EC (CD31) markers (Figure 3G, S3E). Among iCPC-differentiated cells, a majority stained positive for CM-markers (80-90%) and only a fraction stained for SM (5-10%) and EC markers (1-5%) (Figure 3G S3F). These results suggest that iCPCs were multipotent, capable of differentiating into three types of cardiovascular lineage cells. We evaluated the differentiation potential of psg 5 and 30 iCPCs and observed that multipotency was comparable across psgs (Figure S5A). The finding of comparable potency across passages argues against contaminating cell types in the starting AC Fibs confounding the results. Most MLC-2v positive CMs also labeled for MLC-2a, indicating that they were relatively immature. However, we did observe cells that exclusively stain for MLC-2v or MLC-2a (<5%) suggesting that iCPCs can differentiate into both atrial-like and ventricular-like CMs (Figure S5B-C).
Even after attaining highly organized sarcomeres following extended culture periods under low serum conditions, the iCPC-derived CMs did not exhibit spontaneous contractions. We reasoned that co-culturing iCPC-CMs with mESC-derived CMs may provide additional mechanical, electrical and paracrine stimulation to induce further maturation and contraction. Co-culturing with rat CMs has been previously shown to induce contraction in iCMs (Wada et al., 2013). Hence, we infected iCPC-CMs with a constitutive GFP expressing lentivirus to identify reprogrammed cells and co-cultured them with mESC-derived CMs expressing td-tomato. We did not detect cells that co-expressed both GFP and td-tomato (Figure 4A), suggesting that cell fusion between iCPC-CMs and mESC-CMs was unlikely. We immunostained the co-cultured cells for CM markers as well as GFP and noticed that GFP+ iCPC-CMs and GFP- mESC-CMs both stained positive for CM markers and grew side by side as monolayers (Figure S5D). Moreover, immunostaining for Cx43 revealed that iCPC-CMs developed abundant gap junctions with both mESC-CMs as well as other iCPC-CMs (Figure 4B). After 10-14 days of co-culturing, 5-10% of iCPC-CMs started synchronously contracting with mESC-CMs (Movie S2). The contracting iCPC-CMs also showed spontaneous calcium transients that were similar to those in mESC-CMs in frequency and amplitude (Figure 4C-D, Figure S5E-F and Movie S3).
Figure 4. iCPC-CMs Show Contraction, Calcium Transients upon Co-culture with mESC-CMs and Single Cell Derived iCPC Clones Exhibit Cardiovascular Potency.
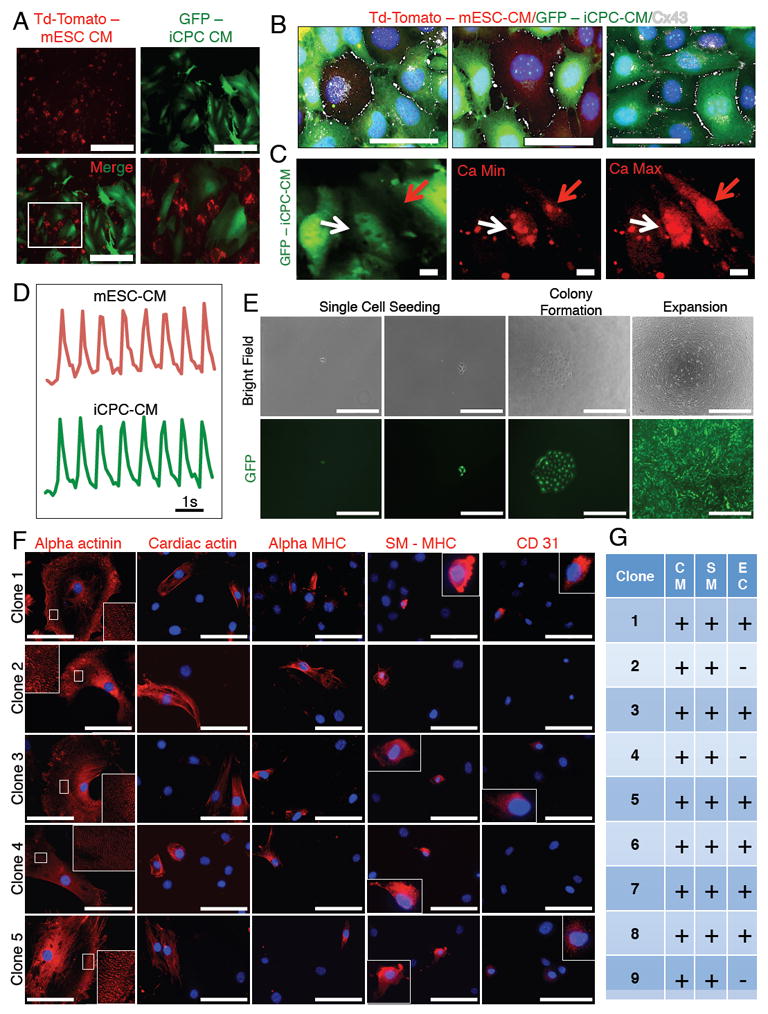
(A) iCPC-CMs infected with a GFP expressing lentivirus co-cultured with mESC-CMs that expressed td-tomato. No cell fusion was detected. (B) Cx43 immunolabeling showed that iCPC-CMs developed gap junctions with mESC-CMs and other iCPC-CMs. (C) iCPC-CMs showed synchronous calcium transients with mESC-CMs 3 weeks after co-culture. White arrow = iCPC-CM, red arrow = mESC-CM. (D) Time course of calcium transients. (E) Images show clonal expansion of single cell F5 iCPCs. F5 iCPCs derived from AC Fibs were seeded in low-density cultures to obtain isolated single cells. Single cell derived colonies were then picked and expanded. (F) Differentiation of iCPC clones followed by immunostaining for cardiac lineage markers revealed that that iCPC clones were either tripotent (differentiated into CMs, SMs and ECs) or bipotent (differentiated into CMs, SMs). (G) Table showing the cardiac lineage potency of various iCPC clones. CM=cardiomyocytes, SM=smooth muscle cell, EC=endothelial cell. Scale bars = 200 μm in A, 50 μm in B, 10 μm in C, 1 second in D, 400 μm in E (single cell seeding), 1000 μm in E (colony formation, expansion), 100 μm in F. See also Figure S3, S5, Movie S2 and S3.
