Abstract
A subset of patients with chronic lymphocytic leukemia (CLL) and nearly all patients with classic hairy cell leukemia (HCL) harbor somatic BRAF activating mutations. However, the pathological role of activated BRAF in B-cell leukemia development and progression remains unclear. In addition, although HCL patients respond well to the BRAFV600E inhibitor vemurafenib, relapses are being observed, suggesting the development of drug resistance in patients with this mutation. To investigate the biological role of BRAFV600E in B-cell leukemia, we generated a CLL-like B-cell line, OSUCLL, with doxycycline-inducible BRAFV600E expression. Microarray and real-time PCR analysis showed that ABCB1 mRNA is upregulated in these cells, and P-glycoprotein (P-gp) expression as well as function were confirmed by immunoblot and rhodamine exclusion assays. Additionally, pharmacological inhibition of BRAFV600E and MEK alleviated the BRAFV600E-induced ABCB1/P-gp expression. ABCB1 reporter assays and gel shift assays demonstrated that AP-1 activity is crucial in this mechanism. This study therefore uncovers a pathological role for BRAFV600E in B-cell leukemia, and provides further evidence that combination strategies with inhibitors of BRAFV600E and MEK can be used to delay disease progression and occurrence of resistance.
Keywords: BRAF, vemurafenib, B-cell, ABCB1, P-glycoprotein, leukemia, lymphoma
Introduction
Multiple solid tumors, particularly melanoma, have been found to carry somatic BRAF mutations that result in constitutive BRAF protein activation and cell transformation. Since the initial description of BRAF mutations in 20021, over 40 distinct point mutations affecting the BRAF kinase domain have been identified2. Among these, a mutation altering valine (V) to glutamic acid (E) at amino acid 600 in the activation segment of the kinase domain shows the highest incidence. While earlier studies found little or no occurrence of BRAF mutations in hematologic diseases, rarer disease types were not studied. More recently, Tiacci et al. found that nearly all classic hairy cell leukemia (HCL) cases bear the BRAFV600E mutation3, and Dietrich and others now report that HCL can be successfully treated with the BRAFV600E-selective inhibitor vemurafenib4. A subset of chronic lymphocytic leukemia (CLL) patients also show mutated BRAF5–7, and BRAF mutations were identified as one of the acquired initiating mutations in early hematopoietic cells of CLL, leading to deregulation of B-cell receptor (BCR) signaling8. Furthermore, a BRAF pseudogene transcript is aberrantly expressed in human diffuse large B-cell lymphoma, positively correlates with BRAF expression, and results in MAPK activation. Expression of this pseudogene in a murine model results in aggressive B-cell lymphoma9. Together, these findings clearly implicate BRAFV600E in the development of a subset of B-cell malignancies.
Although not all BRAF mutations identified to date are V600E, most are activating mutations and result in MAPK stimulation. BRAF is upstream of MEK and ERK, which are involved in regulating cell proliferation, survival, differentiation and senescence following external signals10. Because BRAFV600E is active in the absence of external stimuli, it constitutively activates the MAPK pathway to promote cell transformation through enhanced transcription (e.g. via c-Fos, Elk-1) and translation (e.g. via RSK, eIF4E) of factors that subsequently drive survival and proliferation (e.g. cyclin D1, c-myc). BRAFV600E inhibitors including vemurafenib and dabrafenib show clinical responses in many cases of BRAFV600E mutated cancers. However, resistance to these agents commonly develops11,12. Emerging data also indicate that BRAFV600E HCL patients relapse following vemurafenib treatment that was initially effective13. Interestingly, it was recently reported that a patient with BRAFV600E-driven melanoma who responded to vemurafenib developed CLL-like disease, possibly due to paradoxical BRAF inhibitor-associated ERK activation in B-cells via the BCR/SYK/RAS/RAF axis14. ERK is also a key downstream effector of the BCR pathway, and inhibition of this pathway by the BTK inhibitor ibrutinib leads to loss of ERK phosphorylation both in vitro15 and in samples from ibrutinib-treated patients16. Thus in B-cells, ERK is a point of convergence of the MAPK pathway and the critical BCR pathway, further supporting the relevance of MAPK signaling in B-cell malignancies.
Despite the prevalence of BRAFV600E in HCL and the importance of mutant BRAF in leukemia development and potentially its treatment, downstream targets of this pathway in B-cells remain unclear. Here, we sought to identify transcriptional events resulting from constitutive BRAF activation in transformed B-cells. We demonstrate that BRAFV600E induces the expression of the multi-drug resistance (MDR) gene ABCB1 and its product, P-glycoprotein (P-gp). Further, we determined that MAPK pathway-mediated induction of AP-1 could be a potential mechanism for this effect. This finding may have clinical implications for the long-term use of MDR substrate agents in patients with BRAF-mutated cancers.
Materials and Methods
Cells, cell culture, and reagents
The OSUCLL cell line, previously described, has characteristics similar to the donor’s CLL cells17. OSUCLL cells were retrovirally infected using the Tet-On 3G inducible expression system (OSUCLL-Tet), then with pRetroX-tight-pur vectors (Clontech, Mountain View, CA) expressing wild-type BRAF (OSUCLL-BRAF) or mutant BRAF (OSUCLL-BRAFV600E). The BRAFV600E cDNA construct was purchased from Addgene (Cambridge, MA). For constitutive expression, the pBABE-puro retroviral vector was used (Cell Biolabs, San Diego, CA). Cells were cultured at 37°C, 5% CO2 in RPMI1640 with 10% fetal bovine serum (FBS), 100 U/mL penicillin, 100 µg/mL streptomycin, and 2 mM L-glutamine (Sigma, St. Louis, MO). Vemurafenib (PLX-4032) was purchased from Selleck (Houston, TX), and CI-1040 (PD184352) was synthesized as described18. Doxycycline (dox) was purchased from Clontech, and verapamil, vincristine and rhodamine 123 from Sigma.
