Abstract
Rationale
Only a small portion of the known heritability of cardiovascular diseases such as heart failure can be explained based on single gene mutations. Chromatin structure and regulation provide a substrate through which genetic differences in non-coding regions may impact cellular function and response to disease, but the mechanisms are unknown.
Objective
We conducted genome-wide measurements of DNA methylation in different strains of mice that are susceptible and resistant to isoproterenol-induced dysfunction to test the hypothesis that this epigenetic mark may play a causal role in the development of heart failure.
Methods and Results
BALB/cJ and BUB/BnJ mice, determined to be susceptible and resistant to isoproterenol-induced heart failure respectively, were administered the drug for 3 weeks via osmotic minipump. Reduced representational bisulfite sequencing was then used to compare the differences between the cardiac DNA methylome in the basal state between strains and then following isoproterenol treatment. Single base resolution DNA methylation measurements were obtained and revealed a bimodal distribution of methylation in the heart, enriched in lone intergenic CpGs and depleted from CpG islands around genes. Isoproterenol induced global decreases in methylation in both strains; however, the basal methylation pattern between strains shows striking differences that may be predictive of disease progression prior to environmental stress. The global correlation between promoter methylation and gene expression (as measured by microarray) was modest and revealed itself only with focused analyses of transcription start site and gene body regions (in contrast to when gene methylation was examined in toto). Modules of co-methylated genes displayed correlation with other protein-based epigenetic marks supporting the hypothesis that chromatin modifications act in a combinatorial manner to specify transcriptional phenotypes in the heart.
Conclusions
This study provides the first single base-resolution map of the mammalian cardiac DNA methylome and the first case-control analysis of the changes in DNA methylation with heart failure. The findings demonstrate marked genetic differences in DNA methylation that are associated with disease progression.
Keywords: DNA methylation, epigenetics, chromatin, heart failure, genetics
INTRODUCTION
Common heart failure has a large heritable component,1 however few individual causal gene mutations have been revealed.2 In addition to the role of multiple rare variants,3 another possible mechanism contributing to the heritability of heart failure is epigenetic modifications, which control genomic structure and gene expression in a cell type-specific manner.4, 5 DNA methylation is the covalent addition of a methyl group to the 5’ carbon of cytosine nucleotides. Previous studies have shown that DNA methylation contributes to normal development in plants and animals by influencing gene expression.6 Traditionally, DNA methylation is thought to be a marker of heterochromatin because condensed chromatin has a higher level of DNA methylation, for example as observed during X-chromosome inactivation. More recently, it has been suggested that the effect of DNA methylation on gene expression is context dependent, with promoter/transcription start site methylation being associated with silencing and gene body methylation associated with activation.7 Unlike other protein-based chromatin marks, DNA methylation has long been considered a stable modification of DNA and expected to remain unchanged throughout the life of the fully differentiated somatic cell—recent studies have challenged this concept.8 Aberrant DNA methylation has been linked to many diseases, in particular different types of cancer,9–13 enabling the success of methylation inhibitors as anticancer drugs.14 The role of DNA methylation in other common diseases, such as cardiovascular disease, is less well understood.
Recent studies using human samples have discovered changes in DNA methylation in the setting of heart diseases.15–18 These investigations open a new avenue of research into regulation of cardiac physiology and disease, but there are several unanswered questions. First, human studies lack a case-control format to precisely determine the effects of environment and rule out other co-morbid conditions that are pervasive with human heart failure and potentially undetected. Second, most studies in the heart have not employed methods to give single base resolution for the analysis of DNA methylation, which precludes determination of the genomic loci (be they coding or non-coding) under regulation by the modification. Finally, human studies lack the ability to control for genetic variability, which can be a confounding factor in determining the role of chromatin modifications.19
To overcome these limitations, we have carried out what we believe to be a first of its kind study in cardiovascular epigenomics: mouse strains that are either susceptible (BALB/cJ) or resistant (BUB/BnJ) to isoproterenol-induced heart failure were administered the β-adrenergic receptor agonist followed by genome-wide bisulfite sequencing to measure DNA methylation. Because these mice are inbred and have fully characterized genetic diversity (i.e. their polymorphisms are known) and environmental conditions are constant, this approach enables investigation of the effects of DNA methylation on disease phenotype in a pure manner. Our study has several major findings: the DNA methylome in the mouse heart is characterized for the first time with single base resolution; genetic differences play a large role in basal DNA methylation differences between strains; DNA methylation appears to coordinate with histone-based chromatin marks; and, most importantly, DNA methylation is predictive of susceptibility to heart failure prior to exposure to disease-inducing stress.
METHODS
Please see the Online Material for detailed Methods.
RESULTS
Measurement of cardiac CpG methylation with single base resolution by RRBS
Results on sequencing depth and alignment are provided in Table 1. Given the size of the mouse genome (~2.65 billion bp), ~265 million uniquely aligned 100-bp reads would be needed to achieve a ten-fold coverage of the entire genome using traditional shotgun sequencing. To lower the sequencing demand and data size while maintaining data quality, we used RRBS instead of shotgun sequencing. This method enabled very high coverageof the CpGs within RRBS libraries (Table 1), allowing for confident assignment of methylation status.
Table 1.
RRBS sequencing depth for each sample
| Mouse strain |
Treatment | Batch | # of reads 100bp/read |
# of mapped reads |
# of reads after QC* |
Mapability |
|---|---|---|---|---|---|---|
| BALB/cJ | Control | Batch 1 | 196,132,876 | 108,834,401 | 108,172,874 | 55.15% |
| BALB/cJ | ISO | Batch 1 | 190,915,791 | 113,543,242 | 112,792,817 | 59.08% |
| BUB/BnJ | Control | Batch 1 | 209,747,249 | 105,732,683 | 105,046,961 | 50.08% |
| BUB/BnJ | ISO | Batch 1 | 182,087,726 | 101,999,903 | 100,609,652 | 55.25% |
| BALB/cJ | Control | Batch 2 | 83,224,678 | 43,990,632 | 43,989,246 | 52.86% |
| BALB/cJ | ISO | Batch 2 | 89,028,320 | 46,538,221 | 46,536,900 | 52.27% |
| BUB/BnJ | Control | Batch 2 | 90,112,603 | 47,023,872 | 47,022,943 | 52.18% |
| BUB/BnJ | ISO | Batch 2 | 87,836,780 | 47,242,872 | 47,241,990 | 53.78% |
| BALB/cJ | Control | Batch 3 | 26,750,907 | 13,599,388 | 13,587,484 | 50.79% |
| BALB/cJ | ISO | Batch 3 | 24,447,253 | 14,831,117 | 14,829,851 | 60.66% |
| BUB/BnJ | Control | Batch 3 | 29,104,605 | 16,941,795 | 16,939,936 | 58.20% |
| BUB/BnJ | ISO | Batch 3 | 28,631,652 | 18,342,525 | 18,342,070 | 64.06% |
Reads not fully converted by bisulfite treatment were removed, as indicated by an XS=1 tag in the output of BS Seeker2.
