Abstract
Use of multi-walled carbon nanotubes (MWCNT) is growing which increases occupational exposures to these materials. Their toxic potential makes it important to have an in-depth understanding of the inflammation and disease that develops due to exposure. Epigenetics is one area of interest that has been quickly developing to assess disease processes due to its ability to change gene expression and thus the lung environment after exposure. In this study, promoter methylation of inflammatory genes (IFN-γ and TNF-α) was measured after MWCNT exposure using the pyrosequencing assay and found to correlate with initial cytokine production. In addition, methylation of a gene involved in tissue fibrosis (Thy-1) was also altered in a way that matched collagen deposition. In addition to using epigenetics to better understand disease processes, it has also been used as a biomarker of exposure and disease. In this study, global methylation was determined in the lung to ascertain whether MWCNT alter global methylation at the site of exposure and if those alterations coincide with disease development. Then, global methylation levels were determined in the blood to ascertain whether global methylation could be used as a biomarker of exposure in a more easily accessible tissue. Using the LuUminometric Methylation Assay (LUMA) and 5-Methylcytosine (5-mC) Quantification assay, we found that MWCNT lead to DNA hypomethylation in the lung and blood, which coincided with disease development. This study provides initial data showing that alterations in gene-specific methylation correspond with an inflammatory response to MWCNT exposure. In addition, global DNA methylation in the lung and blood coincides with MWCNT-induced disease development, suggesting its potential as a biomarker of both exposure and disease development.
Keywords: IFN-γ, inflammation, LUMA, 5-mC, methylation, MWCNT, Thy-1, TNF-α
Introduction
Multi-walled carbon nanotubes (MWCNT) are a class of engineered nanomaterials being developed for a wide variety of medical, engineering and personal products and have many potential benefits. However, exposure to MWCNT has been shown to cause adverse health effects in animal studies, raising concerns that there could be increased risk for disease development with increased occupational and environmental exposures (Hamilton et al., 2012, 2013; Lam et al., 2006; Muller et al., 2005). Given the increasingly widespread use of MWCNT and significant potential for exposures to these materials, it is imperative that we gain a better understanding of the disease process associated with this material. In addition, the development of biomarkers of exposure and disease development from easily accessed tissues would be an important advance in evaluating health impacts from exposures.
We previously reported that MWCNT (FA-21) induced inflammation and lung fibrosis in murine models (Hamilton et al., 2012). The mechanism of initiation of inflammation has been linked to release of inflammatory cytokines from alveolar macrophages. These cytokines can cause systemic inflammation and signal chronic inflammation leading to fibrosis (Kolb et al., 2001; Latz et al., 2013). It is possible that epigenetics, which is the study of heritable changes in gene expression that occur without changes in the primary DNA sequence, are in part responsible for changes in gene expression that affect disease development. Aberrant epigenetic regulation, such as hyper- and hypo-methylation in promoter regions of specific genes, underlies a wide variety of pathologies, including cancer and cardiovascular disease (Martin et al., 2011; Webster et al., 2013). Fibrotic lung diseases have been shown to lead to epigenetic alterations within the lung (Sanders et al., 2012; Weigel et al., 2015). Exposure to fine particulate matter (PM) and ozone leads to alterations in DNA methylation of IFN-γ and other genes (Bind et al., 2014), implying that DNA methylation may play a role in inflammatory gene expression after inhalation exposures. Recently, studies have suggested that, alterations in DNA methylation occur in response to materials with at least one dimension in the size range of 1–100 nm. Specifically, Gong et al. reported that engineered SiO2 nanoparticles induced global DNA hypomethylation in HaCaT cells (Gong et al., 2010). More recently, the same group observed hypermethylation of the PARP-1 gene that encodes for a DNA repair protein following exposure to SiO2 (Gong et al., 2012). To the best of our knowledge, there have no studies investigating gene specific methylation changes following MWCNT exposure.
In addition to looking at how epigenetics affect disease development, there is a great deal of potential for the use of epigenetics as biomarkers for disease development (Herceg & Vaissiere, 2011). Studies have reported changes in the methylation status for markers of global methylation including Alu, and LINE1 coinciding with increased inflammation and endothelial dysfunction following exposure to PM 2.5 (Bind et al., 2012). Furthermore, loss of global DNA methylation is associated with chronic disease development (Jones & Baylin, 2002; Komatsu et al., 2012) and is being used as a biomarker of exposure to air pollutants, heavy metals and persistent organic pollutants (Baccarelli et al., 2009; Bollati et al., 2007; Pilsner et al., 2009; Rusiecki et al., 2008; Tarantini et al., 2009).
One study reported that global methylation in cell lines is altered after exposure to nanomaterials (Lu et al., 2015). This evidence provides further justification for determining the effect of MWCNT exposure on epigenetic alterations using an in vivo model. In addition, this data suggests that after exposure to an inhaled particle, epigenetic alterations are likely to occur in the lung. These changes may also be reflected systemically. Systemic alterations in epigenetics, such as changes in global methylation in the blood, would be useful as a biomarker for disease development, as the blood is easily accessible.
The aim of this study was to elucidate the effect of MWCNT on the methylation of genes associated with inflammation and fibrosis in C57BL/6 mice as well as alterations in global methylation. This study was designed to evaluate the changes in gene specific and global DNA methylation levels following exposure to MWCNT in acute (24 h post-exposure) and subchronic exposures (7 days post-exposure). C57BL/6 mice were exposed to 50 μg of MWCNT via oropharyngeal instillation. The type and degree of inflammation and lung pathology were determined using histology. For quantification of DNA methylation, DNA was first extracted from whole lung tissue. We used a pyrosequencing assay to analyze CpG islands located in the promoter region of genes important for inflammation (IFN-γ and TNF-α) and fibrogenesis (thymus cell antigen 1 – Thy-1). We determined global methylation using LUminometric Methylation Assay (LUMA) and by quantifying 5-methylcytosine (5-mC) levels in the lung and blood.
Materials and methods
Animals
C57BL/6 mice (2-month-old) were housed in controlled environmental conditions (22 ± 2 °C; 30–40% humidity, 12 h light:12 h dark cycle) and provided food and water ad libitum. All procedures were performed under protocols approved by the IACUC of the University of Montana.