Viability and proliferation
MTS assays were performed per manufacturer instructions (CellTiter 96, Promega, Madison WI). Cells were incubated 48 hr in 96-well plates with or without dox (1 µg/ml) and other agents, and MTS reagent was added for an additional 2 hr before analysis. To assess growth rate, cells were cultured in 96-well plates for 20 hr with or without dox. BrdU was added and measured 4 hr later per manufacturer instructions (Millipore, Billerica, MA).
Microarray
RNA from OSUCLL-Tet and OSUCLL-BRAFV600E cells incubated 48 hr with or without dox was analyzed using U133 plus 2.0 GeneChips (Affymetrix, Santa Clara, CA) in collaboration with the OSU Comprehensive Cancer Center Genomics Shared Resource.
Flow cytometry
Cells were incubated 48 hr with or without dox and labeled using anti-CD69 or isotype antibodies (BD Biosciences, San Jose, CA). Cells were analyzed on a FC500 flow cytometer and results processed using Kaluza software (Beckman Coulter, Brea, CA). To measure P-gp activity, cells were cultured 48 hr with dox then incubated 1 hr with 2.6 µM rhodamine 123. After two washes in media, cells were incubated 90 min with or without verapamil (10 µg/ml). Rhodamine-positive cells were detected by flow cytometry.
Real-time reverse transcription–polymerase chain reaction (RT-PCR)
Total RNA was extracted by TRIzol (Invitrogen, Carlsbad, CA) and analyzed on an ABI ViiA 7 system (Applied Biosystems, Foster City, CA), using GAPDH as an endogenous control. TaqMan Universal Master Mix, primers, and labeled probes were used per manufacturer instructions (Applied Biosystems). Mean threshold cycle (Ct) values were calculated by ABI ViiA 7 software to determine fold differences.
Immunoblot
SDS-PAGE and immunoblotting were performed using standard procedures. Antibodies against phospho-MEK, MEK, phospho-ERK, ERK, phospho-c-Fos, c-Fos, FosB/B2, phosphor-Fra-1, Fra-1, phospho-c-Jun, c-Jun, JunB and JunD were obtained from Cell Signaling Technology (Danvers, MA), and BRAF, P-gp, and GAPDH from Santa Cruz (Santa Cruz, CA). Human specific BRAFV600E antibody was obtained from Spring Bioscience (Pleasanton, CA). Each immunoblot analysis was repeated a minimum of three times.
ABCB1 promoter activity
NEK-293T cells were transiently transfected with the ABCB1 promoter luciferase reporter construct pTL-MDR1 (Affymetrix) and pBABE-puro plasmid using FuGENE6 reagent (Promega), then incubated 16 hr with vehicle (DMSO), vemurafenib (2 µM), CI-1040 (1 µM), or both. Luciferase activity (relative luciferase units, RLU) was normalized to protein amount as determined by BCA assay (ThermoFisher, Waltham, MA).
Electrophoretic mobility shift assay (EMSA)
AP-1 double-stranded probes [5’-CGCTTGATGAGTCAGCCGGAA-3’ for wild-type and 5’-CGCTTGATAGTAGTGCCGGAA-3’ for mutant19,20] were end-labeled with 32P dCTP using Klenow DNA polymerase (Life Technologies, Grand Island, NY). Nuclear lysates (10 µg) were incubated 1 hr with antibodies to c-Jun, c-Fos, JunB, JunD, and Fra-1 (Santa Cruz Biotechnology, Santa Cruz, CA) or MEK, phospho-c-Jun, and phospho-c-Fos (Cell Signaling). Labeled probes were added, incubations were continued at room temperature for another 30 min, and complexes were separated on native polyacrylamide gels. Gels were dried and signals detected using a phosphor screen (GE Healthcare, Pittsburgh, PA).
Statistical analysis