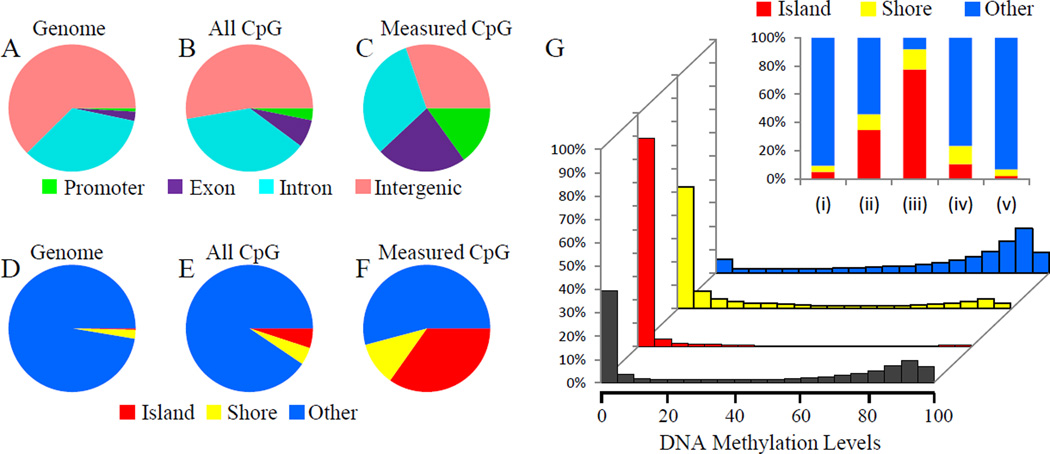
The restriction endonuclease MspI, which recognizes the CCGG motif and generates a -CGG sticky end, was used to digest genomic DNA into fragments during library preparation. To quantify how this step affects CpG measurement, we compared the annotation of CpG sites covered by our RRBS libraries with the total CpGs in the reference genome. Figure 1 shows the location of CpGs covered by our RRBS libraries compared to all the CpGs in the mouse genome. If one looks at these CpGs on the basis of localization in promoter, exon, intron or intergenic region, it is apparent that CpGs are naturally enriched in coding regions (compare panel B to panel A). This enrichment is further accentuated by the RRBS measurements (compare panel C to B; this step, therefore, may influence the portion of the methylome examined in our RRBS studies, but is consistent with that observed by other studies20). Panels D-F of Figure 1 further demonstrate that our measurements enrich for detection of CpGs in and around CpG islands, which are thought to be sites of dynamic regulation by methylation. The CpG islands are also preferentially demethylated as compared with other regions of the genome (Figure 1G), yet all regions we sampled in the heart demonstrate a bimodal distribution. This distribution may be different from other cells/tissues. For example, Jiang, et al. reported a global hypermethylation in oocytes and sperms from zebrafish.21 When examining the degree of methylation in a non-binary manner according to genomic elements, it is apparent that of the genomic regions with <5% methylation, most occur in CpG islands and shores (Figure 1G, inset), whereas the genomic regions methylated >80% of the time tend to distribute according to the functional annotation of genome and do not enrich around islands or shores. Most CpGs outside of any CpG islands (“lone” CpGs) are hypermethylated. These observations were true for both mouse strains and were not fundamentally altered by ISO treatment. In our datasets, the methylation characteristics (i.e. methylated or not methylated) of a given CpG in a fragment tends to be conserved—that is, if one CpG on a fragment is methylated, the other ones present on the same fragment tend also to be methylated, and vice versa. We therefore present all the forthcoming data analyses on a per fragment basis to reduce the complexity of the data. All single base resolution data are available in the Online Material.
Figure 1. Distribution of myocardial DNA methylation measured by bisulfite sequencing.
(A–F) Annotation of CpGs covered by RRBS libraries. CpGs are annotated based on location in promoter, exon, intron or intergenic regions (A–C), or CpG island, island shore or far from CpG island (D–F). A & D, Annotation of mouse reference genome. B & E, Annotation of CpGs within the whole genome. C & F, Annotation of CpGs covered by our RRBS libraries. Promoter, regions within 2kb upstream of transcription start site. CpG island shore, regions within 2kb flanking CpG island. G. Histograms of DNA methylation levels within CpG islands (red), CpG island shores (yellow) and non-CpG island regions (blue). x-axis, DNA methylation level. y-axis, the proportion of total RRBS CpGs. Upper-right panel, stacked histogram of DNA methylation distribution showing the percentage of CpGs located in CpG islands, shores or non-CpG island regions. CpGs are grouped based on their methylation level. (i), all CpGs within the whole genome. (ii), RRBS CpGs. (iii) RRBS CpGs with lower than 5% methylation. (iv) RRBS CpGs with > 5% and < 80% methylation. (v) RRBS CpGs with more than 80% methylation.
DNA methylation varies genetically and is affected by isoproterenol treatment
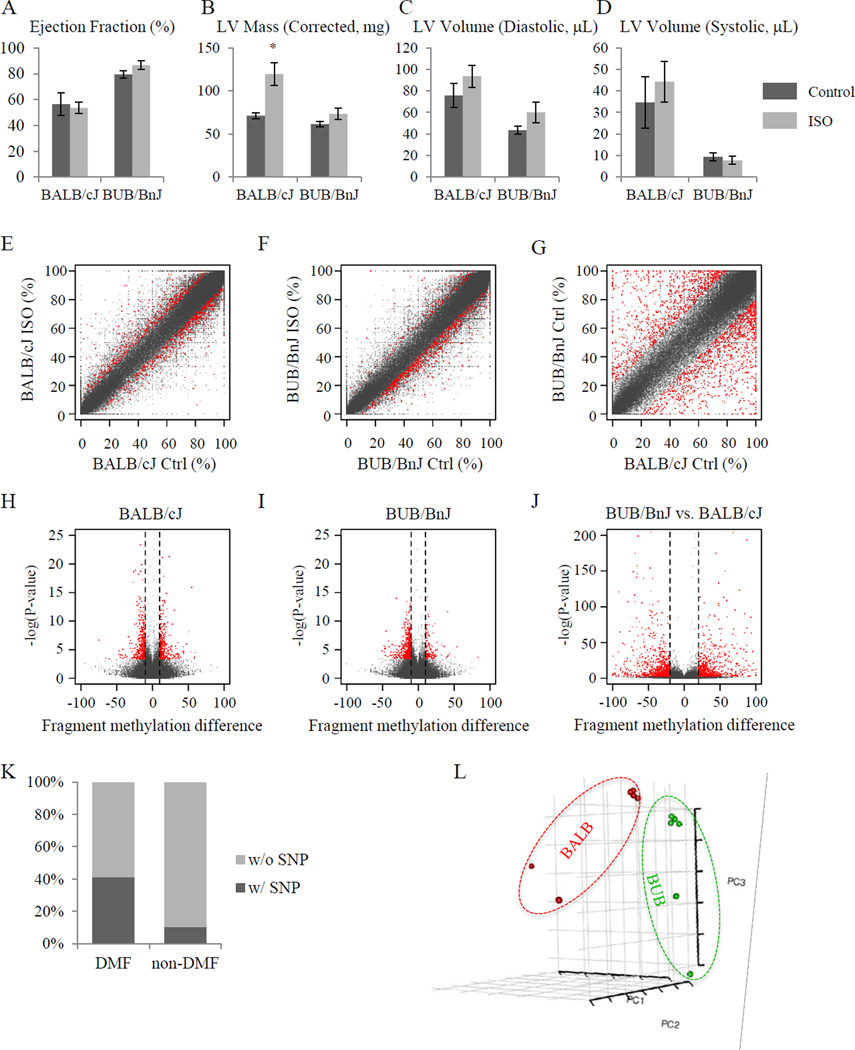
As part of a comprehensive phenotyping study on multiple mouse strains—the results of which were published22 while this manuscript has been under review—we determined the inbred mouse strains BALB/cJ and BUB/BnJ to be susceptible and resistant to ISO-induced cardiac remodeling, respectively. BALB/cJ mice develop left ventricular chamber dilation (Figures 2C&D) 3 weeks after ISO pump implantation, commensurate with a modest decrease in ejection fraction (Figure 2A) and substantial hypertrophy Figure 2B. BUB/BnJ mice, in contrast, are largely resistant to these changes (with the exception of increased LV volume in diastole, Figure 2C). While the strains did not exhibit significant differences in ejection fraction (Figure 2A) it is noteworthy that these measurements were made when the animals were still under constant beta-adrenergic stimulation because of the ISO pump. Furthermore, we cannot totally rule out that the effects of the surgery or pump implantation affected CpG methylation; however, as discussed below, the conspicuous enrichment of DMF in cardiac genes, combined with the 3 week period between pump implantation and experiment, mitigate this concern.
Figure 2. ISO-treatment induces differential DNA methylation.
(A–D) Isoproterenol induced cardiac pathology in mice of BALB/cJ and BUB/BnJ strains indicating strain-specific responses to beta-adrenergic receptor agonist treatment. The heart function as indicated by ejection fraction (A) and size of heart as indicated by left ventricular mass (B), diastolic left ventricular volume (C) and systolic left ventricular volume (D), were measured using M-mode echocardiography. BALB/cJ mice showed a dramatic increase of LV mass and volume after three-week ISO treatment, while BUB/BnJ mice showed only a slight increase of ejection fraction. (n=5–13/group for panels A–D. *P<0.05 vs. control. Vertical bars represent Standard Error) (E–G) Scatter plots showing differential DNA methylation per fragment, i.e. MspI digestion product with length of 50bp to 300bp. Axes indicate DNA methylation level in percentage. N=3/group for all methylation studies. (E) ISO-induced differential DNA methylation in BALB/cJ strain. (F) ISO-induced differential DNA methylation in BUB/BnJ strain (see Online Figure I for individual CpG data). (G) Basal difference between BALB/cJ and BUB/BnJ. (H-J) Volcano plot showing ISO-induced differentially methylated fragments in BALB/cJ (H) and BUB/BnJ (I) strains, as well as basal differences between BALB/cJ and BUB/BnJ (J). x-axis, difference of DNA methylation level between control and ISO treated mice. y-axis, −log10 of p-value. Red, differentially methylated DNA fragments with FDR < 5%. For a CpG methylation event to be considered different in panels H & I (red data points), the site must exhibit a >10% (>20% for basal comparison, panel J) difference with FDR < 0.05. (K) Stacked column plot showing the percentage of DNA fragments harboring at least one genomic variance (SNP) between BALB/cJ and BUB/BnJ. DNA fragments from the whole genome (left column) and with differential methylation levels between two strains at basal level (right column) are shown. P-value was calculated using Pearson’s Chi-squared test with Yates’ continuity correction, and is smaller than 2.2×10−16. (L) Principal component analysis of fragment CpG methylation is shown with the first three principal components, demonstrating a strain-specific DNA methylome profile.