We wanted to determine the cardiovascular potency of single cell iCPCs. Hence, iCPCs were seeded at low-density and cultured to obtain single cell-derived colonies that were picked, expanded and subsequently differentiated (Figure 4E). We found that 6/9 clones exhibited tripotency (differentiated into CMs, SMs and ECs), whereas 3/9 clones were bipotent (differentiated into CMs and SMs) (Figure 4 F-G). This indicates that there is heterogeneity among iCPCs clones regarding cardiac lineage potency.
Cardiac Factors Stably Reprogram Adult Lung and Tail-tip Fibroblasts to iCPCs
To determine whether iCPCs could be reprogramed from non-cardiac sources of fibroblasts, we isolated adult lung fibroblasts (AL Fibs) and adult tail tip fibroblasts (AT Fibs) from Nkx2.5-EYFP/rtTA transgenic mice. Both AL Fibs and AT Fibs stained negative for CPC TFs as well as cardiac lineage differentiation markers and had no EYFP expression. We infected AL Fibs and AT Fibs with either 11 or 5 factors and cultured them in iCPC induction medium. AL Fibs infected with 11 factors or 5 factors produced 9 or 10.5 EYFP+ colonies, respectively (per 50,000 cells). AT Fibs infected with 11 factors or 5 factors produced 5 or 4 EYFP+ colonies (per 50,000 cells), respectively (Figure 5A). Lung-derived iCPCs could be stably expanded in iCPC maintenance medium for at least 10 psgs, stained positive for CPC markers, and differentiated into CMs (50-60%), SMs (10-15%) and ECs (5-10%) (Figure 5B-H). Three lung-derived iCPC lines underwent karyotype analysis, which demonstrated that 2 of 3 lines exhibited normal karyotypes while the third line was near tetraploid (Figure 5G). Similarly, AT Fibs derived iCPCs could be stably expanded in iCPC maintenance medium for at least 5 psgs, stained positive for CPC markers, and differentiated into CMs (30-40%), SMs (5-10%) and ECs (1-5%) (Figure 5B-I).
Figure 5. Cardiac Factors Stably Reprogram Adult Lung and Adult Tail-tip Fibroblasts into Proliferative and Multipotent iCPCs.
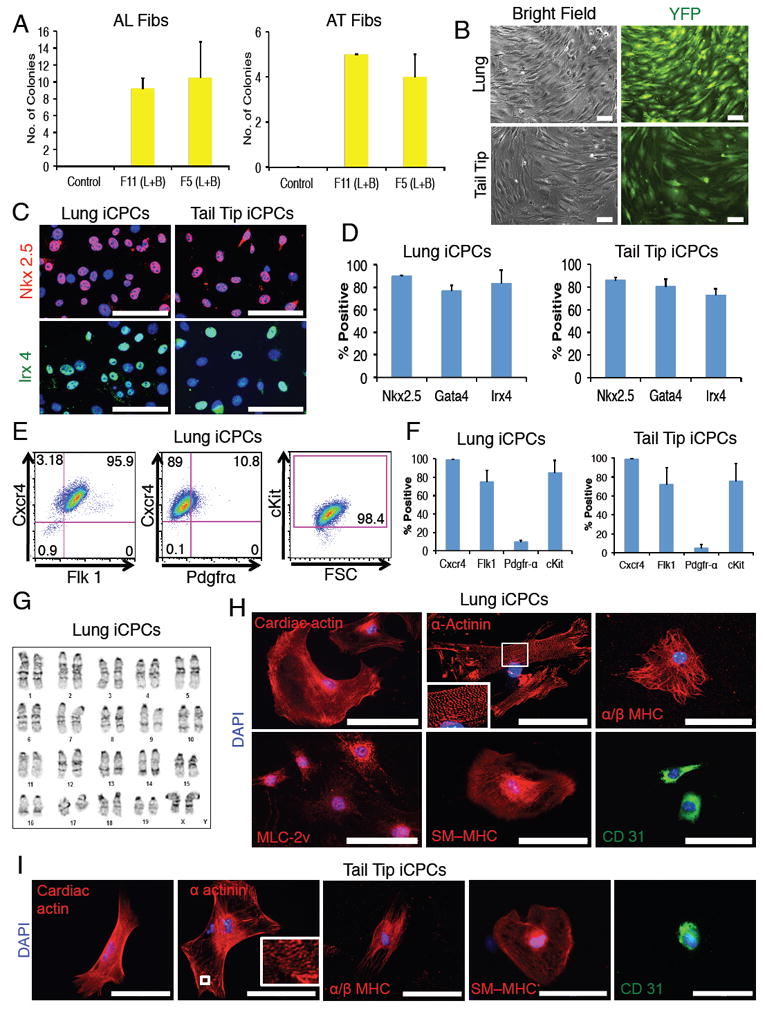
(A) Number of Nkx2.5-EYFP+ colonies produced (per 50,000 seeded cells) after infection of adult lung & adult tail-tip fibroblasts with 11 or 5 factors and culture in iCPC induction medium for 3 weeks (n=4). (L=LIF, B=BIO). (B) EYFP+ cells reprogrammed using 5 factors could be stably expanded without doxycycline for at least 10 psgs (Lung) and 5 psgs (Tail). (C) Immunolabeling revealed F5 lung-iCPCs & tail-tip-iCPCs had nuclear localization of CPC TFs (merged images are depicted, Red = Nkx2.5, Green = Irx4, Blue = DAPI), quantified in (D). (E&F) Flow cytometry analysis revealed that F5 lung-iCPCs & tail-tip-iCPCs expressed cell surface markers associated with CPCs (n=3). (G) F5 lung-iCPCs showed a normal diploid karyotype. (H) Lung-iCPCs were multipotent and differentiated into CMs (cardiac actin, α-actinin, MLC-2v, α/β MHC), SMs (SM-MHC) and ECs (CD31). Note highly organized sarcomere staining for α-actinin. (I) Tail-tip-iCPCs were multipotent and differentiated into cardiomyocytes (cardiac actin, α-actinin, α/β MHC), smooth muscle cells (SM-MHC) and endothelial cells (CD31). Note organized sarcomere staining for α-actinin. Data presented as mean, error bars = SEM. Scale bars = 100μm. See also Figure S2, S3.