MWCNT identification, suspension and instillation
High Ni-MWCNT (FA-21; Sun Innovations Inc., Fremont, CA) were used in this study. The details of characterization for these particles were described previously (Hamilton et al., 2012). The nanotubes were weighed and suspended in dispersion medium (DM, PBS containing 0.6 mg/ml mouse serum albumin and 0.01 mg/ml 1,2-dipalmitoyl-sn-glycero-3-phosphocholine). Nanotube suspensions were sonicated for 30 s at half maximum power in a Masonix cup-horn sonicator (XL2020, Farmingdale, NY) at 550 W and 20 Hz (8000 J) attached to a Forma circulating water bath, at a stock concentration of 2 mg/ml.
Mice were exposed to nanoparticles or dispersion media (DM) by a single oropharyngeal instillation. Briefly, the mice were anesthetized using inhaled isoflurane. A volume of 25 μl of particle suspension (50 μg) or DM was delivered into the back of the throat. By holding the tongue to the side, the solution was aspirated into the lungs. For each experiment three mice per group (DM or MWCNT) were exposed and the experiment was repeated three times. MWCNT-induced pathological changes were first observed 7 days after instillation and these changes lasted for a few weeks. Therefore, 7 days after instillation, mice were euthanized by sodium pentobarbital (Euthasol™) and blood and lung tissue were collected for DNA methylation and histological analysis.
Cytokine release
Bronchoalveolar lavage was performed using 1 ml of ice-cold PBS (pH 7.4) to collect whole lung lavage fluid for analysis. Meso Scale Discovery (MSD), which uses a proprietary combination of electrochemiluminescence detection and patterned arrays, was used to detect cytokines. The Mouse V-Plex Pro-Inflammatory Panel 1 kit (MSD, Rockville, MD) was used to determine cytokine concentrations according to manufacturer’s instructions.
Histology and tri-chrome staining
The lungs from each mouse were inflation-fixed through the trachea with 4% paraformaldehyde (PFA)-PBS and placed in 50 ml conical tubes containing 20 ml PFA overnight at 4 °C. The lungs were rinsed the following day with PBS thrice and then placed in 70% ethanol. The trachea and heart were removed and the lung tissues were placed in labeled cassettes. Processing was completed in a Leica ASP 300 tissue processor (Buffalo Grove, IL) in a 7.25 h program: 30 min in 70 and 95% ethanol, two 1 h changes in 100% ethanol, three changes of Xylene for 30 min each and three paraffin changes, at 45 min for the first bath and 1 h in the second and third changes under vacuum. Tissue was sectioned at a thickness of 5 microns using a Leica RM2235 microtome (Buffalo). The hematoxylin and eosin (H&E) along with the trichrome staining were done in a Shandon 24-4 autostainer (GMI, Ramsey, MN). Mayer’s hematoxylin (Richard-Allan Scientific, Kalamazoo, MI) and alcoholic eosin (Thermo Shandon Limited, Runcorn, UK) were used for the H & E program. Weigert’s hematoxylin (Electron Microscopy Sciences, Hatfield, PA) and Gomori Trichrome (Harleco, EMD, VWR, Randor, PA) were used for the trichrome staining. Histological analysis including lung pathology scoring, airway area and thickness, and assessment of collagen deposition were obtained from stained lung tissue.
Lung pathology scoring
Mouse lung tissue sections were imaged at 100× using a Zeiss Axioskop attached to a Zeiss digital camera and processed using Zeiss AxioVision software (Thornwood, NY). Two expert observers scored the degree of lung disease visible in the lung sections using a 5-point scale (0, 1, 2, 3 and 4) with zero being no effect and 4 being evidence of extreme lung pathology, including lung lesions, inflammatory cell infiltration and increased collagen deposition, as previously described (Hamilton et al., 2012). Both scorers were blind to the experimental conditions. There were six exposed mice per condition. The values shown are the two scorers’ median value for each condition.
Collagen deposition
“Phantom” contours were used to divide up the tissue into very small (8 μm diameter) circles. If a contour showed blue staining, indicating collagen deposition via the Trichrome stain, it was counted as a collagen-positive event. A percentage of positive contours over total tissue contours was derived to compare one tissue to the next. Given that airways and blood vessels naturally have collagen in them for structural purposes, all airways and blood vessels were isolated and removed from the calculation of total collagen deposition. Two sections using one lobe of lung tissue, 21–28 microns apart, were analyzed and averaged. There were six mice per exposure condition.
DNA extraction
Genomic DNA was isolated from snap frozen lung tissue and fresh blood using the DNeasy DNA extraction kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions.
Primer design
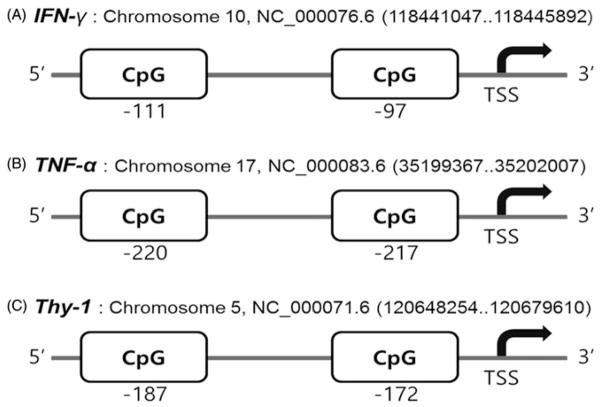
Methylation levels were measured in the promoter regions of selected genes (IFN-γ, TNF-α and Thy-1) using the pyrosequencing assay. Gene-specific primers for these genes were designed using the Pyro-Mark assay design software, version 2.0 (Qiagen, Valencia, CA). In brief, we entered the gene sequences within a 500 bps promoter region for the desired primers and the software found all relevant CpG islands. Then, the program automatically generated primer sets that included both PCR and sequencing primers based on selected target sequences. Each primer set was assigned a score and quality, reflecting its suitability for pyrosequencing analysis. By default, the set with the highest score was selected as the final primer set. Primers were designed for analyzing 2 CpGs within the promoter regions. One of the primers was biotin-labeled to enable immobilization to streptavidin-coated beads (Streptavidin Sepharose High Performance; Figure 1). Gene promoter regions were located using the most recent database of GenBank (NCBI). Table 1 shows the details of the primer and PCR conditions used in this study.
Figure 1.
Position of target genes and CpG sites in their promoters that were analyzed in the study. Pyrosequencing reactions were performed to investigate CpG sites on the IFN-γ, TNF-α and Thy-1 gene promoter regions.
Table 1.