While individual CpG methylation status is necessary to understand locus-specific regulation, recent studies have shown that segments of DNA tend to exhibit similar methylation status when one examines several hundred bases—the size scale of the MspI digested fragments sequenced in the present study. To explore this phenomenon, we binned our methylation measurements according to MspI fragments and plotted behavior per fragment in Figures 2E–J. While the majority of MspI sites had no substantial methylation difference between strains, a surprising number of CpGs exhibited markedly different methylation status: that is, they were methylated in one strain and not the other at baseline (Figures 2G & J; note: cytosines that were variable between the two strains [i.e. cytosine in one strain and another base in the other strain] were omitted from this analysis). In contrast, the number of DNA fragments modified following 3 weeks of ISO treatment is quite small in each strain, as shown in Figures 2E and 2F. As expected, the trends were similar to what is observed with individual CpG analyses (Online Figures IA&B), with the caveat that the differences between strains and following treatment are muted by the fragment-binning step. An advantage of analyzing on the basis of fragments, however, is that one gains a further buffer against experimental variability (i.e. a CpG being modified in one experiment and not another) while increasing quantitative rigor. Because the fragments display the same trend between strains as is seen with individual CpGs, this is an additional level of validation that the strain-dependent differences are genetically linked, in that the chances of difference entire fragments being detected (or not) in a spurious manner between experiments is much lower than the same possibility for individual CpGs. As shown in the volcano plots (Figures 2H & I), most CpGs did not change with statistical significance after ISO. However, a reproducible subset of fragments (red spots) were increased or decreased in methylation after the drug.
To test whether genomic variances contribute to the basal difference of DNA methylation between two strains, we calculated the percentage of DNA fragments with or without SNPs. After removing all CpG dinucleotide whose cytosines overlap with known or imputed SNPs, we found that while in the whole genome 10.2% (20,590 out of 201,515) non-DMF DNA fragments have at least one SNP, this ratio increased to 41.1% (789 out of 1,919) when DNA fragments with different basal methylation levels between two strains were considered (Figure 2K; note: in contrast to the analyses elsewhere, this SNP analysis in Figure 2K refers to variable bases anywhere in the fragment; as described above, whenever the base varied between strains, it was thrown out for consideration of methylation status at that residue). This result demonstrates that DNA fragments with SNPs are more likely to have different DNA methylation levels.
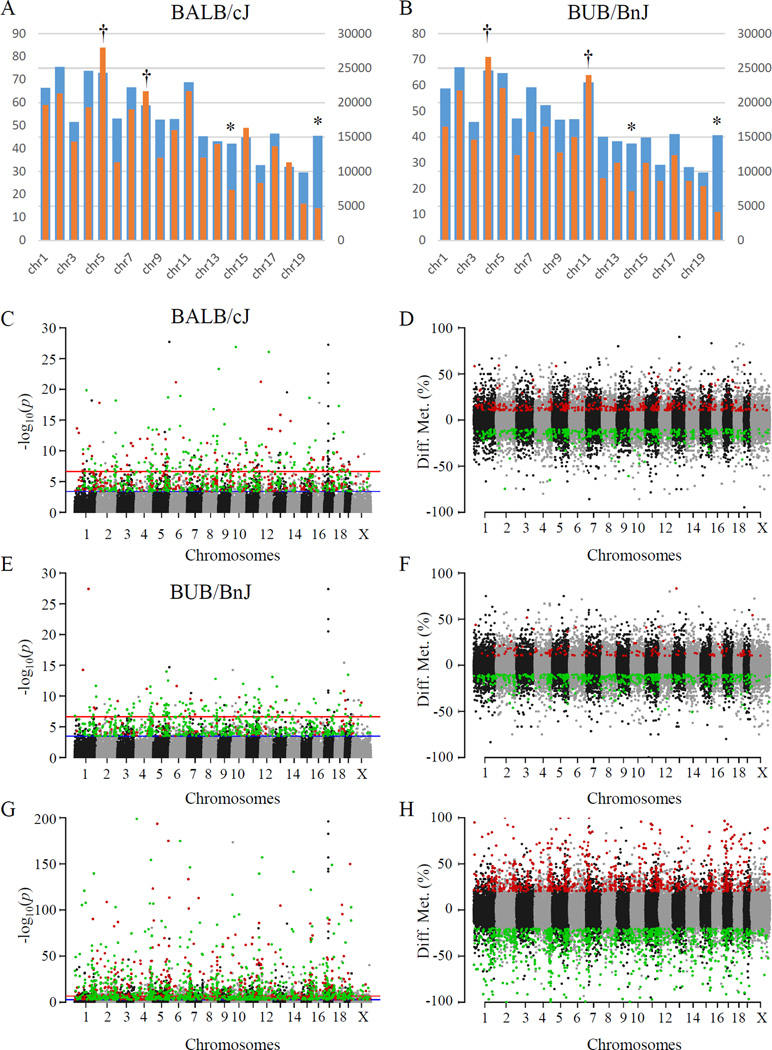
Principal component analyses were also used to investigate the reproducibility of the methylation measurements and to interrogate the relationship between genetics and DNA methylation. Figure 2L shows that strains grouped together according to the first three principal components. These findings reflect that genetic background plays a significant role in determining the DNA methylome. Lastly, a preferential demethylation was observed after ISO in both strains especially in BUB/BnJ (Figures 3A–H and Online Table I). The regions affected by ISO treatment appeared to cluster, with these clusters then distributed across the genome, occurring on all chromosomes with a slight preference on chromosome 5 and reduction on chromosome X (Figures 3A & B; because female mice were studied, no data are available for the Y chromosome). To our knowledge this is the first demonstration that the phenotypic differences observed between these strains may result in part from DNA methylation.
Figure 3. Genomic location of DMFs following ISO treatment.
(A, B) Counts of DMFs by chromosome for BALB/cJ (A) and BUB/BnJ (B). Orange: counts of DMFs. Blue: counts of total RRBS fragments. Chromosomes with significantly fewer DMFs than background are labeled with asterisk, while chromosomes with significantly more DMFs with dagger. Chromosomes are labeled left to right 1 through 19 plus X. (C–H) Genomic location of DMFs induced by ISO treatment in BALB/cJ (C & D) and BUB/BnJ (E & F) strains, as well as basal comparison between two strains (G & H). y-axis, -log10 of p-value for each DNA fragment (C, E & G) or difference of DNA methylation (D, F & H). x-axis, chromosome names. Red: up-regulated DMFs or hypermethylated in G&H. Green: down-regulated DMFs or hypomethylated in G&H.
Differential methylation targets specific genomic regions and types of chromatin domains
Online Table I shows the differentially methylated fragments following ISO treatment in the BALB/cJ and BUB/BnJ mice. Among the 337,027 DNA fragments measured in both strains, 1,509 of them show differential methylation after ISO treatment in at least one strain. Despite the similar volcano plot shape in each strain (Figures 2 H&I), the overlap of differentially methylated fragments between two strains is very small. Only 31 fragments show uniform behavior in both strains according to our statistical threshold (Table 2). To understand the potential function of this differential methylation, we investigated genes associated with differentially methylated fragment(s). Gene Ontology and other functional annotation revealed the genes from the sensitive strain BALB/cJ to be enriched in heart disease-associated functions, while genes from the resistant strain BUB/BnJ are involved in abnormal growth and cardiac muscle contractility (Online Table II).
Table 2.