We have generated over 40 iCPC lines from 3 different fibroblast sources (AC Fibs – F11 (10 lines) & F5 (20 lines), AL Fibs – F11 (4 lines) & F5 (4 lines), AT Fibs – F11 (2 lines) & F5 (2 lines)). All iCPCs lines had comparable morphologies, EYFP expression and proliferation rates. Many of these lines were extensively characterized for expression of CPC markers (TFs and cell surface proteins) and cardiac lineage potency at various passages (ranging for psg 5 to 30). It is interesting to note that various iCPC lines showed reproducible expression pattern for CPC markers and possessed cardiac lineage potency regardless of factor combination (F11 or F5) and tissue of origin. These results indicate that adult fibroblasts from diverse tissues of origin can be reproducibly and stably reprogrammed into proliferative and multipotent iCPCs.
iCPCs Differentiate into Cardiomyocytes when Injected into the Cardiac Crescent of Mouse Embryos
After demonstrating iCPC reprograming from various tissues of origin and defining their cardiovascular potency in vitro, we wanted to assess iCPC potency in the developing heart at a stage when the Nkx2.5-EYFP reporter used to isolate iCPCs would be expressed. We reasoned that if the iCPCs truly mimic native CPCs, they would likewise respond to the rich cardiogenic signaling environment in the native cardiac crescent (CC) (Abu-Issa and Kirby, 2007) and rapidly differentiate to cardiomyocytes. iCPCs were first infected with a constitutive GFP expressing lentivirus to track their progeny, and then injected into the CC of mouse conceptuses (E7.75 - headfold-6 somite pairs stage). We cultured operated and un-operated control conceptuses in a whole embryo ex vivo culture system for 24 or 48 hrs. During this culture period, the CC undergoes a morphogenic shift to develop into a beating linear heart tube. Thus, the endogenous CPCs contained in the CC (E 7.75) differentiate into contracting CMs within 24hrs (E8.5).
We injected 200-500 iCPCs per embryo in a total of 20 embryos in two separate experiments and performed live imaging on injected embryos to determine the location of the GFP+ cells at the end of 24 and 48 hrs of whole embryo culture. In 3/20 embryos no GFP+ cells were detected, possibly due to leakage of cells out of the injection site during the injection. In 15/17 of the remaining embryos (88%), GFP+ cells localized exclusively to the developing heart and appeared to contract along with the endogenous CMs (Figure 6A-B, Figure S6E, Movie S4-5). In contrast, AC Fibs injected into the CC were completely excluded from the developing heart tube. The majority of injected AC Fibs were also excluded from the embryo proper, and localized to the ectoplacental cone (extra-embryonic region) (Figure 6C, S6A-B). The presence of iCPCs in the heart tube suggests that they were able to respond to cardiac-morphogenetic signaling in the developing embryo and localize/differentiate along with host CPCs to the beating heart tube. To assess whether the iCPC-derived cells could integrate with host cells, some of the injected embryos were sectioned and immunostained with a GFP antibody. We observed that iCPC-derived cells integrated with host cells within the heart tube (Figure 6D).
Figure 6. iCPCs Localize to Developing Heart Tube and Differentiate into Cardiomyocytes Upon Injection into the Cardiac Crescent of Mouse Embryos.
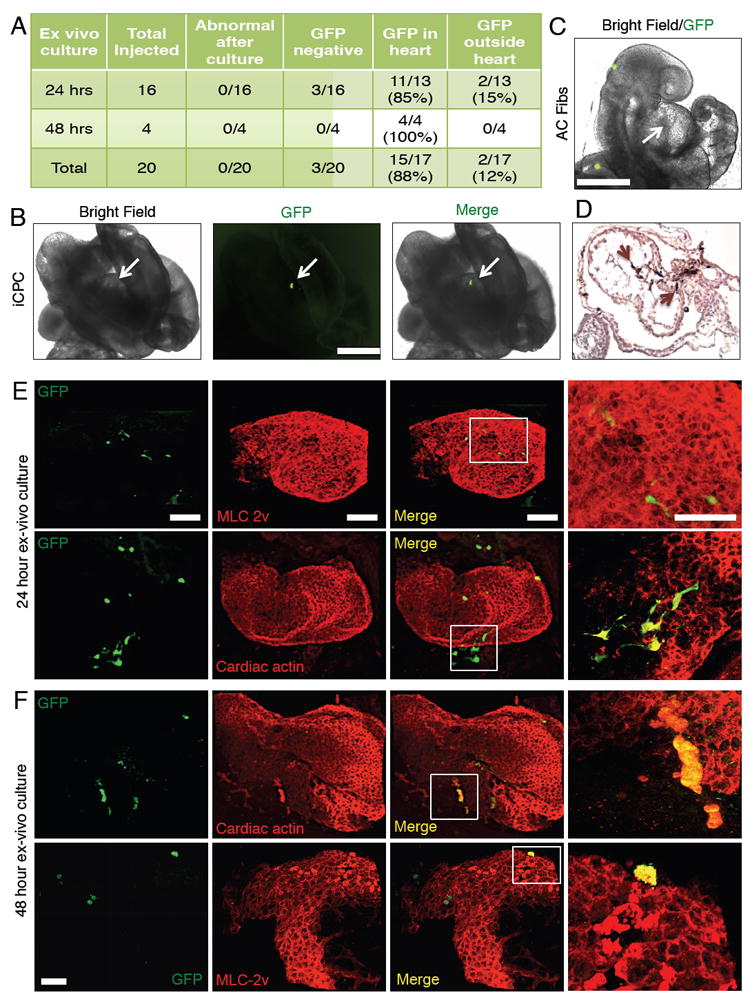
(A) Shows the number of embryos injected with F11 iCPCs and the location of iCPC-derived cells 24 or 48 hrs after whole embryo culture. (B) F11 iCPCs (labeled with GFP expressing lentivirus) injected into the cardiac crescent of mouse embryos colonized the developing heart tube as assessed after 24 hrs of whole embryo culture. See also Movie S4 for 24 hr cultured embryos and Movie S5 for 48 hr cultured embryos. (C) AC Fibs (labeled with GFP expressing lentivirus) injected into the cardiac crescent were excluded from the developing heart tube (arrow) as assessed after 24 hrs of whole embryo culture. (D) Histological sections of iCPC-injected embryos were stained for GFP antibody (dark brown color). iCPC-derived cells (brown arrows) integrated with host cell in the developing heart tube. (E) 24 hrs cultured iCPC injected embryos were immunostained in whole mount preparations for CM markers and GFP. 3D reconstruction images show iCPCs differentiated into CMs, as indicated by co-expression of CM markers and GFP. (F) iCPC-CMs attained shape/size similar to native CMs after 48 hrs culture. Scale bar = 500 μm in B-C, and100 μm in E-F. See also Figure S6, Movie S4-S7.