Differences in BrdU incorporation, ABCB1 mRNA expression, percentage of rhodamine-positive cells, percentage of mitochondrial activity, and relative luciferase activity versus controls were assessed using mixed effects models. Log transformations were applied as appropriate before modeling to stabilize variances. All analyses were performed using SAS/STAT software, version 9.3 (SAS Institute, Inc., Cary, NC).
Results
Generation of B-cells with inducible BRAFV600E expression
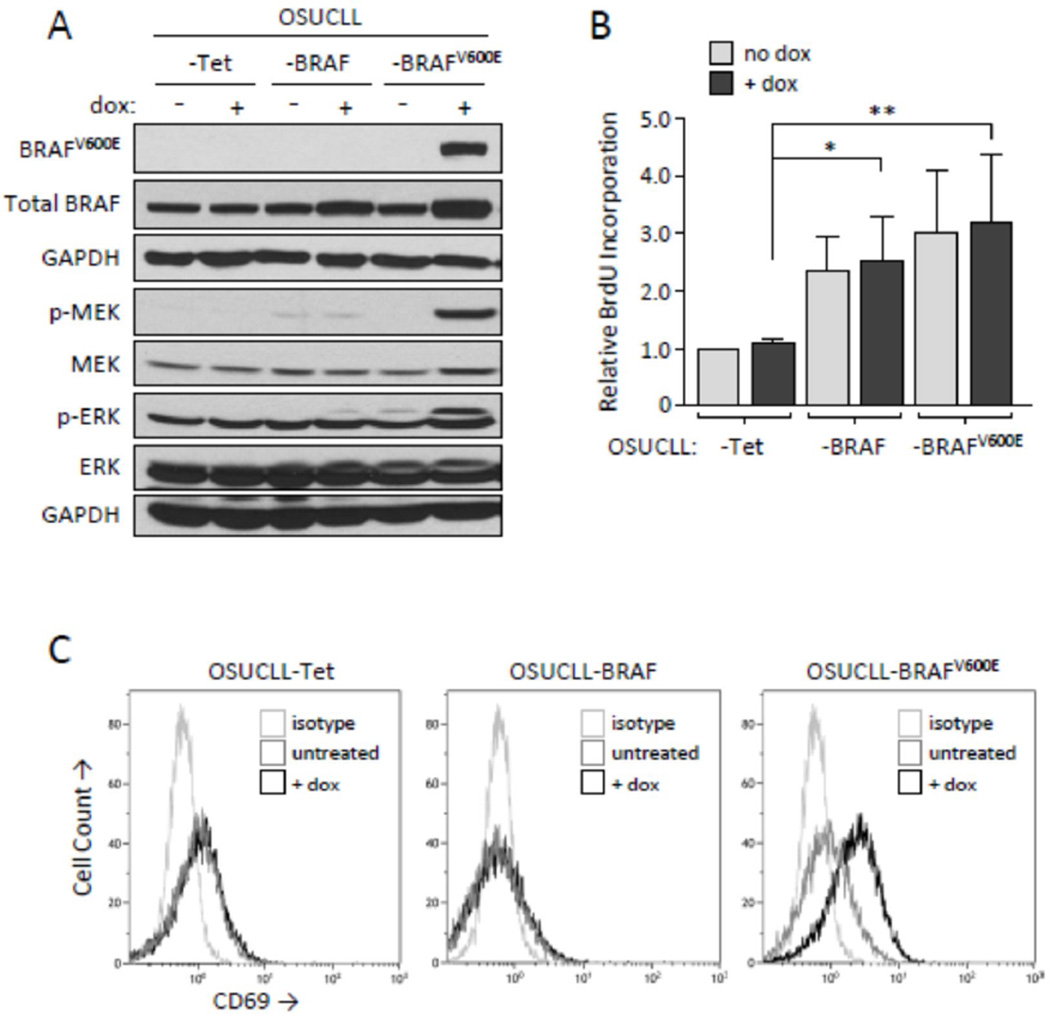
BRAF mutations are found in a subset of CLL, and are early acquired events that may contribute to disease development or progression8 and potentially drug resistance. To investigate effects of this mutant protein in malignant B-cells, we stably transfected OSUCLL cells with a dox-inducible vector system to generate OSUCLL-Tet. This cell line was infected with retroviral vectors containing wild-type or mutant BRAF to produce OSUCLL-BRAF and OSUCLL-BRAFV600E. OSUCLL-BRAFV600E cells strongly upregulated mutant BRAF following 24 hr dox treatment, as detected by immunoblot using a BRAFV600E-specific antibody (Figure 1A). A moderate but consistent increase in wild-type BRAF was observed in dox-treated OSUCLL-BRAF versus OSUCLL-Tet cells. Under these conditions, dox-mediated BRAFV600E induction was accompanied by increased MEK and ERK phosphorylation, indicating functional transfected protein. Minimal effects on MEK and ERK phosphorylation were detected in dox-treated OSUCLL-BRAF cells. However, a slight increase in ERK phosphorylation was detected in OSUCLL-BRAFV600E cells in the absence of dox, and a low level of BRAFV600E could be detected in these cells with prolonged exposures (Figure 2B), indicating transcription from this promoter is not completely absent in the absence of dox. In comparison with OSUCLL-Tet, both OSUCLL-BRAF and OSUCLL-BRAFV600E cells showed a significant increase in growth rate as determined by BrdU proliferation assays (Figure 1B). OSUCLL-BRAFV600E cells appeared to grow slightly faster than OSUCLL-BRAF cells, but this difference was not significant. Interestingly, the addition of dox did not further increase proliferation, suggesting that whatever effects transfected BRAF or BRAFV600E have on growth rate occur at very low expression levels. These results demonstrate that transfected BRAFV600E produces the expected downstream biochemical changes in OSUCLL cells, but its impact on proliferation relative to normal BRAF is limited.
Figure 1. Effects of transfected BRAFV600E on the MAPK pathway and cell growth.
A. Immunoblot analysis of wild-type and mutant BRAF expression and MAPK signaling in transfected OSUCLL cells following 24 hr incubation without or with dox (1 µg/ml). B. Proliferation in OSUCLL-Tet, OSUCLL-BRAF, and OSUCLL-BRAFV600E cells without or with dox treatment (1 µg/ml) for 24 hr. BrdU was added for the last 4 hr of the incubations. Data are shown relative to the OSUCLL-Tet control cell line without dox treatment (N=4, *p<0.05; **p<0.005). C. OSUCLL-Tet, OSUCLL-BRAF, and OSUCLL-BRAFV600E cells were incubated 48 hr with or without dox, then stained with anti-CD69 or isotype antibodies and assessed by flow cytometry. Data are representative of three independent experiments.
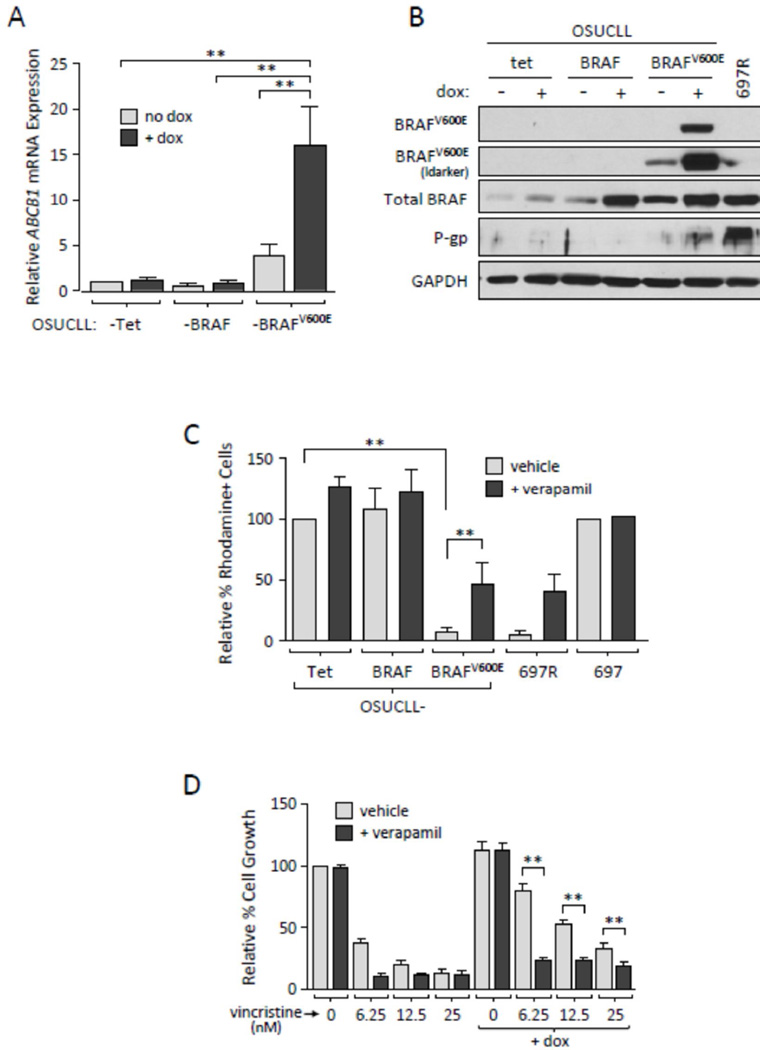
Figure 2. BRAFV600E induces the expression of ABCB1 and functional P-gp in OSUCLL cells.