PCR and sequence primers used in pyrosequencing assay.
| Target (Gene ID) | Primer | Sequence (5′–3′) | Annealing temperature (°C) | PCR product (bp) |
|---|---|---|---|---|
| IFN-γ (15978) | Forward | AATGGTGTGAAGTAAAAGTGTTTTTAGA | 53.4 | 108 |
| Reverse | AAAATTTCCTTTCCACTCCTTAAACTCTCa | |||
| Sequencing | ATGGTATAGGTGGGTA | |||
| TNF-α (21926) | Forward | GATAATGATTAGTTTTGGAGGATAGAGA | 51.9 | 178 |
| Reverse | ACACCCAAACATCAAAAAATCTa | |||
| Sequencing | TGGTTTTAGATTGTTATAGAATT | |||
| Thy-1 (21838) | Forward | AAAGAAAGGGTTATAATTTTAAAAGGAGAGa | 52.2 | 154 |
| Reverse | ACTACTAAAAAAACACTCTCAATCCTATA | |||
| Sequencing | ACACTCTCAATCCTATAAT |
bp, base-pair.
Biotinylated labeled primer.
Pyrosequencing assay
After sodium bisulfite conversion, genomic DNA was analyzed by the pyrosequencing assay as previously described (Yang et al., 2004). In brief, 50 ng of bisulfite-treated DNA was amplified using the PyroMark PCR kit (Qiagen, Valencia, CA) under the following conditions; 95 °C for 5 min, 45× (95 °C for 30 s, annealing temperature of each primer sets for 30 s, 72 °C for 30 s), 72 °C for 5 min. Following amplification, PCR products were checked with electrophoresis on a 2% agarose gel (Seakem® LE Agarose, Lonza, Rockland, ME). In order to run the pyrosequencing, the biotinylated PCR products were mixed with 1 μl streptavidin-coated Sepharose® beads (GE Healthcare, Piscataway, NJ), 40 μl PyroMark binding buffer (Qiagen) and 19 μl purified water for a total volume of 80 μl. This complex was captured using a PyroMark Vacuum Workstation and then purified and denatured with 70% ethanol, PyroMark denaturing solution and wash buffer. The purified single stranded PCR products were then added to the annealing buffer, which contained the corresponding sequencing primer. After annealing, the plate was loaded into the PyroMark Q96 MD instrument (Qiagen). Predetermined variable positions of the CpG regions were chosen for respective markers using the assay software. PyroMark-CpG software automatically generates a dispensation order of dNTPs and control dispensations based on the sequence to analyze. Control dispensations are included in the dispensation order to check the performance of the reactions. For quality control, all runs also included a no template control. Following the sequencing reaction, we analyzed the data with the PyroMark software for quantification of CpG methylation. The percentage methylation within a sample was subsequently determined by averaging across all interrogated CpG sites.
LUMA
Global DNA methylation was determined using the LUMA, which is based on the ability of two isoschizomers (MspI and HpaII) to digest sequences differentially depending on the methylation status of the CpG site contained within the sequence as described in Karimi et al. (2006a). In brief, 300 ng of genomic DNA were cleaved at 37 °C for 4 h and 80 °C for 20 min with HpaII/EcoRI or MspI/EcoRI in two separate 20 μl reactions containing 2 μl of Tango buffer (Thermo Scientific, Waltham, MA) and five units of each restriction enzyme (NEB, Ipswich, MA). Fifteen microliters of annealing buffer (20 mM TRIS-acetate, 2 mM Mg-acetate pH 7.6) were mixed with the digested samples and placed in a PyroMark Q96 MD system. The dispensation order used for sequencing was: GTGTCACATGTGTG. For calculations, the peak heights of dispensations 9 and 10 were used. Samples with peaks lower than 3, the cut-off value for DNA quality, were discarded. Percentage of DNA methylation was expressed as [1 – (HpaII/EcoRIΣG/ΣT)/(MspI/EcoRIΣG/ΣT)] × 100. The samples were analyzed in technical duplicates and each plate included a positive, negative and water control. Lambda DNA (NEB) was used as a negative control and methylated Lambda DNA using M.SssI methyltransferase was used as a positive control. All samples used in the final analysis had an intra-assay coefficient of variation of ≤5%.
5-mC quantification assay
We determined global DNA methylation levels using ELISA-based Methylflash™ Methylated DNA Quantification Kit (Colorimetric) (Epigentek, Farmingdale, NY). The kit measures the 5-mC content as a percentage of the total cytosine in DNA sample. The assay was performed in duplicate according to manufacturer’s instructions with 100 ng of total DNA. The 5-mC in DNA was detected using capture and detection antibodies and then quantified colorimetrically by reading the absorbance at 450 nm in a microplate spectrophotometer. The percentage of 5-mC in genomic DNA was calculated using the following formula: cytosine methylation % = [(ODsample – ODnegative control)/amount of input sample]/[(ODpositive control – ODnegative control) × 2/amount of positive control] × 100. The negative control is an unmethylated polynucleotide containing 50% cytosine, the positive control is a methylated polynucleotide containing 50% 5-methylcytosine, and 2 is a factor used to normalize 5-methylcytosine in the positive control to 100%.
Quantitative real-time PCR
Total RNA was isolated from lung tissue utilizing Trizol LS (Life Technologies, Grand Island, NY) according to the manufacturer’s protocol. After samples were treated with DNase I (Life Technologies), 1 μg of total RNA was transcribed to cDNA using a qScript™ cDNA Synthesis Kit (Quanta BioSciences, Gaithersburg, MD). Thy-1 expression was assayed by CFX Connect™ Real-Time PCR (Bio-rad, Hercules, CA) using SYBR-based detection (Quanta PerfeCTa SYBR Green SuperMix). Primers (Integrated DNA Technologies, Coralville, IA) were as follows: GADPH forward sequence (5′-GAAGGTGAAGGTCGGAGTC-3′), reverse sequence (5′-GAAGATGGTGATGGGATTTC-3′), and Thy-1 primers were derived from Thy-1 sequence (accession number: NM_009382.3), forward primer sequence (5′-CAACTTCACCACCAAGGATG-3′), reverse primer sequence (5′-TCTGAACCAGCAGGCTTATGC-3′). The amplification profile was as follows: 95 °C for 1 min, followed by 40 cycles at 95 °C for 15 s, 57 °C for 1 min and 72 °C for 1 min. Expression levels were normalized to GAPDH expression, and fold changes were calculated using the ΔΔCt method.