To determine if injected iCPCs differentiated into CMs in vivo, whole mount embryos were co-immunostained for GFP and CM markers. Specimens were imaged as optical sections (1μm) using multi-photon excitation microscopy, and 3D reconstructions of the z-stack images were performed. In the 24-hr cultured embryos, we detected multiple GFP+ cells in the heart tube that co-stained for CM markers such as MLC-2v and cardiac actin. In 24-hr cultured embryo samples the iCPC-derived CMs had an elongated appearance and looked morphologically distinct from the native CMs (Figure 6E, Movie S6). However, iCPC-derived CMs in the 48-hr cultured embryos had a rounder morphology and appeared similar in shape and size to the native CMs (Figure 6F, Figure S6F, Movie S7). iCPC-CMs were observed in developing atria, ventricles as well as outflow track, showing no spatial preference within the heart tube. In contrast, injected AC Fibs failed to differentiate into CMs as indicated by absence of staining for CM markers (Figure S6C-D).
Although we observed endothelial differentiation from iCPCs in vitro, we were unable to convincingly detect iCPC-derived CD31+ cells in vivo. Due to the limited endothelial potency of iCPCs (only 1-5% detected during in vitro differentiation), we may have missed rare CD31+ cells within embryos. The whole embryo culture technique used here cannot be extended beyond E9.75 as the embryo becomes increasingly dependent upon formation of a chorio-allantoic placenta and interaction with its mother (Lawson et al., 1991). Hence, we were unable to assay embryos for smooth muscle as the onset of smooth muscle differentiation is after E10.5 (Miano et al., 1994), which exceeded our whole embryo culture duration (E7.75-E9.75).
iCPCs Injected into Adult Heart Post Myocardial Infarction Engraft Within Scar and Differentiate into Cardiac Lineage Cells In vivo
In order to examine the applicability of the iCPC technology for cardiac regenerative medicine, we tested the potency of iCPCs in an adult heart injury model. To induce myocardial infarction we permanently ligated the left coronary artery in 8 week old C57BL/6J male mice. Two days post MI we injected 1-1.5 million GFP labeled iCPCs (F5 iCPCs reprogrammed from AC Fibs) or PBS in the border zone of the infarct and monitored the animals for 4 weeks. Kaplan-Meier survival analysis revealed that 75% of the animals that received iCPC injection survived to 4 weeks, which was significantly higher compared to 11% of mice that survived after PBS injection (Figure 7A). To determine if injected iCPCs could differentiate into cardiac lineage cells we harvested the hearts of injected animals and performed immunohistochemistry on tissue sections with cardiac lineage differentiation markers and GFP. We first analyzed the hearts of animals 4 days after iCPC injection. A majority of surviving iCPCs were localized at the border of the infarct zone and showed faint expression for cardiac actin (Figure 7B). This suggested that some iCPCs were differentiating into CMs. When we analyzed the hearts of animals 28 days after injection we noticed that some iCPC-derived cells migrated and engrafted more than 1mm deep within the scar tissue (Figure 7C, S7A). Moreover, the iCPC-derived cells exhibited strong expression of cardiac actin indicating their differentiation into CMs. Even though iCPC-derived CMs did not completely resemble host CMs in morphology they showed organized cardiac actin staining (Figure 7C, S7A). Some iCPCs-CMs attained rod shaped morphology and appeared highly aligned, a characteristic of adult CMs (Figure 7D, S7A). The intensity of cardiac actin staining in iCPC-derived CMs (yellow arrow) is comparable to that in host CMs (red arrow) suggesting that iCPCs had differentiated into functional CMs when assessed 28 days after injection. Some of the injected iCPCs differentiated into smooth muscle cells (Figure 7E, S7B) and endothelial cells (Figure 7F, S7C) as indicated by expression of smooth muscle actin and CD31, respectively. During the 4-week time course of the study we did not observe any tumor formation. These data indicate that iCPCs can survive, engraft and differentiate into cardiac lineage cells in the post-MI heart and improve survival in treated mice.
Figure 7. iCPCs Differentiate into Cardiac Lineage Cells In vivo and Improve Survival in Mice Post Myocardial Infarction.