A. ABCB1 mRNA levels were assessed in OSUCLL cells using real-time RT-PCR (N=3, **p<0.001). B. Immunoblot of normal BRAF, mutant BRAF, and P-gp in OSUCLL cells incubated 48 hr without or with dox. C. OSUCLL cells cultured 48 hr with dox were incubated 1 hr with the fluorescent P-gp substrate rhodamine 123, then transferred to rhodamine-free media with or without 10 µM verapamil. Retained rhodamine was assessed by flow cytometry after 90 min. Parental 697 cells and drug-resisant 697-R cells were included as controls. Results were averaged from three identical experiments (**p<0.005) and are shown as percent rhodamine-positive cells relative to vehicle-treated OSUCLL-Tet (far left). D. OSUCLL-BRAFV600E cells were incubated 48 hr with or without dox, vincristine, and/or verapamil, and mitochondrial activity was evaluated by MTS assay. Results shown are averaged from three identical experiments. At each concentration of vincristine, comparisons were made between the dox and no dox conditions, as well as between the verapamil and control conditions in the presence of dox (**p<0.001).
Based on the increased signaling in BRAFV600E transfected cells, we examined transcriptional changes resulting from the presence versus absence of BRAFV600E expression using microarray. The top 30 genes increased or decreased in dox-treated OSUCLL-BRAFV600E cells relative to dox-treated OSUCLL-Tet cells are shown in Supplementary Table S1. Among these, the lymphocyte activation marker CD69 and the multidrug resistance gene ABCB1 were observed to be increased in OSUCLL-BRAFV600E cells versus the control. In validation of these results, CD69 expression was found to be markedly increased in dox-treated OSUCLL-BRAFV600E compared to OSUCLL-Tet and OSUCLL-BRAF cells (Figure 1C).
BRAFV600E induces ABCB1 mRNA and P-gp protein expression in OSUCLL cells
To confirm BRAFV600E-mediated ABCB1/P-gp induction, OSUCLL-Tet, -BRAF, and -BRAFV600E cells were incubated with or without dox and examined by real-time RT-PCR and immunoblot. As early as 24 hr, but consistently by 48 hr, ABCB1 mRNA and P-gp protein were clearly up-regulated in dox-treated OSUCLL-BRAFV600E cells (Figures 2A, 2B, 3A). Drug-resistant 697R cells that have increased P-gp expression21 were included as a positive control. As previously noted, a low level of BRAFV600E protein could be detected in OSUCLL-BRAFV600E cells in the absence of dox treatment (Figure 2B), and ABCB1 expression in these cells was slightly higher relative to OSUCLL-Tet cells (Figures 2A, 3A). However, increased P-gp expression was not clearly detected in OSUCLL-BRAFV600E cells in the absence of dox, nor was ABCB1 mRNA increased in dox-treated OSUCLL-BRAF cells (Figure 2A). As an additional validation, OSUCLL cells were transfected with cDNAs driven by a constitutive promoter. Cells transfected with BRAFV600E again showed increases in MAPK signaling and in P-gp expression relative to cells transfected with normal BRAF or the empty vector (Supplementary Figure S1).
Figure 3. BRAFV600E and MEK inhibition block ABCB1/P-gp expression in OSUCLL cells.
A. Real-time RT-PCR analysis of ABCB1 expression in OSUCLL cells in the presence or absence of BRAFV600E or MEK inhibitors. Cells were incubated without or with dox 24 hr, then inhibitors (2 µM vemurafenib and/or 1 µM CI-1040) were added for an additional 16 hr. Inhibitor comparisons in the presence of dox were performed versus BRAFV600E + dox (N=5, *p<0.05; **p<0.001). B. Immunoblot analysis of P-gp, normal BRAF, and BRAFV600E expression in OSUCLL cells treated as in A. Results shown are representative of three individual experiments.
As P-gp expression may not necessarily correlate with its function22, efflux of the fluorescent P-gp substrate rhodamine 123 was examined by flow cytometry in dox-induced OSUCLL cells. Within 90 minutes of removal from rhodamine-containing media, OSUCLL-BRAFV600E cells were essentially negative for rhodamine compared to OSUCLL-Tet and -BRAF cells (Figure 2C, black bars; p = 0.0003). This effect was significantly reversed by the P-gp inhibitor verapamil (Figure 2C, grey bars; p = 0.0025). As a further determination of P-gp function, cells were examined for sensitivity to the P-gp substrate vincristine. Cells were incubated 24 hr without or with dox, then treated with vincristine in the presence or absence of verapamil. After an additional 48 hr, cell proliferation was evaluated by MTS assay. As shown in Figure 2D, dox-mediated induction of BRAFV600E resulted in a significant increase in resistance to vincristine (p<0.001). Furthermore, the addition of verapamil significantly reduced this vicristine resistance (p<0.001). Together, these data indicate that BRAFV600E induces P-gp expression and function in OSUCLL cells.