Statistical analysis
Statistical analyses involved comparison of means or medians using an unpaired t-test for parametric scale level endpoints and a one-tailed Mann–Whitney U test for subjective ordinal level endpoints. Statistical power was greater than 0.8. Statistical significance was defined as a probability of type I error occurring at less than 5% (p<0.05). The minimum number of experimental replications was 3. Graphics and analyses were performed on PRISM version 5.0 (GraphPad Software Inc., San Diego, CA).
Results
Cytokine production
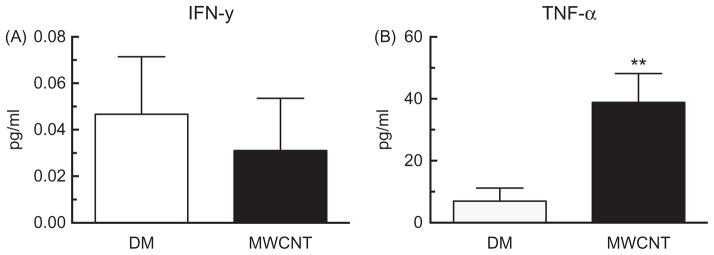
Cytokine production was measured in whole lung lavage 24 h after MWCNT to assess the type and degree of inflammatory response to MWCNT. The level of the IFN-γ production was assessed, but remained at basal levels less than 1 pg/ml, for both the DM control and the MWCNT exposed groups (Figure 2A). However, we observed a significant increase in TNF-α after MWCNT exposure (Figure 2B).
Figure 2.
Inflammatory cytokine release in whole lung lavage after 24 h MWCNT exposure. Alteration in level of (A) IFN-γ, (B) TNF-α. Data are expressed as mean ± SEM pg/ml. Asterisks indicate significance **p<0.01, *p<0.05 compared to the DM controls. n = 9 mice per condition.
Lung pathology
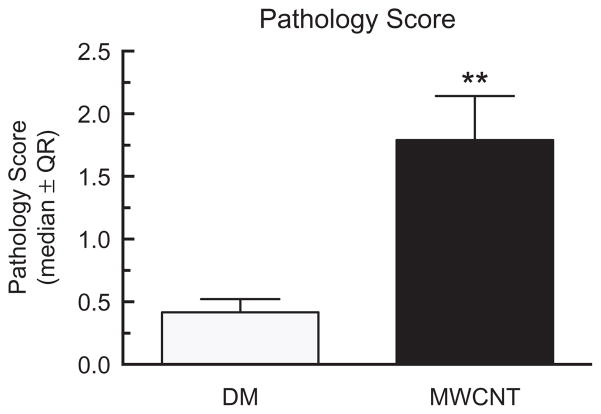
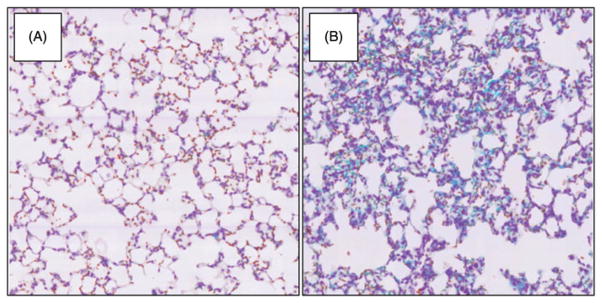
Lung tissue sections were obtained from mice exposed to MWCNT for 7 days. We used H&E and trichrome staining to determine the degree of lung pathology in these mice. Consistent with previous studies, a significant increase in disease pathology due to MWCNT exposure compared to DM control was observed (Figure 3). Specifically, the lung interstitium showed an increase in cellularity in MWCNT-exposed mice (Figure 4B) compared to the DM controls (Figure 4A) along with an increase in collagen deposition, as indicated by the punctate blue areas due to the trichrome stain.
Figure 3.
Lung pathology score in mice exposed to MWCNT for 7 days. Median ± interquartile range for all pathology scores on day 7 samples compared to the DM controls versus the MWCNT-exposed mice. Asterisks indicate significance at **p<0.01 compared to the DM controls by a non-parametric one sided Mann–Whitney test. n = 5 for the DM control and n = 6 for MWCNT treatment.
Figure 4.
Lung pathology in mice exposed to MWCNT for 7 days using hemotoxylin and trichrome stained lung tissue sections. Interstitium of (A) DM control and (B) MWCNT-exposed lung. The representative photomicrographs were taken at 40× magnification for each group.
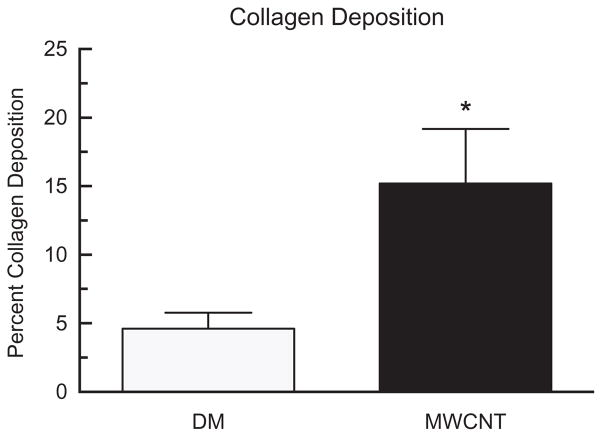
Collagen deposition
Collagen deposition can be used as an indicator of tissue fibrosis in the lungs. Laser scanning cytometry was used to determine the percent collagen deposition in the lungs (Figure 5) as determined by the presence of blue staining after the sections had been stained using trichrome. The results demonstrated a significant increase in collagen deposition 7 days after MWCNT exposure compared to the DM controls.
Figure 5.
Collagen deposition in mice exposed to MWCNT for 7 days. Alteration in level of percent collagen deposition after 7 days of MWCNT exposure. Data expressed as mean ± SEM collagen deposition. Asterisks indicate significance at *p<0.05, compared to the DM controls. n = 5 for DM control and n = 6 for MWCNT treatment.
Gene specific alterations in DNA methylation in response to MWCNT exposure
Changes in the methylation of genes associated with inflammation and fibrosis were examined in C57BL/6 mice 24 h or 7 days following exposure to MWCNT. Figure 1 shows a schematic of genes assessed for alterations in methylation, including the CpG sites in their promotor regions that were analyzed.