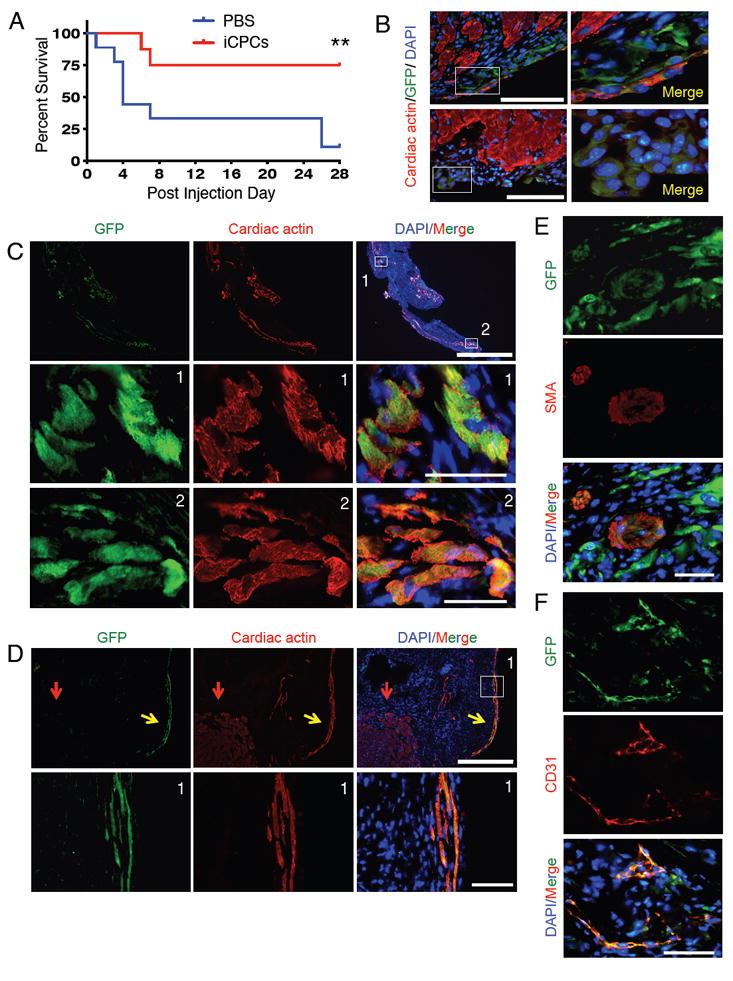
(A) Kaplan-Meier survival analysis revealed that iCPC injection significantly improved survival in treated animals as compared to control. (n=8 for iCPCs, n=9 for PBS) **p<0.01, Mantel-Cox test. (B) AC Fibs derived F5 iCPCs (labeled with GFP expressing lentivirus) were injected into the MI border zone and hearts were analyzed 4 days post injection. Immunolabeling of tissue sections revealed that a majority of surviving cells were localized on the edge of the scar tissue and showed faint expression of cardiac actin. Merged images are depicted. Scale bars = 100μm. (C) Injected iCPCs differentiated into cardiomyocytes based on organized cardiac actin immunolabeling (28 days after injection). Lower two rows of images are from insets in the top row. Scale bars = 1000μm, 50μm in insets. Tissue section represents the apex region of the left ventricular wall. (D) Immunolabeling for GFP and cardiac actin revealed that some iCPC-derived cardiomyocytes aligned and attained rod-shaped morphology 28 days after injection. Lower row of images are insets from top row. Red arrow = host CM, yellow arrow = iCPC-derived CM. Scale bars = 400μm, 50μm in insets. (E) iCPCs differentiated into smooth muscle cells within scar tissue based on co-expression of GFP and smooth muscle actin (SMA), as assessed 28 days after injection. Scale bar = 50μm. (F) iCPCs differentiated into endothelial cells within scar tissue based on co-expression of GFP and CD31, as assessed 28 days after injection. All images are from Mouse 1. Scale bar = 50μm. See also Figure S7 (Mouse 2 and 3).
DISCUSSION
Here we demonstrate that a combination of 11 (Mesp1, Mesp2, Gata4, Gata6, Baf60c, SRF, Isl1, Nkx2.5, Irx4, Tbx5, Tbx20) or 5 (Mesp1, Tbx5, Gata4, Nkx2.5, Baf60c) cardiac factors, in conjunction with Wnt and JAK/STAT signaling can stably reprogram adult fibroblasts from three different tissue origins into proliferative and multipotent iCPCs. We screened defined factors and culture conditions to induce reprogramming based on two criteria: the capability to activate the Nkx2.5-EYFP reporter and the ability of resulting EYFP+ cells to maintain a proliferative state on extended passaging without forced expression of reprogramming factors. This stringent approach allowed screening for factors/culture conditions that induced stably reprogrammed iCPCs. The iCPCs differentiate into the major cell lineages found in the heart including SMs, ECs and CMs.
Most lineage reprogramming studies to date utilized embryonic or neonatal fibroblasts as a starting cell source for reprogramming. These cells, though easier to reprogram, are clinically less relevant since comparable cell types for humans are not available. Hence, we opted to use adult fibroblasts in our study. It is well documented that adult cells are more challenging to reprogram as compared to embryonic or neonatal cells (Takahashi and Yamanaka, 2006; Yu et al., 2007). As cells mature their epigenetic state becomes progressively more rigid and thus less amenable to reprogramming. Hence, the efficiency of iCPC reprogramming was low (0.01-0.02%), but comparable to efficiencies reported for iPSC reprogramming (Takahashi and Yamanaka, 2006). We reprogrammed fibroblasts from both male and female mice that ranged from 1 to 3 months in age, and we did not detect differences in reprogramming efficiency. Although reported in vitro iCM reprogramming efficiencies range from 5-20%, it should be noted that these values were based on percentage of cells showing reporter activity 1-2 weeks after infecting with iCM factors. Completely reprogrammed iCMs represent a small fraction of the cells that initially showed reporter activity. In spite of low reprogramming efficiencies we were able to generate millions of stably reprogrammed iCPCs owing to their proliferative capacity.