DNA fragments with altered CpG methylation in both strains
| Chromosome | Start | End | Diff. Met (BALB/cJ) |
Diff. Met (BUB/BnJ) |
Associated Gene |
|---|---|---|---|---|---|
| Up in both | |||||
| chr1 | 189873628 | 189873810 | 10.85% | 10.05% | Smyd2 (+48644), Ptpn14 (+145451) |
| chr2 | 27182325 | 27182514 | 11.71% | 10.49% | Dbh (+17186), Sardh (+64847) |
| chr4 | 134316356 | 134316481 | 13.37% | 16.03% | Slc30a2 (−26788), Trim63 (+1298) |
| chr7 | 128540107 | 128540257 | 13.72% | 15.52% | Inpp5f (−71178), Bag3 (+16599) |
| chr8 | 120582053 | 120582171 | 10.49% | 10.14% | Gins2 (+7192), Gse1 (+93665) |
| chr8 | 122238337 | 122238506 | 15.54% | 14.03% | Gm22 (−31148), Banp (+287877) |
| chr8 | 122238507 | 122238633 | 17.16% | 14.12% | Gm22 (−30999), Banp (+288026) |
| chr18 | 37842431 | 37842627 | 10.70% | 10.98% | Pcdhga11 (+22983), Diap1 (+92947) |
| chr18 | 84192436 | 84192578 | 10.18% | 14.11% | Zadh2 (+104349), Zfp407 (+396997) |
| Down in both | |||||
| chr1 | 89992857 | 89993101 | −17.99% | −17.27% | Gbx2 (−59689), Asb18 (+21598) |
| chr1 | 131010735 | 131010911 | −14.65% | −12.38% | Il19 (−71582), Il10 (−9022) |
| chr1 | 182108412 | 182108579 | −12.25% | −11.32% | Enah (−88515), Srp9 (−16255) |
| chr5 | 63936930 | 63937056 | −30.98% | −27.08% | Rell1 (+31912), 0610040J01Rik (+124498) |
| chr5 | 119680671 | 119680792 | −14.89% | −15.65% | Tbx5 (−153932), Tbx3 (+9713) |
| chr5 | 119841821 | 119842008 | −10.13% | −14.21% | Rbm19 (−274596), Tbx5 (+7251) |
| chr5 | 135621905 | 135622044 | −16.49% | −12.05% | Ccl24 (−48931), Rhbdd2 (−10644) |
| chr6 | 91965207 | 91965382 | −19.10% | −14.33% | Fgd5 (−21816), 4930590J08Rik (+50535) |
| chr7 | 25645126 | 25645303 | −10.19% | −10.14% | Bckdha (+13547), B3gnt8 (+17590) |
| chr8 | 83668524 | 83668653 | −12.78% | −12.31% | Ptger1 (+1725), Pkn1 (+25472) |
| chr10 | 66770619 | 66770763 | −17.70% | −11.02% | Reep3 (+326297) |
| chr10 | 68268862 | 68269022 | −10.68% | −11.80% | Arid5b (+9784), Rtkn2 (+289344) |
| chr11 | 106659519 | 106659639 | −10.27% | −11.17% | Tex2 (−46649), Pecam1 (+55680) |
| chr11 | 117353468 | 117353611 | −10.96% | −15.55% | Gm11733 (−130829), Sept9 (+153878) |
| chr13 | 44534613 | 44534790 | −14.24% | −16.57% | Jarid2 (−196570), Cd83 (+749594) |
| chr14 | 44865790 | 44865946 | −13.20% | −12.50% | Ptger2 (−122327), Ptgdr (−6493) |
| chr15 | 76188602 | 76188749 | −15.50% | −11.39% | BC024139 (−62133), Plec (+17646) |
| chr15 | 102106728 | 102106852 | −14.85% | −13.63% | Tenc1 (+3802), Spryd3 (+29444) |
| chr16 | 8562615 | 8562745 | −11.76% | −15.22% | Pmm2 (−74994), Abat (+49251) |
| chr16 | 37681813 | 37681987 | −11.87% | −12.73% | Fstl1 (−94973), Ndufb4 (−27447) |
| chr17 | 65744154 | 65744265 | −18.76% | −16.28% | Txndc2 (−102005), Rab31 (+28543) |
| chr19 | 5569310 | 5569437 | −14.86% | −16.70% | Rnaseh2c (−32500), Ap5b1 (+1299) |
Diff. Met: Difference of CpG methylation after ISO treatment.
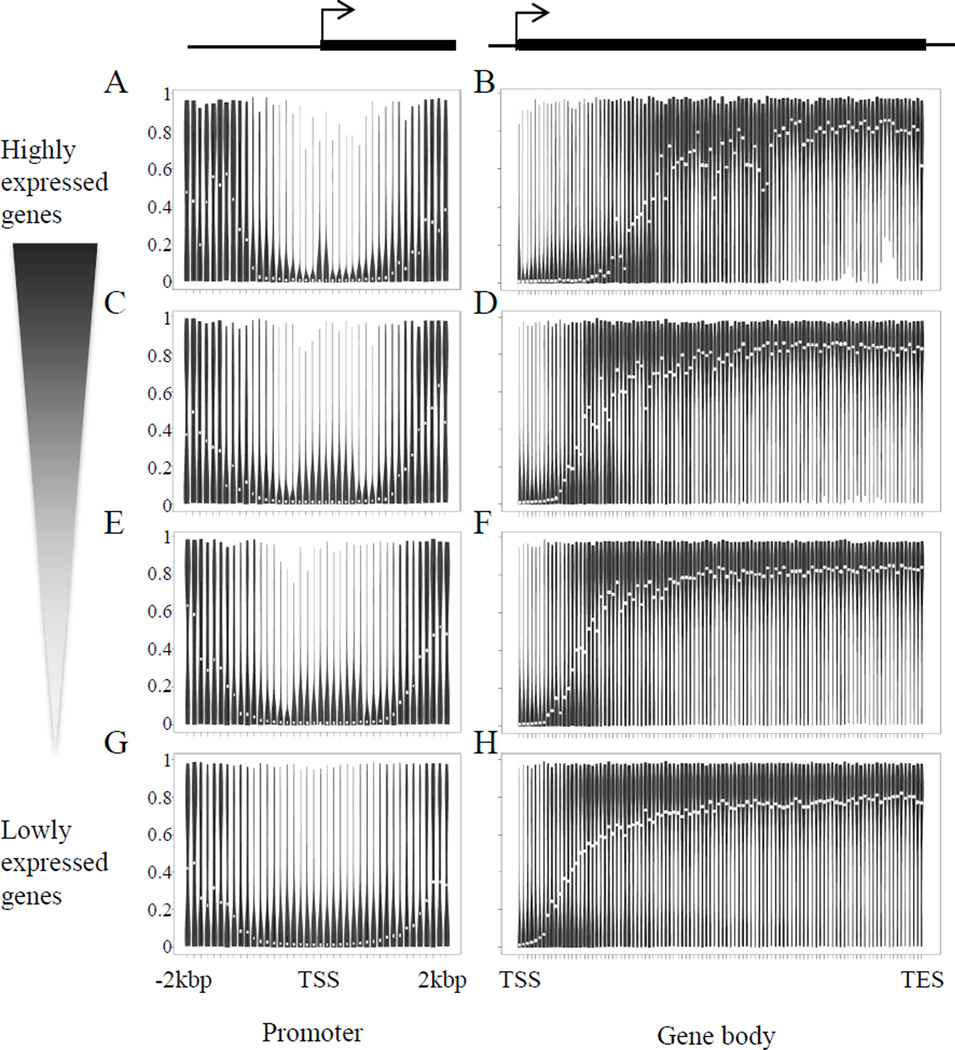
Previous investigations have suggested DNA methylation in the promoter region is negatively correlated with gene expression. To test this phenomenon in the heart, we conducted gene expression analyses using microarrays. When examining the methylation of promoters defined by the sum of 500bp up- and down-stream of the transcription start site for all genes, there is no correlation between DNA methylation and gene expression (Online Figures IIA–C and III; note: these data refer to genes for which we have both expression and CpG methylation data). To explore this relationship in more depth, we subdivided the groups of genes based on their relative expression levels (Online Figure IV) and investigated the DNA methylation profile across the promoter, rather than taking the sum behavior. Interestingly, despite all transcription start sites favoring demethylation, a variable pattern of regulation revealed itself with this analysis (Figures 4A,C,E,G): highly expressed genes tended to have less variability in the methylation at the transcription start site (the thick region of the violin plot is compacted towards the bottom in the vertical dimension). Genes with lower expression tended to have greater variability in this region (the thick region extends higher in the violin plot). The same analysis of the gene body also revealed a variable pattern of DNA methylation between` genes of difference expression levels, with highly expressed genes having the track of demethylation extend further into the gene body than lowly expressed genes (Figures 4B,D,F,H; Online Figure V shows the average behavior around transcription start sites and in gene bodies for all genes measured on the array).