BRAFV600E and MEK inhibition blocks P-gp induction in OSUCLL cells
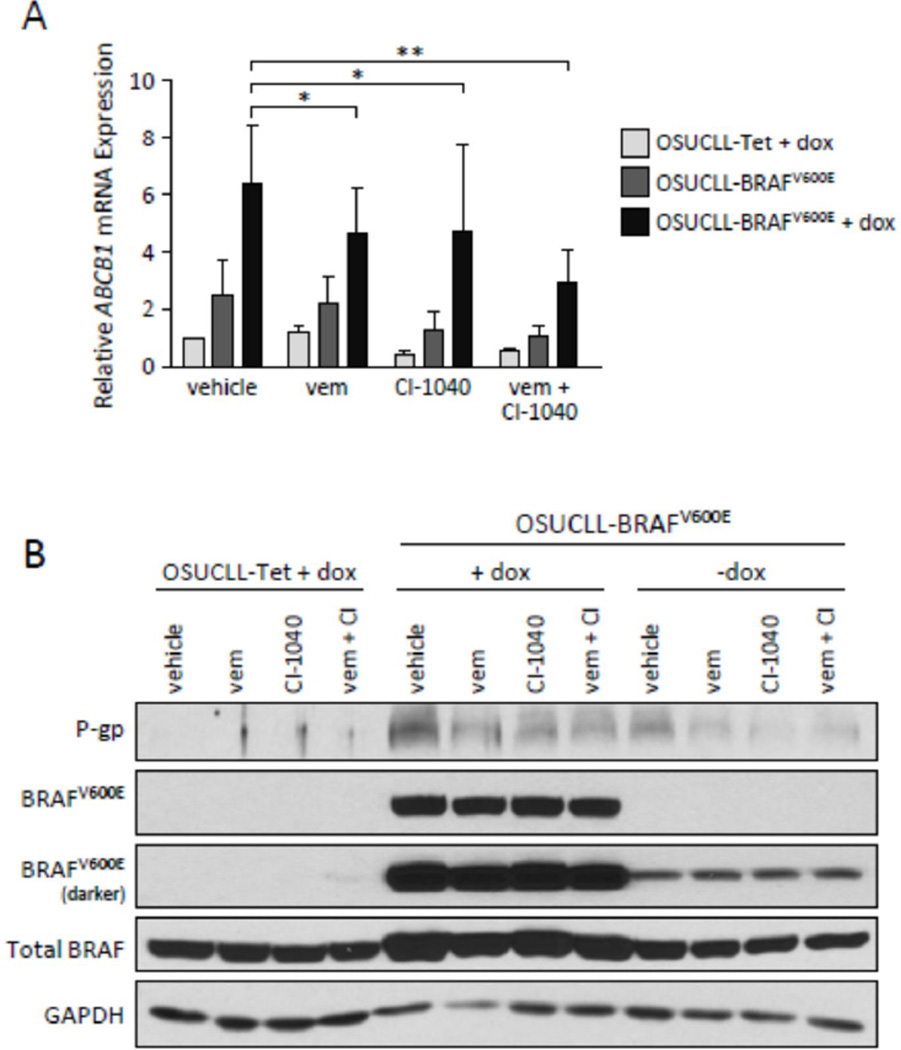
Previous studies using various tumor cell lines have shown that ABCB1 expression can be regulated through the MAPK pathway23,24, involvement of the transcription factor AP-125 or reactive oxygen species26, and/or NF-κB signaling and CRE transcriptional activity27. As the induction of BRAFV600E causes MEK-ERK activation in our model, we first examined this pathway using the BRAFV600E inhibitor vemurafenib and the MEK inhibitor CI-1040. Following a 24-hr incubation with dox, OSUCLL cells were treated with vehicle (DMSO), vemurafenib (2 µM), and/or CI-1040 (1 µM) for 16 hr. As shown in Figures 2A–B and 3A–B, the induction of BRAFV600E by dox caused a notable increase in P-gp protein as well as ABCB1 mRNA. This effect was inhibited by vemurafenib or CI-1040, and stronger inhibition was noted with the combination.
BRAFV600E enhances ABCB1 promoter activity
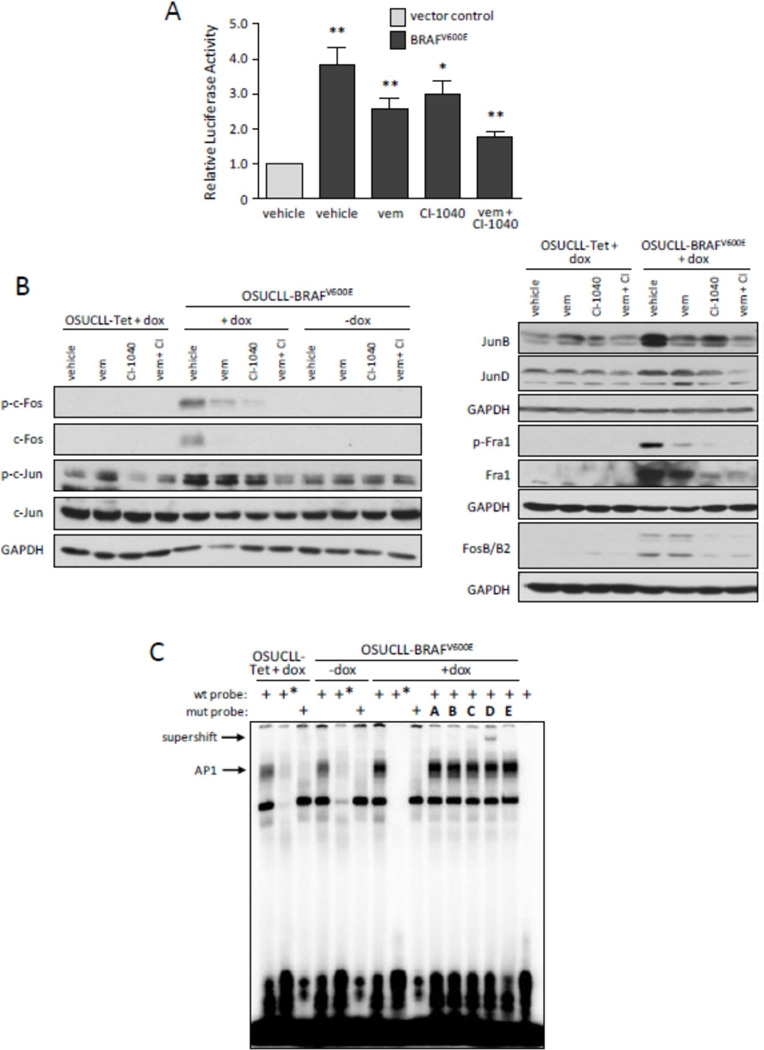
To investigate the mechanism of BRAFV600E-mediated ABCB1 induction, a construct containing 1 kb of the ABCB1 promoter driving a luciferase reporter (pTL-ABCB1) was transiently co-transfected with either an empty vector (pBABE-puro) or one containing BRAFV600E (pBABE-puro-BRAFV600E) into HEK293T cells. Cells were then treated without or with vemurafenib and/or CI-1040, and lysates were measured for luciferase activity (Figure 4A). ABCB1 promoter activity was notably increased with BRAFV600E expression, and BRAFV600E and MEK inhibition reduced this effect. Based on data demonstrating the role of AP-1 in ABCB1 regulation25, a panel of AP-1 proteins were assessed by immunoblot in OSUCLL cells following 24-hr dox treatment. As shown in Figure 4B, c-Fos, FosB/B2, Fra1, c-Jun, JunB, and JunD all appeared to be upregulated and/or phosphorylated upon BRAFV600E induction, and these effects could be blocked by vemurafenib and/or CI-1040.