Based on current knowledge, transdifferentiation is thought to be a long and stochastic process in which cells transition through intermediate states before achieving a stable reprogrammed state (Jaenisch and Young, 2008). The first EYFP+ cells were detected 3-5 days after dox induction and over the course of 3 weeks underwent a dramatic morphological change to develop into proliferative colonies. The cells continued to undergo morphological change for the first 2-3 psgs. Gene expression analysis of psg 1 cells revealed that a relatively small 2-8 fold increase in cardiac marker expression was sufficient to initiate reprogramming towards an iCPC state, whereas late psg cells demonstrated a further increase in expression of cardiovascular genes and decrease in expression of fibroblasts genes. This indicates that even though the Nkx2.5-EYFP reporter is activated very early (3-5 days), iCPC reprogramming progressively continues over the initial 2-3 psgs (35-40 days), after which the cells reach a stable reprogrammed state. This is in congruence with reprogramming processes for iCMs (Qian et al., 2013) as well as iPSCs (Brambrink et al., 2008).
RNA-seq analysis revealed that iCPCs have cardiac mesoderm-restricted gene expression. Interestingly, Nanog was the only pluripotency-associated gene upregulated in iCPCs. Nanog is also expressed in the epiblast and prevents Bmp-induced differentiation (Shin et al., 2011). Correspondingly, Bmp genes (4/6/7) were downregulated in iCPCs and negative regulators of Bmp signaling such as smad6/7 and Grem1, were upregulated. Recently, it was shown that inhibition of Bmp signaling, along with Wnt activation can maintain CPCs derived from human PSCs in a stem cell state (Cao et al., 2013).
Ieda et al. (2010) elegantly showed that iCMs are directly reprogrammed from fibroblasts without transitioning through a progenitor state. SM genes are also not expressed during iCM reprogramming. During iCM reprogramming cell proliferation is staunchly suppressed along with the activation of CM differentiation-specific genes (Qian et al., 2013; Wada et al., 2013). Conversely, during iCPC reprogramming cell proliferation is activated, cardiac mesoderm associated genes are turned on and cardiac lineage differentiation genes are not expressed. This indicates that the reprogramming route fibroblasts follow during conversion to iCPCs is distinct from the route for iCM reprogramming.
The iCPC-CMs progressively mature with extended low serum culture based on myofilament immunolabeling and organization. However, unlike ES- or iPSC-derived CMs, iCPC-CMs did not exhibit spontaneous contractions in culture. Adult heart derived CPCs have also been differentiated into CM-like cells in vitro which failed to spontaneously contract. However, we were able to induce contractions in iCPC-CMs by co-culturing with mESC-CMs. iCPC-CMs developed gap junctions with mESC-CMs and showed synchronous calcium transients. This result demonstrates the ability of iCPC to differentiate into fully functional CMs and couple with other CMs. When injected into the cardiac crescent of mouse embryos, iCPCs not only colonized the developing heart tube, but also integrated and differentiated into CMs. We noticed that iCPC-CMs appeared to mature in the cultured embryos and attain the shape and size similar to native CMs. It is interesting to note that iCPCs could differentiate into CMs 24-48 hrs after in vivo injection. This suggests that iCPCs in the appropriate microenvironment can rapidly differentiate into functional CMs.
Lineage tracing studies have shown that the mammalian heart is derived from progenitors of the first and second heart fields as well as the epicardium and neural crest (Abu-Issa and Kirby, 2007). Some of these progenitor populations have been identified in vivo as well as during in vitro differentiation of PSCs. iCPCs share common properties with other defined CPC populations such as ability to self-renew and the potency to differentiate into cardiac lineage cells (CMs, SMs, ECs). The iCPCs exhibit some heterogeneity as detected by cell surface markers and gene expression, so it possible that iCPCs contain subsets of both first heart field and second heart field CPCs, among others. Further characterization is necessary to determine if iCPCs resemble one or more of these native CPC populations. It is possible that the appropriate combination of CPCs, can generate cardiac organ buds which can be used for both drug discovery and regenerative medicine applications (Takebe et al., 2015). Also, the adult heart consists of distinct specialized CMs including atrial, ventricular and conduction system cells. If iCPCs differentiate into a particular type of CM or a mixture of all three requires further experimentation. However, current data examining MLC2v expression suggest a majority of iCPC-CMs to be ventricular-like.
iCPC reprogramming provides a promising cell source for cardiac regenerative therapy. Direct reprogramming to iCPCs avoids transitioning through an iPSC state, and thus theoretically reduces the tumorigenic risk associated with pluripotent cells. Also, iCPCs can be readily expanded (9.14 × 1015 million to 1 × 1016 million cells in 20 psgs) and hence cardiac cell therapy applications, that require millions to billions of cells (progenitors or differentiated progeny), are possible (Lalit et al., 2014). iCPCs injected into the adult mouse heart post-MI significantly improved survival in treated mice. Some of the injected iCPCs survived and differentiated into CMs, SMs and ECs repopulating the scar tissue with relevant cardiac lineage cells. iCPC-derived CMs showed mature-like rod shaped morphology, alignment as well as organized cardiac actin staining. Importantly, iCPC-derived cells contributed to the vasculature within the scar area by differentiating into SMs and ECs. These results provide promise for future studies, which can examine effects of iCPC therapy on cardiac functionality as well as optimize cell delivery strategies including tissue-engineered scaffolds. In vivo reprogramming post-MI of native cardiac fibroblasts to iCMs has shown promise in mouse models (Qian et al., 2012; Song et al., 2012), and it is possible that in vivo reprogramming to iCPCs may show benefit. The scalability of iCPCs also enables in vitro applications such as disease modeling, drug discovery, and basic cardiovascular research. Future studies reprogramming human cells to iCPCs will be an important future step to advance these applications.