Figure 4. Relationship of promoter and gene body DNA methylation to gene expression level.
(A,C,E,G) Each promoter is divided into 40 bins with each bin 100bp length. (B,D,F,H) Each gene body is divided into 100 bins. Violin plot at each bin represents the distribution of CpG methylation levels. White circle represents median methylation level at that bin. Methylation profiles for subsets of genes as grouped in Online Figure IV. Upper panel, genes with higher expression level. Lower panel, genes with lower expression level. Y-axis, DNA methylation levels from 0 (no methylation) to 100% (fully methylated).
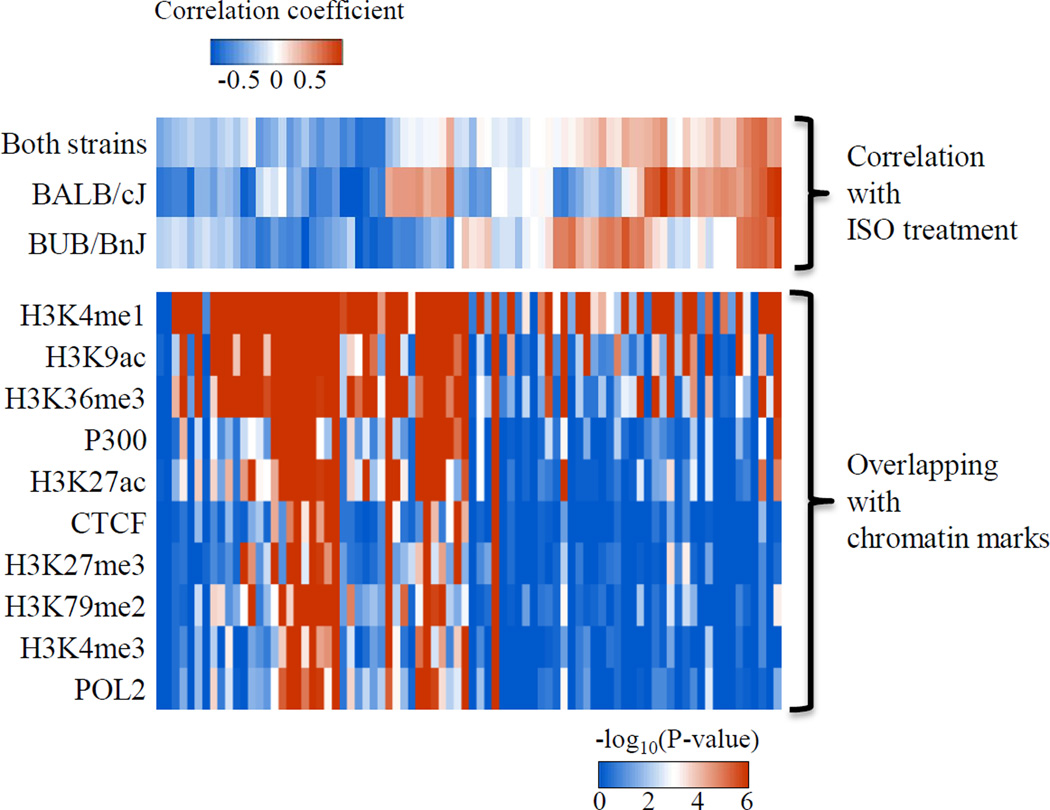
Inspection of the raw DNA methylation data revealed that some CpGs were highly correlated with each other in terms of their changes in methylation after ISO. To quantify this phenomenon across the measured methylome, we calculated the correlation among the top 15,000 DNA fragments with variable methylation and built a weighted network based on the correlation matrix (Online Figure VI). In this analysis, DNA fragments are grouped in a module when they exhibit similar behaviors and one “EigenCpG” is assigned to each module to represent the group behavior. To show the higher-order topology, we correlated the EigenCpG network with the physiological traits after ISO treatment. Interestingly, we found that most modules have some level of correlation with ISO treatment: some modules show a uniform response to ISO treatment in both BALB/cJ and BUB/BnJ strains, while others show a different response between two strains (Figure 5, top panel and Online Figure VI). We also found a few modules that, despite no significant responses to ISO treatment, show distinct methylation levels between two strains at the basal level indicating methylation polymorphisms between the two strains.
Figure 5. Association of co-methylation modules with distinct protein-based chromatin signatures.
Top panel, heatmap showing the correlation between EigenCpG methylation and ISO treatment for both strains, BALB/cJ or BUB/BnJ strain. Each column represents one module of CpGs. Bottom panel, heatmap of −log10 of P values showing the significance of overlapping between certain CpG modules and cardiac epigenetic markers.
Network analysis allowed analysis of interactions between DNA methylation signatures and other chromatin marks. Histone modifications act in a combinatorial manner to specify chromatin states, including transcriptional outcomes. To determine the relationship between the above DNA methylation modules and chromatin states, we investigated the presence of known cardiac protein-level features revealed by chromatin immunoprecipitation and DNA sequencing and deposited in ENCODE. To rule out the possibility that some chromatin marks favor CpG rich regions independent of methylation status, we chose the set of 15K DNA fragments used for network construction (rather than the whole genome) as the background to calculate the P value for the overlap amongst these subsets of loci. Surprisingly, we observed (Figure 5) modularity within the heatmap according to both cardiac epigenome track direction (row-wise) and the DNA methylation module direction (column-wise). Moreover, we found that most of DNA methylation modules that exhibit a decrease of methylation after ISO treatment overlap with H3K4me1 tracks, suggesting that these epigenetic marks may coordinately regulate cardiac gene expression.
To move beyond the correlative nature of these chromatin associations, we next sought to further examine the mechanisms through which DNA methylation may alter cardiac signaling networks. We hypothesized that despite the fact that the pattern of DNA methylation in and around transcription start sites was similar across genes, strain-specific epigenetic regulation may operate by differential targeting of transcription factor motifs. Interestingly, although the group of transcription factors regulated by methylation was similar, the two strains showed different preference for the transcription factors whose target genes were most enriched for altered methylation (Online Table III; significance values compare enrichment of transcription factor binding motifs in the differentially methylated subset versus enrichment across the entire genome; only those factors with significant p values, expressed as –log, are shown): for instance, BUB/bnJ had preferential targeting of E2F-4 signaling, whereas BALB/cJ showed greater regulation of Egr-1 targets. As a transcriptional repressor,23 E2F-4 would be expected to normally inhibit cell cycle/proliferative genes, whereas the Egr-1 family has been recently implicated in cardiac pathologies including hypertrophy,24 although the mechanisms by which these factors choose their genomic targets is incompletely understood. Previous studies had implicated histone regulation25 in altering transcription factor targeting, but this is the first evidence, to our knowledge, that either of these transcription factors are regulated by DNA modification. Future experiments will be required to determine whether, and by what mechanisms, DNA methylation alters transcription factor binding.