Figure 4. BRAFV600E enhances ABCB1 promoter activity via MAPK and AP-1.
A. HEK293T cells were transiently co-transfected with 1 µg ABCB1 reporter construct (pTL-MDR1) and 1 µg empty vector (pBabepuro) or mutant BRAF plasmid (pBabepuro-BRAFV600E). After 8 hr, inhibitors were added as in Figure 3A. After an additional 16 hr, luciferase activity was assessed in total cell lysates. Results are shown normalized to the amount of lysate; each inhibitor was compared to control, and in addition, the control was compared to empty vector (N=3, *p<0.05; **p<0.001). B. The immunoblot from Figure 3B was additionally analyzed for c-Fos and c-Jun proteins. C. Nuclear extracts were prepared from OSUCLL cells incubated 24 hr with or without dox, then mixed with 32P-labeled wild-type (wt) or mutant (mut) AP-1 probes, 100× cold probe (*), and antibodies as indicated: A. c-Fos; B. c-Jun; C. JunB; D. JunD; E. MEK (irrelevant control). The final lane is free probe without nuclear extract. Mixtures were separated by native acrylamide electrophoresis and bands detected by autoradiography. Results shown are representative of three individual experiments.
NF-κB and CRE have also been implicated in ABCB1 regulation27. Here, phosphorylation of NFκB p65 was observed with BRAFV600E induction. However, this effect was inconsistent despite the consistent increase in ABCB1 mRNA. Also, phosphorylation of CREB was unaltered upon BRAFV600E induction (Supplementary Figure S2). These results further support that BRAFV600E-induced ABCB1 expression in OSUCLL cells occurs via the MEK/ERK/AP-1 pathway. To identify AP-1 factors involved in ABCB1 regulation in OSUCLL cells, EMSAs were performed using AP-1 elements from the ABCB1 promoter as previously identified19,20. Following 24-hr dox incubation of OSUCLL cells, nuclear extracts were prepared, incubated with labeled probe in the absence or presence of antibodies to AP-1, and separated on a non-denaturing acrylamide gel (Figure 4C). A mobility shift was observed upon induction of BRAFV600E (lanes 1 vs. 7) that appeared to be specific for AP-1, as it was reduced following addition of excess cold probe or substitution of a mutated probe (lanes 2 and 3, respectively). While most of the antibodies to AP-1 family members produced no effect, an anti-JunD antibody resulted in a clear supershift in the AP-1 complex. Overall, these results indicate that BRAFV600E mediates ABCB1/P-gp expression in OSUCLL cells, and that JunD is likely to be an important component of this regulatory mechanism.
Discussion
A subset of B-cell malignancies carry activating BRAF mutations, but the role of activated BRAF in B-cell disease is unclear. We investigated the pathological role of BRAFV600E in B-cell leukemia using a model system in which we could control BRAFV600E expression to better assess specific signaling events and outcomes. While the list of genes affected by increased BRAF signaling includes several interesting candidates, here we focused on ABCB1 because many melanoma patients with BRAFV600E are either resistant to therapy or relapse following treatment, hinting at a correlation between BRAFV600E and drug resistance. The protein product of ABCB1, P-gp, is a member of a class of glycoproteins that export xenobiotic agents across the cytoplasmic membrane. Thus, its presence in certain cell types acts to reduce intracellular accumulation of anti-cancer drugs, resulting in relative resistance to those agents. P-gp is normally expressed in the liver, pancreas, kidney, and gut, but is also aberrantly upregulated in certain solid tumors and hematologic malignancies by a variety of mechanisms24,28. Separately, drug resistance in BRAFV600E mutated cancers has also been reported to result from concurrent activation of the PI3K pathway through PTEN loss, amplification of cyclin D1, and feedback activation of EGFR29–35. However, the vast majority of these resistance studies have focused on increased gene expression as a consequence of chronic exposure to chemotherapeutic agents, and few studies describe the involvement of P-gp in de novo BRAFV600E-mediated drug resistance. Here, we report that mutationally activated BRAF drives ABCB1 transcription via AP-1 activity in a model of B-cell malignancy, leading to enhanced P-gp expression and function.