EXPERIMENTAL PROCEDURES
Isolation of Primary Fibroblasts
For isolation of adult fibroblasts, the respective organs were derived from 1-3 month-old double transgenic mice (Nkx2.5-EYFP/rtTA). The organs were washed with PBS and minced in fibroblast medium (DMEM, 10% FBS, 1% NEAA/L-glutamine/Pen/strep) to obtain tissue pieces around 1mm3 in size. These were briefly trypsinized (0.25% trypsin-EDTA) for 10 minutes. Explants were then plated on 0.1% gelatin coated dishes in fibroblasts medium for 10-12 days. Fibroblasts migrated from explants were harvested, filtered through a 40uM cell strainer (BD Biosciences) to avoid contamination of heart tissue, passaged 1-2 times and frozen or used for experiments.
Infecting Fibroblasts with Lentiviruses
Lentivirus particles were produced as detailed in extended experimental procedures. Primary fibroblasts were seeded in a gelatinized 12 well plate at density of 50,000 cells/well 2 days prior to infection. Cells were then fed with lentivirus infection media (lentivirus supertatant+8ug/ml Polybrene, Sigma) and infection was continued for 48 hrs.
iCPC Culture/Differentiation Conditions
Lentivirus infection media was replaced with iCPC induction media (DMEM, 10% FBS, 1% NEAA, 1% L-glutamine, 1% Pen/strep, 4ug/ml dox (Sigma), 2.5 uM BIO (Cayman chemical), 103 units/ml LIF (Millipore). After reprogramming was achieved, iCPC were maintained in iCPC maintenance medium (iCPC induction medium without dox). The iCPCs are split 1:6 on a 10 cm dish for which a confluent dish yields 2.5-3.0 million cells. To differentiate, iCPC were aggregated in 24 well low attachment plates (Corning) for 4-6 days in cardiac differentiation medium (Fibroblast medium, 5uM IWP4 (Stemgent), 50 ng/ml Bmp4 (RD Systems), 10 ng/ml Vegf (RD Systems), 30 ng/ml bFgf (RD Systems). Aggregates were then plated on gelatin-coated dishes and cultured in fibroblast medium containing 1% serum for 10-50 days.
Immunocytochemistry and Flow Cytometry
Cells were platted, fixed and stained with various antibodies via a standard protocol detailed in the extended experimental procedures.
Quantitative RT-PCR
Total RNA was isolated using RNAqueous Kit (Invitrogen). Reverse transcription was performed using iScriptTM Reserve Transcription Supermix (Bio Rad). qRT-PCR was performed on CFX96TM Real Time Systems (Bio Rad) using SsoFastTM EvaGreen Supermix (Bio Rad). MIKEQ guidelines were followed in designing qPCR experiments. mRNA levels were normalized by comparison to β-actin (Δ CT) and data are presented as fold change w.r.t expression in AC Fibs (ΔΔ CT).
RNA-seq and Bioinformatics Analysis
RNA was extracted as above from AC Fibs derived iCPCs either at low psg (1-3) or high psg (8-10). Gene expression data for mESC-CPCs and mESC-CMs was obtained from (Wamstad et al., 2012) and used as positive control. For heat map analysis, gene expression was compared with uninfected AC Fibs. RNAseq was performed using HiSeq 2500 (Illumina) in duplicates from independent biological samples. Sequencer outputs were processed using CASAVA-1.8.2 (Illumina), and each sample’s reads were processed using RSEM version 1.2.3 to obtain expression measures for genes. Differential analysis was done using EBSeq version 1.5.3., and targeted false discovery rate for each run was 0.05. Transcripts per million values were used for all calculations. STRING database was used for GO analysis (Franceschini et al., 2013). See extended experimental procedures for more details.
Ca2+ Imaging
Cells were loaded with Rhod-2, AM (Invitrogen) for 20 mins at 37°C in fibroblast medium, then washed and incubated for additional 30 mins at 37°C to allow for de-esterification of the dye. Rhod-2 loaded cells were analyzed by Nikon epifluorescence microscope with NIS elements software.
Embryo Injections, Immunostaining and Imaging
Cardiac crescent stage mouse embryos were obtained by timed matings. iCPCs and AC Fibs were infected with a GFP lentivirus (Addgene #17448) to trace cells in vivo. Approximately 200-500 iCPCs or AC Fibs were introduced into the cardiac crescent of dissected mouse embryos in dissection medium via a mouth-held glass capillary (~20μm opening). Operated and stage-matched unoperated embryo samples were then placed into whole embryo culture medium (Downs, 2006), and cultured for 24 or 48 hours. At the end of the culture period, embryos were imaged using a Nikon epifluorescence microscope to determine the location of injected GFP+ cells. Embryos were then fixed with 4% PFA. Immunofluorescence of whole mount embryos was done and imaging was performed using a custom built multi-photon microscope. Imaris software (Bitplane) was used to make 3D reconstructions. See extended experimental procedures for more details.
Mouse MI Model and iCPC Injections
MIs were surgically induced in 8-week-old male C57BL/6J mice by permanent ligation of the left coronary artery. Two days following MI surgery, in mice with echocardiographically proven large infarctions, a total of 1-1.5 million GFP labeled iCPCs (reprogrammed using 5-factor and AC Fibs) or PBS were injected in three injections of approximately 25 μl in the border zone. Animals were monitored for 4 weeks, and the hearts of surviving animals were excised for immunohistochemistry. See extended experimental procedures for more details.
Statistical Analysis
Differences between multiple groups were tested for statistical significance using one-way ANOVA followed by Tukey’s post hoc analysis. For comparison of two groups, unpaired Student’s t-test was used. p values of <0.05 were regarded as significant.
Supplementary Material
Acknowledgments
We would like to thank the UWCCC Experimental Pathology Core (especially Joe Hardin) for their help with immunohistochemistry experiments. We would also like to thank the Graduate School at the University of Wisconsin-Madison (MRS, WCC). The research was supported by NIH U01HL099773 (TJK, JAT), R01 HL129798 (TJK, GEL), AHA pre-doctoral fellowship 12PRE9520035 (PAL).