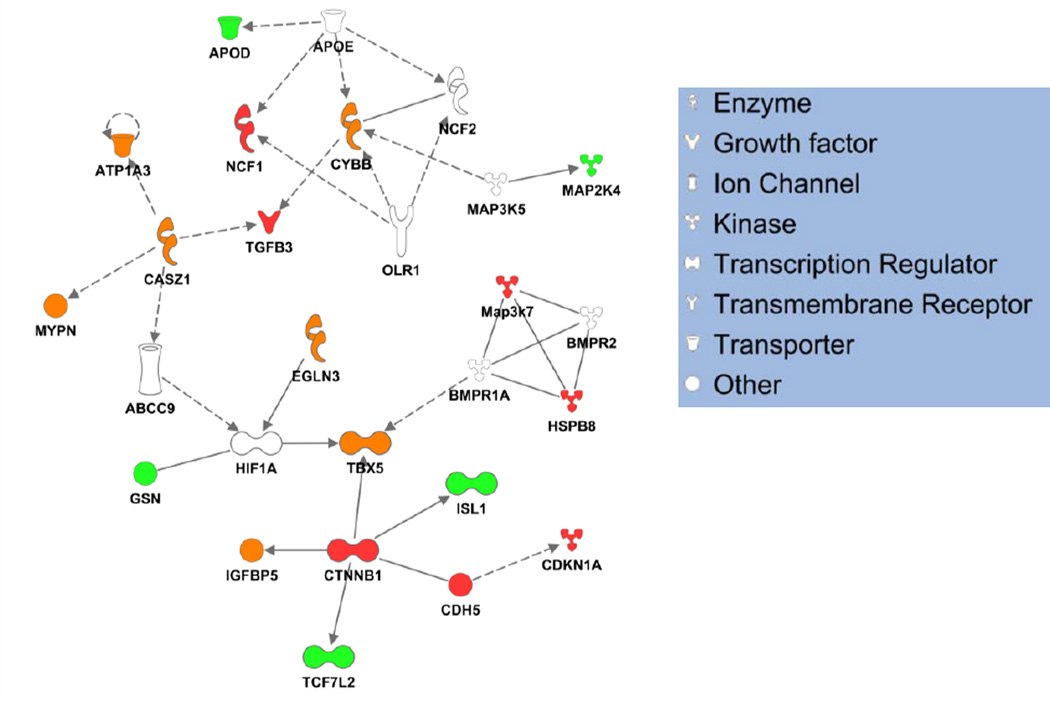
Next, we examined whether differentially methylated genes constituted a sub-network that may functionally interact in vivo. Ingenuity Pathway Analysis was used to interrogate relationships amongst genes from both strains with altered promoter methylation following ISO treatment, revealing a cardiac interactome that is highly connected based on known experimental literature (Figure 6, cardiac interactome data used; note that when the total interactome is used without reference to cell type, the sampled sub-network loses its structure and becomes dominated by a massive hub, Online Figure VIII). Interestingly, this sub-network is populated with nodes regulated in both strains, as well as those regulated in only one strain, implying that the effects of the methylation machinery at the level of the genome are coordinated with the ultimate protein interaction networks, yet this coordination, while having the potential to alter the flow of information through the network, does not change its structure.
Figure 6. Functional annotation reveals mechanisms linking differential methylation after ISO to cardiac phenotype.
Cardiac-specific pathways were examined for enrichment in differentially methylated genes using Ingenuity Pathway Analytics, revealing cardiac development to be physiological process most targeted by differential methylation after ISO. Interestingly, several known cardiac genes underwent altered regulation, allowing relationships amongst the signaling network to be mapped. Red nodes are differentially methylated only in BALB/cJ; green nodes only in BUB/BnJ; orange in both and white in neither. Solid lines indicated validated direct relationships; dashed lines, validated indirect interactions.
Lastly, we asked the question whether regions of genetic variation, shown in our recent study to associate with cardiac traits across the HMDP, were also host to dynamic regulation by DNA methylation. As shown in Online Table IV, GWAS hits (that is, genes where variation correlated with a heart failure phenotype by genome wide association study22) exhibited alterations in DNA methylation, with an apparent preference for demethylation in these regions that was significant for BALC/cJ (p value based on binomial distribution of 0.016) but not for BUB/bnJ (p value=0.618), likely due to a more pronounced overall hypomethylation in the latter strain. These findings further support the intriguing possibility that epigenetic variation in the form of DNA methylation may also contribute to the altered traits observed across the HMDP.
DISCUSSION
Genetic differences between inbred mouse strains have been shown to contribute to various cardiac26 and other27 phenotypes, although the role of common genetic variation in cardiac hypertrophy and failure is unknown. Furthermore, the mechanisms through which common variation predisposes for or against heart disease have not been explored in the context of chromatin. Our study provides the first cardiac epigenomic analysis of DNA methylation between individuals with fully characterized (and stable) genetic differences: the data demonstrate, with single base resolution, variations in basal DNA methylation between mouse strains before stress that correlate the phenotypic response to ISO. These investigations significantly advance our understanding of how the cardiac epigenome may integrate genetic and environmental signals. These studies also have fundamental implications for disease prediction.
How might the differences in methylation alter subcellular processes to produce a differential phenotypic response to ISO? Our studies do not support dynamic alteration of promoter methylation as a pervasive mechanism of gene regulation in hypertrophy (in cis), although effects of DNA methylation in trans are certainly possible within and between chromosomes. CpGs with distinct methylation status may contribute to altered functionality of enhancers, thereby influencing transcription indirectly. In general, we posit that the altered DNA methylation landscape leads to a global shift in chromatin accessibility, such that the plasticity of the genome differs in a heritable way that manifests through the epigenome and that has powerful effects on phenotype (in this study: response to isoproterenol).
We have no evidence for altered expression of DNA methylating/demethylating enzymes between these strains, but the possibility exists that SNPs in genes for proteins that participate in DNA methylation and its reading (e.g. transcription factors that use methylated DNA as a cue for binding) may result in altered transcript and protein expression, thereby altering the DNA methylome between strains. Another potential mechanism involves altered chromatin accessibility at specific loci associated with response to stress. In this model, the susceptible and resistant strains would respond differently to ISO-induced gene expression because the signaling pathways activated by the drug would impinge on a genome with different propensity to pathologic gene expression in one mouse versus another. This conjecture clearly requires further experimental evidence, including (1) evaluation of whether the DNA methylome differences in the hearts of these strains are the same differences observed in other organs; (2) whether the correlation between DNA methylation and heart failure phenotypes is robust across multiple genetic backgrounds; and (3) whether DNA methylation differences lead to altered protein binding.
Despite recent studies using patient samples15–18 and pigs,28, 29 the molecular mechanisms for how altered DNA methylation contributes to the progression of cardiomyopathy remains unclear. Such clinical studies are important to demonstrate the involvement of DNA methylation in cardiovascular disease, yet are limited due to the genetic, environmental and disease heterogeneity of human samples. Our study utilized a strict case-control experimental design, including consistent housing conditions and diet. Furthermore, the genome-wide RRBS method we used here samples randomly from the entire mouse genome, providing more information than a microarray-based method that measures only promoter regions.
Previous studies of mouse genetics focused on how to explain strain-specificity through genetic variation27 and such studies are similar to Genome Wide Association Studies (GWAS) in humans. It is also very likely that DNA methylation acts as an amplifier of the effects of primary DNA sequence differences.30 DNA methylation polymorphism is highly dependent on genetic polymorphism31. One piece of evidence for this is that the F1 hybrids of two different strains maintain the strain-specific methylation pattern from their parental allele.32 Functional annotation of regions associated with altered DNA methylation after ISO identified different pathways in the susceptible and resistant strains. For example, when studying the DNA regions with up-regulated methylation after ISO, we found genes associated with heart weight and size enriched in the BALB/cJ mice, whereas muscle contractility-associated genes were enriched in the pool of genes with increased methylation in the BUB/BnJ mice. While the present study was in review, a similar genome wide association study using the same experimental model of ISO-induced cardiac pathology in mice was published, identifying several candidate loci controlling cardiac mass and fibrosis22. Whether variation in DNA methylation similarly associates with clinical traits of heart disease in a GWAS approach will require further investigation, as will the question of how genetic and epigenetic variation interact in a genome wide manner.
For a single gene, whether and how DNA methylation regulates its expression depends on the combination of many factors including histone modifications, binding of transcription factors and higher order chromatin structure. A study of DNA methylation and gene expression levels in human blood cells reported that while some modules are highly correlated (in a positive or negative manner) with gene expression, most modules had no correlation between gene expression and DNA methylation.33 A recent study of human placenta indicates that the relationship between DNA methylation and gene expression is highly dependent on the type of gene (i.e. its function in the cell type of interest) and the region in the gene where the methylation occurs.34 One interpretation of the data in Figures 4A,C,E,G is that demethylation of promoters alone does not silence genes (hence the variability in the silenced genes) and other protein-dependent mechanisms are necessary, whereas demethylation is invariably required for high gene expression. This may also explain the greater variability in demethylated gene bodies seen in highly contrasted with lowly expression genes in Figures 4B,D,F,H.
The traditional method of identifying differentially methylated fragments using a statistical hypothesis test is based on the assumption that all methylation measurements are independent and identically distributed (i.i.d.). However, this strong assumption does not account for the reality of biological networks, in which co-dependency of interactions and combinatorial specification are common occurrences. To account for this biological phenomenon, we used DNA fragments to build a co-methylation network, thus circumventing the difficulty that some DNA fragments are located in enhancers or other functional element regions far away from known genes. Next, we built an epigenetic interaction network in the heart, showing that DNA fragments can be clustered into modules, that is, groups of CpG-containing fragments with similar regulation features across multiple samples and experimental conditions (Note: The modularity in the DNA methylation (column-wise, Figure 5) cannot be explained by the membership of DNA fragments in multiple modules, because each DNA fragment was assigned to only one DNA methylation module, disallowing overlap). An implication of this clustering is that the principles underlying regulation of these loci are shared: fragments that exhibit similar dynamics in terms of DNA methylation are predicted to reflect similar chromatin remodeling and transcriptional regulatory mechanisms at these loci. Interestingly, members within the same methylation module can be located far away from each other in the linear position of one chromosome or even located on different chromosomes. We interpret this to indicate hierarchical regulation by epigenetic machinery, rather than progressive actions along a chromosome. It is interesting to speculate that regions in the same DNA methylation module are positioned proximally in three-dimensional space although additional experiments would be required to test this conjecture.