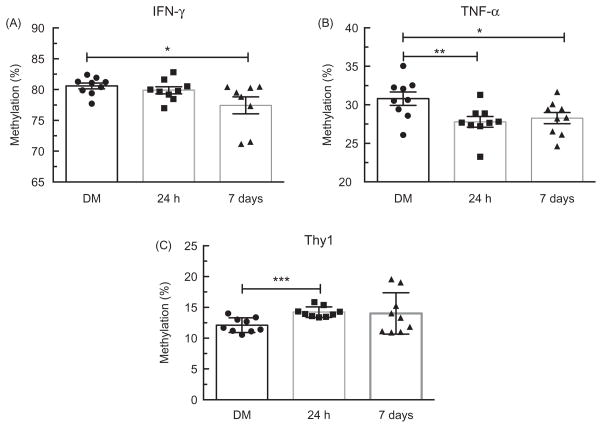
First, we compared isolated lung DNA from animals exposed to MWCNT to DM control mice using the pyrosequencing assay. There were no significant differences in the level of DNA methylation at the IFN-γ promotor between the DM controls and MWCNT-exposed mice 24 h following treatment (82.42% versus 82.82%, Figure 6A). This matches the cytokine data showing no change in IFN-γ production at 24 h post-MWCNT exposure. However, at 7 days following treatment, there was a significant decrease in the level of IFN-γ DNA methylation in the MWCNT group compared to the DM control group (82.42% versus 80.48%, p<0.05; Figure 6A). This suggests that IFN-γ may play a role in MWCNT-induced inflammation at time points later than 24 h. In regards to TNF-α promoter methylation analysis, the average of the two CpG sites showed statistical differences in DNA methylation levels 24 h following exposure as compared to the DM controls (30.80% versus 23.26%, p<0.01; Figure 6B). This is also reflected in the 24 h cytokine data (Figure 2B). TNF-α promotor methylation was still significantly hypomethylated at 7 days compared to the DM controls (30.80% versus 28.28%, p<0.05; Figure 6B). Taken together, the methylation data indicates that MWCNT induce DNA methylation changes in the CpG island region of inflammation-related genes, which could play a major role in activation of the immune response.
Figure 6.
Promoter methylation status in lung tissue of C57BL/6 mice. Change in percent methylation 24 h and 7 days after MWCNT exposure for three genes. (A) IFN-γ at 7 days post-MWCNT exposure was significantly hypomethylated (p<0.05) compared to the DM controls. (B) TNF-α promoter was statistically hypomethylaed at 24 h (p<0.01) and 7 days (p<0.05) post-MWCNT exposure compared to the DM controls. (C) Thy-1 was significantly hypermethylated within the promoter region 24 h post-MWCNT exposure compared with the DM controls (p<0.05). Data expressed as mean ± SEM percent methylation. Asterisks indicate significance at **p<0.01, *p<0.05 compared to the DM controls. n = 9 per condition.
DNA methylation level of a gene related to fibrosis, Thy-1 were also analyzed (Figure 6C). DNA methylation levels of the Thy-1 promoter in mice 24 h following exposure to MWCNT were significantly increased compared to the DM controls (12.12% versus 14.23%, p<0.001; Figure 6C). Furthermore, lung mRNA expression of Thy-1 was significantly decreased at 7 daysafter MWCNT exposure compared to the DM controls (1.01 versus 0.85, p<0.05). Taken together, these data appears to show that changes in DNA methylation are affected by MWCNT exposure and reflective of the inflammatory response.
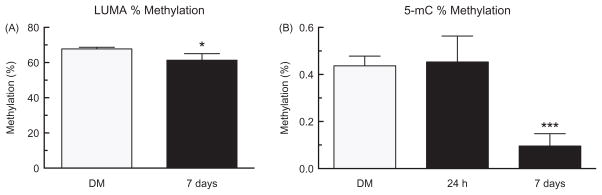
Global hypomethylation in lung tissue
In order to determine alterations in global DNA methylation from MWCNT exposure, LUMA and 5-mC quantification were conducted. LUMA showed significant genomic hypomethylation in the MWCNT-exposed group compared to the DM controls in lung tissue taken from additional mice in the same experiment (Figure 7A). 5-mC also showed significant genomic hypomethylation in the MWCNT-exposed group compared to the DM controls in lung tissue DNA after 7 days, further confirming changes in DNA methylation seen in the LUMA assay (Figure 7B). However, these changes were only seen at 7 days post-exposure and not at 24 h, indicating that some time needs to pass after exposure before these alterations can be observed.
Figure 7.
Global DNA methylation in lung after 24 h or 7 days of MWCNT exposure. Levels of DNA methylation in the lungs of mice after exposure to DM or MWCNT for 7 days using the (A) LUMA and after 24 h and 7 days using the (B) 5-mC assay. Data expressed as mean ± SEM percent methylation. Asterisks indicate significance at *p<0.05, ***p<0.001 compared to the DM controls. n = 10 for DM control and n = 7 for MWCNT treatment for LUMA, and n = 9 in all groups for 5-mC.
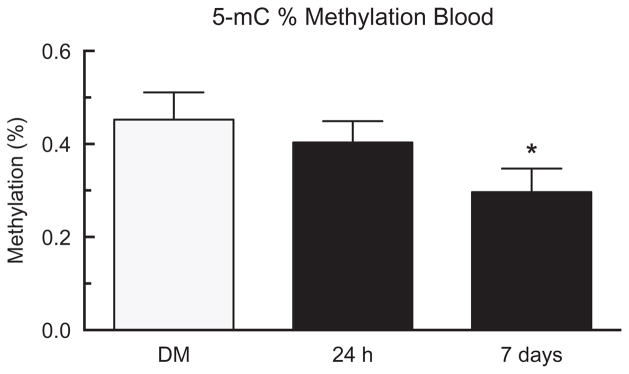
Global hypomethylation in blood
In order to evaluate the systemic inflammatory response, genomic DNA methylation levels were determined in circulating white blood cells (WBC). The results from 5-mC quantification showed significant genomic hypomethylation in the MWCNT-exposed group compared to the DM controls in WBC DNA (Figure 8) at 7 days, matching the changes in DNA methylation that were seen in the lungs. There were no changes in percent methylation in the blood at 24 h, which parallels the lung DNA methylation data. Therefore, these results indicate that WBC DNA methylation levels have the potential to be used as a biomarker for lung inflammation and disease following MWCNT exposure.
Figure 8.