Previous reports indicate that the expression levels of ABCB1 mRNA and P-gp protein do not necessarily correlate with P-gp function22. Furthermore, the size and glycosylation of P-gp make this protein challenging to detect by immunoblot. Therefore, following real-time RT-PCR validation of increased ABCB1 mRNA expression following BRAFV600E induction, we assessed P-gp function via rhodamine exclusion as well as drug sensitivity in the presence and absence of the P-gp inhibitor verapamil. These experiments confirmed increased P-gp function in BRAFV600E-expressing cells. To investigate the relationship between BRAF activity and ABCB1 regulation, we treated cells with the BRAFV600E inhibitor vemurafenib. Interestingly, this agent abolished ERK phosphorylation in OSUCLL cells only transiently (data not shown). This transient effect, which is also seen in melanoma and other BRAFV600E-mutant cell lines36,37, could be the result of P-gp-mediated drug elimination, as vemurafenib has been reported to be a substrate of efflux pumps such as P-gp. In support of this, brain distribution of vemurafenib is diminished via P-gp and BCRP/ABCG2 in patients with metastatic melanoma, and can be enhanced by inhibition of these proteins38,39. Furthermore, Wu et al. showed that vemurafenib blocked the efflux activity not only of P-gp, but also BCRP, and re-sensitized a BRAFV600E and BCRP-expressing melanoma cell line to the BCRP substrates topotecan and mitoxantrone40. These observations further support that elevated MDR protein expression might constitute an important mechanism of resistance to vemurafenib. Although we did not detect increases in other MDR proteins such as BCRP in BRAFV600E-expressing cells, our data support that the BRAFV600E mutation might promote resistance to drugs that target it. In addition to V600E, other mutations in BRAF have been identified (e.g. G466, L597, K601) that result in enhanced BRAF kinase activity. While these are rarer and therefore less characterized, we hypothesize that these mutations would also increase ABCB1 expression to cause resistance to chemotherapies including vemurafenib. However, more experiments are needed to address this.
BRAFV600E kinase inhibitor-induced paradoxical MAPK activation is known to stimulate receptor tyrosine kinases such as Ras or c-Raf to re-induce MAPK signaling. Interestingly, it was recently reported that a patient with BRAFV600E-driven melanoma who responded to vemurafenib developed CLL-like disease, possibly due to paradoxical BRAF inhibitor-associated ERK activation in B-cells via the BCR/SYK/RAS/RAF axis14. To avoid this and enhance drug efficacy, combinations of MEK inhibitors with vemurafenib are now being explored, and studies are also emerging with new inhibitors of ERK. In this study, we found that MAPK pathway-induced AP-1 protein expression results in increased ABCB1/P-gp expression. We observed increased expression and/or phosphorylation of several proteins in the Fos and Jun family including c-Fos, FosB/B2, Fra-1, c-Jun, JunB, and JunD. Although only JunD was positively identified to interact with the ABCB1 promoter element in EMSAs, other AP-1 components could also be involved in ABCB1 expression, and combinations of MAPK pathway inhibitors will likely be needed to effectively prevent this effect in patients.
While we demonstrate that increased P-gp expression and function can result from constitutive BRAF activation in B-cells, the interpretations of these results are limited by the use of cell lines. It will be essential to demonstrate that these effects are noted in tumor cells derived from patients with the BRAFV600E mutation, and such studies are underway. The strategy utilized here also does not address the role of BRAFV600E in disease development, as transfections were performed in a cell line derived from malignant B-cells. Chung et al. reported that the BRAFV600E mutation is expressed in hematopoietic stem cells in HCL patients and that these cells produce an HCL-like phenotype in immune-deficient mice41. However, pan-hematopoietic expression of BRAFV600E via the Mx1 promoter appears to produce a non-lymphoid histiocytic malignant phenotype, rather than HCL-like disease41–43, and BRAFV600E mutations are found in approximately half the cases of histiocytic malignancies Langerhans cell histiocytosis44 and Erdheim-Chester disease45. CD19-restricted BRAFV600E expression in the absence of other mutations does not appear to be sufficient to induce disease41. Thus, exactly how and at what stage BRAFV600E expression promotes the development of B-cell tumors remains unclear. A murine model with B-lineage restricted co-expression of BRAFV600E and a relevant second hit will likely provide a more accurate model to investigate the pathological role of BRAFV600E in B-cell malignancies.
Supplementary Material
Highlights.
A new cell line system with inducible expression of mutant BRAF was used to determine targets of this oncoprotein in B-cells.
These findings indicate that activating BRAF mutations, now reported in several B-cell malignancies, can result in the increased expression of ABCB1/P-gp. Thus, incorporation of relevant BRAF pathway inhibitors into therapies, especially those that include P-gp substrate drugs, may benefit patients with such mutations.
Acknowledgments
The authors would like to thank Byrd laboratory members for assistance with experiments. This work was supported by the National Cancer Institute (P50 CA140158 and P30 CA016058).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: No authors have conflicts of interest to disclose.
Author Contributions:
YTT designed, performed and interpreted experiments and wrote the paper; GL performed flow cytometry and mutation analysis; AL conducted the statistical analyses; EJS assisted with experiments; EH provided expertise and the OSUCLL cell line; SBS and CSC provided expertise and reagents; MRG designed the study and interpreted the data; JCB designed the study, interpreted the data, provided patient samples, and wrote the paper; DML oversaw the work, designed the study, interpreted the data, and wrote the paper. All authors reviewed the manuscript prior to submission.