Our study also revealed that chromosomal regions with similar DNA methylation changes after ISO stimulation share a common pattern of epigenetic marks. In addition to the aforementioned modularity in the DNA methylation measurements, Figure 5 demonstrates clustering of the epigenome tracks (i.e. protein marks, row-wise in Figure 5), which may be the result of correlation between the different epigenetic marks and/or because of the correlation with DNA methylation. Since DNA modules are ordered based on their correlation with ISO treatment, the observation of epigenomic clustering indicates that if the DNA methylation patterns of two genomic regions respond to the environment in a similar way, these two regions may also have a similar histone modification pattern. An open question is the temporal relationship between DNA methylation and epigenomic protein marks at individual loci.
The observation that transcription factor binding motifs are differentially targeted by methylation in the two strains provides a potential mechanism that spans individual locus to entire genome: by altering methylation and thereby transcription factor binding, evolution can tailor the transcriptome and its response to stress on a global level. Although additional experiments will confirm or deny this hypothesis, as with protein-based chromatin signatures, a DNA methylation mechanism at the level of transcription factor binding could explain coordination across multiple loci, enabling rapid, coordinated transcriptional responses tuned by epigenetic programming. To validate our observations in the context of the known cardiac interactome, we additionally examined the sub-network defined by the differentially methylated genes following ISO. Interestingly, the pathologic stressor engaged methylation to regulate a highly connected group of molecules, with strain-specific differences emerging with regard to which nodes were altered. These findings suggest that the proteome-level networks in heart failure are regulated in part by epigenetic factors, such as DNA methylation, and that genetic variation influences gene expression and, by virtue of differentially targeting specific nodes, potentially the flow of information through protein networks. These observation can be further explored in a larger cohort of genetically variable individuals—like the HMDP or a human population—which would further test the relationship amongst the protein network as well as the role of DNA methylation and genetics to modulate this network in the setting of pathologic stimulation.
There are some limitations with the ISO model used in this study, namely that it recapitulates only part of the complex syndrome of heart failure in humans, which is the sequela of genetic risk compounded by environmental factors like ischemia, stress, toxicity, neural hormonal imbalance, and hypertension. We chose this model because we had data on how ISO differentially affects cardiac phenotype and gene expression in a large panel of inbred mice22 and because it is a unitary, well defined stressor, that is relatively operator independent (as opposed to infarction or pressure overload). The presence of the ISO pump throughout the experiment means that the animals are receiving constant beta-adrenergic stimulation. In the strains we examined, EF was different between them but depressed in neither after ISO, suggesting we are studying a form of cardiac pathology that has not progressed to failure according to clinical measurements. It is noteworthy that the EF and fetal gene expression measurements in these mice fall in different ends of the spectrum in terms of genetic susceptibility to disease (Online Figures VI and VII). The timing of analysis is also likely contributory to the DNA methylome features: in addition to the changes induced by ISO infusion, alterations in methylation may also be influenced by the compensatory actions of the heart (and indeed the entire organism) to the enhanced β-adrenergic state.
To understand how our findings from mice may have clinical relevance, we need to consider DNA methylation in the context of endogenous regulation. The difference in DNA methylation observed in this study may come from non-cardiac cells that infiltrate the heart during stress and certainly from non-cardiomyocytes, which make up 50% or more of the mammalian heart by some estimates.35, 36 Sorting this out is difficult and will require further studies, yet our work clearly shows there is a different epigenomic signature associated with heart failure susceptibility and resistance, the first time such an observation has been reported. Chromatin modifications are also known to be regulated by cellular metabolites. Previous studies have found that cardiac metabolism is aberrant in disease. This includes S-adenosylhomocysteine (SAH) / S-adenosylmethionine (SAM) ratio imbalance, as indicated by hyperhomocysteinemia in cardiovascular disease,37 and accumulation of α-ketoglutarate or 2-oxoglutarate, due to inactivation of ketoglutarate dehydrogenase caused by abnormal mitochondrial function and oxidative stress.38 Previous studies of heart failure metabolism mainly focused on how metabolites impair normal cellular functions, such as mitochondrial function or cell signaling pathways. In view of those studies showing the involvement of SAH/SAM39, 40 and α-ketoglutarate41 in DNA methylation, it is possible that metabolic dysfunction in heart failure may alter the epigenome directly. Interestingly, a recent DNA methylation study from the liver20 used a program called glmnet to predict liver phenotypes based on a subset of CpG methylation events. In principle, a similar approach could be taken based on the proof of concept in this study and a larger population of genetically variable subjects, to explore the role of DNA methylation to serve as a biomarker for heart failure susceptibility.
Supplementary Material
Novelty and Significance.
What Is Known?
Common heart failure has a heritable component, leading to differential disease progression in humans.
Epigenetic mechanisms, such as DNA methylation, control gene regulatory networks in the adult heart, including response to disease.
What New Information Does This Article Contribute?
Genome-wide analyses of DNA methylation provide single base resolution of this epigenomic mark in the adult heart.
Analyses of multiple strains of mice, with distinct phenotypic responses to the adrenergic agonist isoproterenol, show that DNA methylation presages cardiac pathogenesis.
Transcriptome analyses conducted in parallel reveal the relationship between DNA methylation and gene expression in the mouse heart, and the mechanisms by which this epigenetic mark controls transcriptional networks.
The relationship between epigenetic marks and gene expression during the onset of heart disease is poorly understood. Likewise, there is a need for molecular diagnostics for non-congenital forms of heart failure that would enable differential treatment. Therefore, we undertook a novel epigenomic study in which DNA methylation was measured by bisulfite sequencing in the hearts of mice subjected to isoproterenol stimulation. Our findings reveal differential patterns of DNA methylation in the promoters versus the bodies of genes and demonstrate that highly expressed genes have distinct DNA methylation features from genes expressed at low levels. We also found that DNA methylation marks discriminate a heart failure- susceptible mouse strain from a resistant one prior to the administration of isoproterenol, suggesting that DNA methylation may be a potential biomarker for disease stratification and might be useful in predicting the severity of disease pathogenesis in humans.
Acknowledgments
SOURCES OF FUNDING
This project was supported by the American Heart Association (IRG18870056 to TMV) and the National Institutes of Health (HL-105699 and HL-115238 to TMV). HC was the recipient of an AHA Predoctoral Fellowship (11PRE7290056).
Nonstandard Abbreviations and Acronyms
- RRBS
reduced representational bisulfite sequencing
- DMF
differentially methylated fragment
- WGCNA
weighted gene co-expression network analysis
- HMDP
hybrid mouse diversity panel
Footnotes
DISCLOSURES:
None.