Global DNA methylation in blood after 24 h or 7 days of MWCNT exposure. Levels of DNA methylation in the blood of mice after exposure to DM or MWCNT for 24 h or 7 days using the 5-mC assay. Data expressed as mean ± SEM percent methylation. Asterisks indicate significance at *p<0.05 compared to the DM controls. n = 9 in all groups.
Discussion
For this study, we used nickel-containing MWCNT (previously referred to as FA-21) that were 5–15 μm long with a diameter of 27 nm and were previously associated with lung pathology following exposure (Hamilton et al., 2012). In fact, they were demonstrated to be one of the most toxic MWCNT that were examined. In the current study, increases in initial inflammation were observed at 24 h by elevation of the TNF-α cytokine (Figure 2B). Increases in pathology scores were noted, as well as increased collagen deposition, indicating some level of tissue fibrosis (Figures 3–5).
The promoter methylation of IFN-γ and TNF-a were determined in this study, and compared to alterations in protein level for the corresponding cytokines to determine if alterations in the degree of methylation reflected changes in protein production. Increases in neutrophil infiltration have been shown to enhance IFN-γ production through the production of B-cell activating factor (Coquery et al., 2014). While changes in IFN-γ production were not observed at 24 h in this study, it is possible that this cytokine is latently activated after neutrophil recruitment. A previous study from our lab showed increases in neutrophil recruitment 24 h after MWCNT exposure (Hamilton et al., 2013). Therefore, we suggest that MWCNT induce latent activation of INF-γ due to neutrophil recruitment following exposure. Increased IFN-γ gene expression has been associated with hypomethylation of the promoter region in various inflammatory diseases (Brand et al., 2012; Gonsky et al., 2009; Wang et al., 2013). We analyzed the DNA methylation status of IFN-γ and observed no changes at 24 h; however, we observed a significant decrease in mice 7 days post-MWCNT exposure compared with the DM controls (Figure 6A), which was consistent with our 24 h cytokine data (Figure 2A). Taken together, our findings along with the results from other studies, support a potential role for the IFN-γ promoter’s methylation status in the inflammatory response following MWCNT exposure.
Exposure to ultrafine particles has been shown to result in increased expression of other inflammatory cytokines, including TNF-α (Brown et al., 2004). Increased serum and gene expression levels of TNF-α, a recognized pro-inflammatory marker (Zulet et al., 2007), are associated with an increased risk of fibrosis (Booth et al., 2004). Therefore, we targeted and measured DNA methylation levels in the TNF-α gene and determined whether increased cytokine release correlated with TNF-α hypomethylation. TNF-α was significantly hypomethylated in mice 24 h and 7 days following exposure to MWCNT compared to the DM controls (Figure 6B). We found a significant increase in TNF-α production 24 h after MWCNT exposure, which would be expected given the methylation data (Figure 2B). Taken together, the methylation and cytokine data suggest that the inflammatory response induced by exposure to MWCNT is associated with methylation changes in the promoter of the TNF-α gene and subsequent secretion of TNF-α. These alterations have the potential to impact disease development including fibrosis.
In order to better understand the role of MWCNT-induced changes in DNA methylation and subsequent effects to lung pathology and fibrosis, we conducted further studies to assess the role of promoter methylation in fibroblast activation and the fibrogenesis related gene, Thy-1. Thy-1 is an important protein involved in the development of tissue fibrosis, specifically within the lung, making it an ideal candidate gene to assess for methylation levels in this study. Silencing of Thy-1 by DNA hypermethylation was found in patients with idiopathic pulmonary fibrosis, suggesting that this may be an important mechanism for pathogenic fibroblast alterations (Hagood et al., 2005; Rege & Hagood, 2006; Sanders et al., 2008). We found a significant increase in Thy-1 methylation at 24 h post-MWCNT exposure compared with the DM controls (Figure 6C). Thus, these data, in combination with what is already known about Thy-1, suggest that hypermethylation of the Thy-1 gene may contribute to MWCNT-induced pulmonary fibrosis in the mice. Overall, the gene specific methylation data support the notation that alterations in DNA methylation coincide with disease development and could be a driver in gene transcription affecting the level of inflammatory response and disease development. We further evaluated Thy-1 gene expression to confirm whether hypermethylation of Thy-1 in its promoter region was associated with changes in expression. mRNA expression of Thy-1 was significantly decreased in lung RNA 7 days after MWCNT exposure, corresponding with the expected outcome given the hypermethylation of Thy-1 in the promoter region.
In order to determine changes in global DNA methylation, two different measures were applied, LUMA and 5-mC assay, to investigate the association between DNA methylation and MWCNT exposure. LUMA targets all CCGG sequences in the genome by using methylation specific enzymes to differentiate between methylated and non-methylated DNA by pyrosequencing, and 5-mC assay covers the whole genome by incorporation of antibody-labeled 5-mC (Lee et al., 2008, Karimi et al., 2006a). These assays differ by targeted genomic regions and have different methodological advantages and limitations, but both assays have been used to determine associations of global DNA methylation and environmental exposures and/or disease outcomes (Deneberg et al., 2010; Karimi et al., 2006b; Lee et al., 2008, 2014; Romermann et al., 2007). In confirmation of our hypothesis, DNA hypomethylation in the lungs as evidenced by both the LUMA and 5-mC assays was demonstrated following MWCNT exposure (Figure 7A and B). This is an important finding because it is critical to understand whether or not epigenetic alterations affecting gene expression following environmental exposures change lung DNA methylation and thus the lung environment, in a long-term manner (Weigel et al., 2015). This lasting alteration could increase the likelihood of disease development, including fibrosis, long after the initial response to the particle.
The DNA methylation data within the lung allows for a better understanding of disease development; however, due to the limited accessibility of lung tissue in population studies, studying changes in lung DNA methylation is not a viable option. There is evidence, however, to suggest that blood DNA methylation as a biomarker for disease development is viable. Changes in blood DNA methylation have been shown with cancer; however, no one has ever looked at its potential as a biomarker for tissue fibrosis (Hsiung et al., 2007; Lofton-Day et al., 2008). Therefore, in this study, we determined not only if DNA methylation changes occur in the lungs after MWCNT exposure, but also if those changes correlate with DNA methylation changes in the blood. The results demonstrated that DNA methylation in the blood followed the same trend as in the lungs (Figure 8). This novel finding has never been shown with any fibrosis-associated disease, and provides the first step toward developing an assay using DNA methylation in the blood as a biomarker for MWCNT-induced disease. Our results may also have larger implications as a biomarker for other types of fibrotic lung disease as well. However, further mechanistic studies would need to be performed to determine if MWCNT alterations in the lung lead to epigenetic alterations in the blood.