References
- Abu-Issa R, Kirby ML. Heart field: from mesoderm to heart tube. Annual review of cell and developmental biology. 2007;23:45–68. doi: 10.1146/annurev.cellbio.23.090506.123331. [DOI] [PubMed] [Google Scholar]
- Birket MJ, Ribeiro MC, Verkerk AO, Ward D, Leitoguinho AR, den Hartogh SC, Orlova VV, Devalla HD, Schwach V, Bellin M, et al. Expansion and patterning of cardiovascular progenitors derived from human pluripotent stem cells. Nature biotechnology. 2015;33:970–979. doi: 10.1038/nbt.3271. [DOI] [PubMed] [Google Scholar]
- Bondue A, Tannler S, Chiapparo G, Chabab S, Ramialison M, Paulissen C, Beck B, Harvey R, Blanpain C. Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J Cell Biol. 2011;192:751–765. doi: 10.1083/jcb.201007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell stem cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao N, Liang H, Huang J, Wang J, Chen Y, Chen Z, Yang HT. Highly efficient induction and long-term maintenance of multipotent cardiovascular progenitors from human pluripotent stem cells under defined conditions. Cell research. 2013;23:1119–1132. doi: 10.1038/cr.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Downs KM. In vitro methods for studying vascularization of the murine allantois and allantoic union with the chorion. Methods in molecular medicine. 2006;121:241–272. doi: 10.1385/1-59259-983-4:239. [DOI] [PubMed] [Google Scholar]
- Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, Elefanty AG, Gramolini A, Keller G. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nature biotechnology. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison GM, Vicinanza C, Smith AJ, Aquila I, Leone A, Waring CD, Henning BJ, Stirparo GG, Papait R, Scarfo M, et al. Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell. 2013;154:827–842. doi: 10.1016/j.cell.2013.07.039. [DOI] [PubMed] [Google Scholar]
- Foshay K, Rodriguez G, Hoel B, Narayan J, Gallicano GI. JAK2/STAT3 directs cardiomyogenesis within murine embryonic stem cells in vitro. Stem Cells. 2005;23:530–543. doi: 10.1634/stemcells.2004-0293. [DOI] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han DW, Tapia N, Hermann A, Hemmer K, Hoing S, Arauzo-Bravo MJ, Zaehres H, Wu G, Frank S, Moritz S, et al. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell stem cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islas JF, Liu Y, Weng KC, Robertson MJ, Zhang S, Prejusa A, Harger J, Tikhomirova D, Chopra M, Iyer D, et al. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:13016–13021. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell stem cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Lalit PA, Hei DJ, Raval AN, Kamp TJ. Induced pluripotent stem cells for post-myocardial infarction repair: remarkable opportunities and challenges. Circulation research. 2014;114:1328–1345. doi: 10.1161/CIRCRESAHA.114.300556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Meneses JJ, Pedersen RA. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- Masino AM, Gallardo TD, Wilcox CA, Olson EN, Williams RS, Garry DJ. Transcriptional regulation of cardiac progenitor cell populations. Circulation research. 2004;95:389–397. doi: 10.1161/01.RES.0000138302.02691.be. [DOI] [PubMed] [Google Scholar]
- Miano JM, Cserjesi P, Ligon KL, Periasamy M, Olson EN. Smooth muscle myosin heavy chain exclusively marks the smooth muscle lineage during mouse embryogenesis. Circulation research. 1994;75:803–812. doi: 10.1161/01.res.75.5.803. [DOI] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Nelson TJ, Faustino RS, Chiriac A, Crespo-Diaz R, Behfar A, Terzic A. CXCR4+/FLK-1+ biomarkers select a cardiopoietic lineage from embryonic stem cells. Stem Cells. 2008;26:1464–1473. doi: 10.1634/stemcells.2007-0808. [DOI] [PubMed] [Google Scholar]
- Noseda M, Abreu-Paiva M, Schneider MD. The Quest for the Adult Cardiac Stem Cell. Circulation journal : official journal of the Japanese Circulation Society. 2015;79:1422–1430. doi: 10.1253/circj.CJ-15-0557. [DOI] [PubMed] [Google Scholar]
- Qian L, Berry EC, Fu JD, Ieda M, Srivastava D. Reprogramming of mouse fibroblasts into cardiomyocyte-like cells in vitro. Nature protocols. 2013;8:1204–1215. doi: 10.1038/nprot.2013.067. [DOI] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qyang Y, Martin-Puig S, Chiravuri M, Chen S, Xu H, Bu L, Jiang X, Lin L, Granger A, Moretti A, et al. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell stem cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- Shin M, Alev C, Wu Y, Nagai H, Sheng G. Activin/TGF-beta signaling regulates Nanog expression in the epiblast during gastrulation. Mechanisms of development. 2011;128:268–278. doi: 10.1016/j.mod.2011.03.001. [DOI] [PubMed] [Google Scholar]
- Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takebe T, Enomura M, Yoshizawa E, Kimura M, Koike H, Ueno Y, Matsuzaki T, Yamazaki T, Toyohara T, Osafune K, et al. Vascularized and Complex Organ Buds from Diverse Tissues via Mesenchymal Cell-Driven Condensation. Cell stem cell. 2015;16:556–565. doi: 10.1016/j.stem.2015.03.004. [DOI] [PubMed] [Google Scholar]
- van Berlo JH, Kanisicak O, Maillet M, Vagnozzi RJ, Karch J, Lin SC, Middleton RC, Marban E, Molkentin JD. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509:337–341. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada R, Muraoka N, Inagawa K, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Kaneda R, Suzuki T, Kamiya K, et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:12667–12672. doi: 10.1073/pnas.1304053110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, He ZY, You P, Han QW, Xiang D, Chen F, Wang MJ, Liu CC, Lin XW, Borjigin U, et al. Reprogramming fibroblasts into bipotential hepatic stem cells by defined factors. Cell stem cell. 2013;13:328–340. doi: 10.1016/j.stem.2013.06.017. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