REFERENCES
- 1.Lee DS, Pencina MJ, Benjamin EJ, Wang TJ, Levy D, O'Donnell CJ, Nam BH, Larson MG, D'Agostino RB, Vasan RS. Association of parental heart failure with risk of heart failure in offspring. N Engl J Med. 2006;355:138–147. doi: 10.1056/NEJMoa052948. [DOI] [PubMed] [Google Scholar]
- 2.Dorn GW., 2nd Genetics of common forms of heart failure. Current opinion in cardiology. 2011;26:204–208. doi: 10.1097/HCO.0b013e328345d336. [DOI] [PubMed] [Google Scholar]
- 3.Marian AJ, Belmont J. Strategic approaches to unraveling genetic causes of cardiovascular diseases. Circ Res. 2011;108:1252–1269. doi: 10.1161/CIRCRESAHA.110.236067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosa-Garrido M, Karbassi E, Monte E, Vondriska TM. Regulation of chromatin structure in the cardiovascular system. Circulation journal : official journal of the Japanese Circulation Society. 2013;77:1389–1398. doi: 10.1253/circj.cj-13-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu H, Zhang Y. Reversing DNA methylation: Mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones PA. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 8.Kohli RM, Zhang Y. Tet enzymes, tdg and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 10.Esteller M. Cpg island hypermethylation and tumor suppressor genes: A booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 11.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. Cpg island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashita K, Dai T, Dai Y, Yamamoto F, Perucho M. Genetics supersedes epigenetics in colon cancer phenotype. Cancer cell. 2003;4:121–131. doi: 10.1016/s1535-6108(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 13.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K. Identification of a cpg island methylator phenotype that defines a distinct subgroup of glioma. Cancer cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell RM, Tummino PJ. Cancer epigenetics drug discovery and development: The challenge of hitting the mark. J Clin Invest. 2014;124:64–69. doi: 10.1172/JCI71605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koczor CA, Lee EK, Torres RA, Boyd A, Vega JD, Uppal K, Yuan F, Fields EJ, Samarel AM, Lewis W. Detection of differentially methylated gene promoters in failing and nonfailing human left ventricle myocardium using computation analysis. Physiological genomics. 2013;45:597–605. doi: 10.1152/physiolgenomics.00013.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haas J, Frese KS, Park YJ, Keller A, Vogel B, Lindroth AM, Weichenhan D, Franke J, Fischer S, Bauer A, Marquart S, Sedaghat-Hamedani F, Kayvanpour E, Kohler D, Wolf NM, Hassel S, Nietsch R, Wieland T, Ehlermann P, Schultz JH, Dosch A, Mereles D, Hardt S, Backs J, Hoheisel JD, Plass C, Katus HA, Meder B. Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO molecular medicine. 2013;5:413–429. doi: 10.1002/emmm.201201553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Movassagh M, Choy MK, Goddard M, Bennett MR, Down TA, Foo RS. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PLoS One. 2010;5:e8564. doi: 10.1371/journal.pone.0008564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Movassagh M, Choy MK, Knowles DA, Cordeddu L, Haider S, Down T, Siggens L, Vujic A, Simeoni I, Penkett C, Goddard M, Lio P, Bennett MR, Foo RS. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124:2411–2422. doi: 10.1161/CIRCULATIONAHA.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner BM. Environmental sensing by chromatin: An epigenetic contribution to evolutionary change. FEBS letters. 2011 doi: 10.1016/j.febslet.2010.11.041. [DOI] [PubMed] [Google Scholar]
- 20.Orozco LD, Morselli M, Rubbi L, Guo W, Go J, Shi H, Lopez D, Furlotte NA, Bennett BJ, Farber CR, Ghazalpour A, Zhang MQ, Bahous R, Rozen R, Lusis AJ, Pellegrini M. Epigenome-wide association of liver methylation patterns and complex metabolic traits in mice. Cell metabolism. 2015;21:905–917. doi: 10.1016/j.cmet.2015.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang L, Zhang J, Wang JJ, Wang L, Zhang L, Li G, Yang X, Ma X, Sun X, Cai J, Zhang J, Huang X, Yu M, Wang X, Liu F, Wu CI, He C, Zhang B, Ci W, Liu J. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 2013;153:773–784. doi: 10.1016/j.cell.2013.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rau CD, Wang J, Avetisyan R, Romay M, Ren S, Wang Y, Lusis AJ. Mapping genetic contributions to cardiac pathology induced by beta-adrenergic stimulation in mice. Circulation Cardiovascular Genetics. 2015;8:40–49. doi: 10.1161/CIRCGENETICS.113.000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhan L, Huang C, Meng XM, Song Y, Wu XQ, Miu CG, Zhan XS, Li J. Promising roles of mammalian e2fs in hepatocellular carcinoma. Cellular signalling. 2014;26:1075–1081. doi: 10.1016/j.cellsig.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 24.Ramadas N, Rajaraman B, Kuppuswamy AA, Vedantham S. Early growth response-1 (egr-1) - a key player in myocardial cell injury. Cardiovascular & hematological agents in medicinal chemistry. 2014;12:66–71. doi: 10.2174/1871525713666150123152131. [DOI] [PubMed] [Google Scholar]
- 25.Carr SM, Poppy Roworth A, Chan C, La Thangue NB. Post-translational control of transcription factors: Methylation ranks highly. The FEBS journal. 2015 doi: 10.1111/febs.13524. [DOI] [PubMed] [Google Scholar]
- 26.Guo Y, Flaherty MP, Wu WJ, Tan W, Zhu X, Li Q, Bolli R. Genetic background, gender, age, body temperature, and arterial blood ph have a major impact on myocardial infarct size in the mouse and need to be carefully measured and/or taken into account: Results of a comprehensive analysis of determinants of infarct size in 1,074 mice. Basic research in cardiology. 2012;107:288. doi: 10.1007/s00395-012-0288-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghazalpour A, Rau CD, Farber CR, Bennett BJ, Orozco LD, van Nas A, Pan C, Allayee H, Beaven SW, Civelek M, Davis RC, Drake TA, Friedman RA, Furlotte N, Hui ST, Jentsch JD, Kostem E, Kang HM, Kang EY, Joo JW, Korshunov VA, Laughlin RE, Martin LJ, Ohmen JD, Parks BW, Pellegrini M, Reue K, Smith DJ, Tetradis S, Wang J, Wang Y, Weiss JN, Kirchgessner T, Gargalovic PS, Eskin E, Lusis AJ, LeBoeuf RC. Hybrid mouse diversity panel: A panel of inbred mouse strains suitable for analysis of complex genetic traits. Mammalian genome : official journal of the International Mammalian Genome Society. 2012;23:680–692. doi: 10.1007/s00335-012-9411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi M, Lee J, Le MT, Nguyen DT, Park S, Soundrarajan N, Schachtschneider KM, Kim J, Park JK, Kim JH, Park C. Genome-wide analysis of DNA methylation in pigs using reduced representation bisulfite sequencing. DNA research : an international journal for rapid publication of reports on genes and genomes. 2015;22:343–355. doi: 10.1093/dnares/dsv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schachtschneider KM, Madsen O, Park C, Rund LA, Groenen MA, Schook LB. Adult porcine genome-wide DNA methylation patterns support pigs as a biomedical model. BMC genomics. 2015;16:743. doi: 10.1186/s12864-015-1938-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK. DNA methylation patterns associate with genetic and gene expression variation in hapmap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, Dempster EL, Ren B. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schilling E, El Chartouni C, Rehli M. Allele-specific DNA methylation in mouse strains is mainly determined by cis-acting sequences. Genome Res. 2009;19:2028–2035. doi: 10.1101/gr.095562.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Eijk KR, de Jong S, Boks MP, Langeveld T, Colas F, Veldink JH, de Kovel CG, Janson E, Strengman E, Langfelder P, Kahn RS, van den Berg LH, Horvath S, Ophoff RA. Genetic analysis of DNA methylation and gene expression levels in whole blood of healthy human subjects. BMC genomics. 2012;13:636. doi: 10.1186/1471-2164-13-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroeder DI, Blair JD, Lott P, Yu HO, Hong D, Crary F, Ashwood P, Walker C, Korf I, Robinson WP, LaSalle JM. The human placenta methylome. Proc Natl Acad Sci U S A. 2013;110:6037–6042. doi: 10.1073/pnas.1215145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jugdutt BI. Ventricular remodeling after infarction and the extracellular collagen matrix: When is enough enough? Circulation. 2003;108:1395–1403. doi: 10.1161/01.CIR.0000085658.98621.49. [DOI] [PubMed] [Google Scholar]
- 36.Banerjee I, Fuseler JW, Price RL, Borg TK, Baudino TA. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am J Physiol Heart Circ Physiol. 2007;293:H1883–H1891. doi: 10.1152/ajpheart.00514.2007. [DOI] [PubMed] [Google Scholar]
- 37.Chen NC, Yang F, Capecci LM, Gu Z, Schafer AI, Durante W, Yang XF, Wang H. Regulation of homocysteine metabolism and methylation in human and mouse tissues. FASEB J. 2010;24:2804–2817. doi: 10.1096/fj.09-143651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunn W, Broadhurst D, Deepak S, Buch M, McDowell G, Spasic I, Ellis D, Brooks N, Kell D, Neyses L. Serum metabolomics reveals many novel metabolic markers of heart failure, including pseudouridine and 2-oxoglutarate. Metabolomics. 2007;3:413–426. [Google Scholar]
- 39.Ulrey CL, Liu L, Andrews LG, Tollefsbol TO. The impact of metabolism on DNA methylation. Human molecular genetics. 2005;14(Spec No 1):R139–R147. doi: 10.1093/hmg/ddi100. [DOI] [PubMed] [Google Scholar]
- 40.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annual review of biochemistry. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 41.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by mll partner tet1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.