This study does not demonstrate how the DNA methylation changes in the lung affect the disease process. Future studies should be done to determine how DNA methylation is involved in the MWCNT response. A dose-response study would provide further information as to whether or not the degree of MWCNT exposure correlates with the degree of alterations in global and gene specific methylation and would show whether or not alterations in methylation correspond to alterations in disease development. Multiple time points should be assessed to determine when in the disease process DNA methylation changes occur, either before, during or after fibrotic development. To determine if epigenetic alterations are responsible for disease development, high and low methyl donor diets should be used to alter DNA methylation, as previously described, prior to MWCNT exposure (Sun et al., 2013). A dose response using a these diets, while assessing multiple time points, would provide mechanistic data needed to determine the relationship between MWCNT exposure, epigenetic alterations and subsequent disease development. The data in this study provide ample evidence that these studies are justified.
These data provide an excellent first step for the development of future diagnostic tools. The results demonstrate that DNA methylation changes also occur in the blood when MWCNT-induced disease occurs in lung tissue. For risk assessment in industry, a diagnostic tool has the potential to identify at risk populations before MWCNT exposure and after exposure, to show the likelihood of disease development based on the degree of DNA methylation. This bioassay would allow for determination of an individual’s over when working with MWCNT due to their individual susceptibility for disease development. Once disease has developed, blood DNA methylation levels could also serve as a biomarker to inform clinicians of its progression.
Conclusions
These studies confirm that MWCNT lead to inflammation in the lungs. Significantly, we describe here for the first time that DNA methylation of IFN-γ and TNF-α appears to be involved in the inflammatory response of laboratory mice exposed to MWCNT. In addition, the development of fibrosis in these mice was linked to the hypermethylation of the Thy-1 gene. Based on our findings, we suggest that DNA methylation may be a potential pathogenic mechanism in progression of murine lung injury caused by MWCNT. Furthermore, we were able to show that MWCNT leads to global DNA hypomethylation within the lung tissue 7 days after exposure. The correlating hypomethylation of the blood to the lung tissue, along with evidence of inflammation and the development of tissue fibrosis, indicates that blood DNA methylation is a promising biomarker for MWCNT-induced disease development.
Acknowledgments
We would like to thank Britten Postma, Mary Buford, Pam Shaw and Lou Herritt for their expert technical assistance with various aspects of this manuscript as well as the Inhalation, Histology and Fluorescence Cytometry Cores within the Center for Environmental Health Sciences at the University of Montana.
Footnotes
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper. This work was supported by National Institute of General Medical Sciences (P30GM10333) and National Institute of Environmental Health Sciences (ES023209). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH, NIGMS or NIEHS.
References
- Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, et al. Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med. 2009;179:572–8. doi: 10.1164/rccm.200807-1097OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bind MA, Baccarelli A, Zanobetti A, Tarantini L, Suh H, Vokonas P, Schwartz J. Air pollution and markers of coagulation, inflammation, and endothelial function: associations and epigene-environment interactions in an elderly cohort. Epidemiology. 2012;23:332–40. doi: 10.1097/EDE.0b013e31824523f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bind MA, Lepeule J, Zanobetti A, Gasparrini A, Baccarelli A, Coull BA, et al. Air pollution and gene-specific methylation in the Normative Aging Study: association, effect modification, and mediation analysis. Epigenetics. 2014;9:448–58. doi: 10.4161/epi.27584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–80. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- Booth M, Mwatha JK, Joseph S, Jones FM, Kadzo H, Ireri E, et al. Periportal fibrosis in human Schistosoma mansoni infection is associated with low IL-10, low IFN-gamma, high TNF-alpha, or low RANTES, depending on age and gender. J Immunol. 2004;172:1295–303. doi: 10.4049/jimmunol.172.2.1295. [DOI] [PubMed] [Google Scholar]
- Brand S, Kesper DA, Teich R, Kilic-Niebergall E, Pinkenburg O, Bothur E, et al. DNA methylation of TH1/TH2 cytokine genes affects sensitization and progress of experimental asthma. J Allergy Clin Immunol. 2012;129:1602–1610e. 1606. doi: 10.1016/j.jaci.2011.12.963. [DOI] [PubMed] [Google Scholar]
- Brown DM, Donaldson K, Borm PJ, Schins RP, Dehnhardt M, Gilmour P, et al. Calcium and ROS-mediated activation of transcription factors and TNF-alpha cytokine gene expression in macrophages exposed to ultrafine particles. American journal of physiology Lung Cell Mol Physiol. 2004;286:L344–53. doi: 10.1152/ajplung.00139.2003. [DOI] [PubMed] [Google Scholar]
- Coquery CM, Wade NS, Loo WM, Kinchen JM, Cox KM, Jiang C, et al. Neutrophils contribute to excess serum BAFF levels and promote CD4+ T cell and B cell responses in lupus-prone mice. PLoS One. 2014;9:e102284. doi: 10.1371/journal.pone.0102284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneberg S, Grovdal M, Karimi M, Jansson M, Nahi H, Corbacioglu A, et al. Gene-specific and global methylation patterns predict outcome in patients with acute myeloid leukemia. Leukemia. 2010;24:932–41. doi: 10.1038/leu.2010.41. [DOI] [PubMed] [Google Scholar]
- Gong C, Tao G, Yang L, Liu J, Liu Q, Li W, Zhuang Z. Methylation of PARP-1 promoter involved in the regulation of nano-SiO2-induced decrease of PARP-1 mRNA expression. Toxicol Lett. 2012;209:264–9. doi: 10.1016/j.toxlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Gong C, Tao G, Yang L, Liu J, Liu Q, Zhuang Z. SiO2 nanoparticles induce global genomic hypomethylation in HaCaT cells. Biochem Biophys Res Commun. 2010;397:397–400. doi: 10.1016/j.bbrc.2010.05.076. [DOI] [PubMed] [Google Scholar]
- Gonsky R, Deem RL, Targan SR. Distinct methylation of IFNG in the gut. J Interferon Cytokine Res. 2009;29:407–14. doi: 10.1089/jir.2008.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagood JS, Prabhakaran P, Kumbla P, Salazar L, MacEwen MW, Barker TH, et al. Loss of fibroblast Thy-1 expression correlates with lung fibrogenesis. Am J Pathol. 2005;167:365–79. doi: 10.1016/S0002-9440(10)62982-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton RF, Jr, Buford M, Xiang C, Wu N, Holian A. NLRP3 inflammasome activation in murine alveolar macrophages and related lung pathology is associated with MWCNT nickel contamination. Inhal Toxicol. 2012;24:995–1008. doi: 10.3109/08958378.2012.745633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton RF, Jr, Wu Z, Mitra S, Shaw PK, Holian A. Effect of MWCNT size, carboxylation, and purification on in vitro and in vivo toxicity, inflammation and lung pathology. Part Fibre Toxicol. 2013;10:57. doi: 10.1186/1743-8977-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herceg Z, Vaissiere T. Epigenetic mechanisms and cancer: an interface between the environment and the genome. Epigenetics. 2011;6:804–19. doi: 10.4161/epi.6.7.16262. [DOI] [PubMed] [Google Scholar]
- Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, Kelsey KT. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–14. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Karimi M, Johansson S, Ekstrom TJ. Using LUMA: a Luminometric-based assay for global DNA-methylation. Epigenetics. 2006a;1:45–8. doi: 10.4161/epi.1.1.2587. [DOI] [PubMed] [Google Scholar]
- Karimi M, Johansson S, Stach D, Corcoran M, Grander D, Schalling M, et al. LUMA (LUminometric Methylation Assay) – a high throughput method to the analysis of genomic DNA methylation. Exp Cell Res. 2006b;312:1989–95. doi: 10.1016/j.yexcr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Kolb M, Margetts PJ, Anthony DC, Pitossi F, Gauldie J. Transient expression of IL-1beta induces acute lung injury and chronic repair leading to pulmonary fibrosis. J Clin Invest. 2001;107:1529–36. doi: 10.1172/JCI12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu Y, Waku T, Iwasaki N, Ono W, Yamaguchi C, Yanagisawa J. Global analysis of DNA methylation in early-stage liver fibrosis. BMC Med Genomics. 2012;5:5. doi: 10.1186/1755-8794-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam CW, James JT, McCluskey R, Arepalli S, Hunter RL. A review of carbon nanotube toxicity and assessment of potential occupational and environmental health risks. Crit Rev Toxicol. 2006;36:189–217. doi: 10.1080/10408440600570233. [DOI] [PubMed] [Google Scholar]
- Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Geli J, Larsson C, Wallin G, Karimi M, Zedenius J, et al. Gene-specific promoter hypermethylation without global hypomethylation in follicular thyroid cancer. Int J Oncol. 2008;33:861–9. [PubMed] [Google Scholar]
- Lee Y, Kim YJ, Choi YJ, Lee JW, Lee S, Cho YH, Chung HW. Radiation-induced changes in DNA methylation and their relationship to chromosome aberrations in nuclear power plant workers. Int J Radiat Biol. 2014;91:142–9. doi: 10.3109/09553002.2015.969847. [DOI] [PubMed] [Google Scholar]
- Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54:414–23. doi: 10.1373/clinchem.2007.095992. [DOI] [PubMed] [Google Scholar]
- Lu X, Miousse IR, Pirela SV, Melnyk S, Koturbash I, Demokritou P. Short-term exposure to engineered nanomaterials affects cellular epigenome. Nanotoxicology. 2015 doi: 10.3109/17435390.2015.1025115. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DI, Cropley JE, Suter CM. Epigenetics in disease: leader or follower? Epigenetics. 2011;6:843–8. doi: 10.4161/epi.6.7.16498. [DOI] [PubMed] [Google Scholar]
- Muller J, Huaux F, Moreau N, Misson P, Heilier JF, Delos M, et al. Respiratory toxicity of multi-wall carbon nanotubes. Toxicol Appl Pharmacol. 2005;207:221–31. doi: 10.1016/j.taap.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Pilsner JR, Hu H, Ettinger A, Sanchez BN, Wright RO, Cantonwine D, et al. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect. 2009;117:1466–71. doi: 10.1289/ehp.0800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rege TA, Hagood JS. Thy-1 as a regulator of cell-cell and cell-matrix interactions in axon regeneration, apoptosis, adhesion, migration, cancer, and fibrosis. FASEB J. 2006;20:1045–54. doi: 10.1096/fj.05-5460rev. [DOI] [PubMed] [Google Scholar]
- Romermann D, Hasemeier B, Metzig K, Schlegelberger B, Langer F, Kreipe H, Lehmann U. Methylation status of LINE-1 sequences in patients with MDS or secondary AML. Verhandlungen der Deutschen Gesellschaft fur Pathologie. 2007;91:338–42. [PubMed] [Google Scholar]
- Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect. 2008;116:1547–52. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders YY, Ambalavanan N, Halloran B, Zhang X, Liu H, Crossman DK, et al. Altered DNA methylation profile in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;186:525–35. doi: 10.1164/rccm.201201-0077OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders YY, Pardo A, Selman M, Nuovo GJ, Tollefsbol TO, Siegal GP, Hagood JS. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2008;39:610–18. doi: 10.1165/rcmb.2007-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liang D, Sahbaie P, Clark JD. Effects of methyl donor diets on incisional pain in mice. PLoS One. 2013;8:e77881. doi: 10.1371/journal.pone.0077881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, et al. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect. 2009;117:217–22. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Xu J, Zhu Q, Qin X, Cao Y, Lou J, et al. Downregulation of IFNG in CD4(+) T cells in lung cancer through hypermethylation: a possible mechanism of tumor-induced immunosuppression. PLoS One. 2013;8:e79064. doi: 10.1371/journal.pone.0079064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster AL, Yan MS, Marsden PA. Epigenetics and cardiovascular disease. Can J Cardiol. 2013;29:46–57. doi: 10.1016/j.cjca.2012.10.023. [DOI] [PubMed] [Google Scholar]
- Weigel C, Schmezer P, Plass C, Popanda O. Epigenetics in radiation-induced fibrosis. Oncogene. 2015;34:2145–55. doi: 10.1038/onc.2014.145. [DOI] [PubMed] [Google Scholar]
- Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucl Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulet MA, Puchau B, Navarro C, Marti A, Martinez JA. Inflammatory biomarkers: the link between obesity and associated pathologies. Nutr Hosp. 2007;22:511–27. [PubMed] [Google Scholar]