References
- 1.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Cantwell-Dorris ER, O'Leary JJ, Sheils OM. BRAFV600E: implications for carcinogenesis and molecular therapy. Mol Cancer Ther. 2011;10(3):385–394. doi: 10.1158/1535-7163.MCT-10-0799. [DOI] [PubMed] [Google Scholar]
- 3.Tiacci E, Trifonov V, Schiavoni G, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364(24):2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dietrich S, Glimm H, Andrulis M, von Kalle C, Ho AD, Zenz T. BRAF inhibition in refractory hairy-cell leukemia. N Engl J Med. 2012;366(21):2038–2040. doi: 10.1056/NEJMc1202124. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, Reis M, Khoriaty R, et al. Sequence analysis of 515 kinase genes in chronic lymphocytic leukemia. Leukemia. 2011;25(12):1908–1910. doi: 10.1038/leu.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langabeer SE, Quinn F, O'Brien D, et al. Incidence of the BRAF V600E mutation in chronic lymphocytic leukaemia and prolymphocytic leukaemia. Leuk Res. 2012;36(4):483–484. doi: 10.1016/j.leukres.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 7.Jebaraj BM, Kienle D, Buhler A, et al. BRAF mutations in chronic lymphocytic leukemia. Leuk Lymphoma. 2013;54(6):1177–1182. doi: 10.3109/10428194.2012.742525. [DOI] [PubMed] [Google Scholar]
- 8.Damm F, Mylonas E, Cosson A, et al. Acquired Initiating Mutations in Early Hematopoietic Cells of CLL Patients. Cancer Discov. 2014;4(9):1088–1101. doi: 10.1158/2159-8290.CD-14-0104. [DOI] [PubMed] [Google Scholar]
- 9.Karreth FA, Reschke M, Ruocco A, et al. The BRAF Pseudogene Functions as a Competitive Endogenous RNA and Induces Lymphoma In Vivo. Cell. 2015;161:319–332. doi: 10.1016/j.cell.2015.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puxeddu E, Durante C, Avenia N, Filetti S, Russo D. Clinical implications of BRAF mutation in thyroid carcinoma. Trends Endocrinol Metab. 2008;19(4):138–145. doi: 10.1016/j.tem.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dietrich S, Pircher A, Andrulis M, et al. BRAF Inhibition in Hairy Cell Leukemia: Multicentre Experience of 21 Patients Treated with Vemurafenib. Blood. 2014;124:3634a. [Google Scholar]
- 14.Yaktapour N, Meiss F, Mastroianni J, et al. BRAF inhibitor-associated ERK activation drives development of chronic lymphocytic leukemia. J Clin Invest. 2014;124(11):5074–5084. doi: 10.1172/JCI76539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman SE, Gordon AL, Hertlein E, et al. Bruton tyrosine kinase represents a promising therapeutic target for treatment of chronic lymphocytic leukemia and is effectively targeted by PCI-32765. Blood. 2011;117(23):6287–6296. doi: 10.1182/blood-2011-01-328484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woyach JA, Smucker K, Smith LL, et al. Prolonged lymphocytosis during ibrutinib therapy is associated with distinct molecular characteristics and does not indicate a suboptimal response to therapy. Blood. 2014;123(12):1810–1817. doi: 10.1182/blood-2013-09-527853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hertlein E, Beckwith KA, Lozanski G, et al. Characterization of a new chronic lymphocytic leukemia cell line for mechanistic in vitro and in vivo studies relevant to disease. PLoS One. 2013;8(10):e76607. doi: 10.1371/journal.pone.0076607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett SD, Bridges AJ, Dudley DT, et al. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg Med Chem Lett. 2008;18(24):6501–6504. doi: 10.1016/j.bmcl.2008.10.054. [DOI] [PubMed] [Google Scholar]
- 19.Miao ZH, Ding J. Transcription factor c-Jun activation represses mdr-1 gene expression. Cancer Res. 2003;63(15):4527–4532. [PubMed] [Google Scholar]
- 20.Chen C, Shen HL, Yang J, Chen QY, Xu WL. Preventing chemoresistance of human breast cancer cell line, MCF-7 with celecoxib. J Cancer Res Clin Oncol. 2011;137(1):9–17. doi: 10.1007/s00432-010-0854-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta SV, Sass EJ, Davis ME, et al. Resistance to the translation initiation inhibitor silvestrol is mediated by ABCB1/P-glycoprotein overexpression in acute lymphoblastic leukemia cells. AAPS J. 2011;13(3):357–364. doi: 10.1208/s12248-011-9276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivy SP, Olshefski RS, Taylor BJ, Patel KM, Reaman GH. Correlation of P-glycoprotein expression and function in childhood acute leukemia: a children's cancer group study. Blood. 1996;88(1):309–318. [PubMed] [Google Scholar]
- 23.Katayama K, Yoshioka S, Tsukahara S, Mitsuhashi J, Sugimoto Y. Inhibition of the mitogen-activated protein kinase pathway results in the down-regulation of P-glycoprotein. Mol Cancer Ther. 2007;6(7):2092–2102. doi: 10.1158/1535-7163.MCT-07-0148. [DOI] [PubMed] [Google Scholar]
- 24.Shen H, Xu W, Luo W, et al. Upregulation of mdr1 gene is related to activation of the MAPK/ERK signal transduction pathway and YB-1 nuclear translocation in B-cell lymphoma. Exp Hematol. 2011;39(5):558–569. doi: 10.1016/j.exphem.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Roy KR, Reddy GV, Maitreyi L, et al. Celecoxib inhibits MDR1 expression through COX-2-dependent mechanism in human hepatocellular carcinoma (HepG2) cell line. Cancer Chemother Pharmacol. 2010;65(5):903–911. doi: 10.1007/s00280-009-1097-3. [DOI] [PubMed] [Google Scholar]
- 26.Nishanth RP, Ramakrishna BS, Jyotsna RG, et al. C-Phycocyanin inhibits MDR1 through reactive oxygen species and cyclooxygenase-2 mediated pathways in human hepatocellular carcinoma cell line. Eur J Pharmacol. 2010;649(1–3):74–83. doi: 10.1016/j.ejphar.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Kim HG, Hien TT, Han EH, et al. Metformin inhibits P-glycoprotein expression via the NF-kappaB pathway and CRE transcriptional activity through AMPK activation. Br J Pharmacol. 2011;162(5):1096–1108. doi: 10.1111/j.1476-5381.2010.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quiney C, Billard C, Faussat AM, Salanoubat C, Kolb JP. Hyperforin inhibits P-gp and BCRP activities in chronic lymphocytic leukaemia cells and myeloid cells. Leuk Lymphoma. 2007;48(8):1587–1599. doi: 10.1080/10428190701474332. [DOI] [PubMed] [Google Scholar]
- 29.Dankort D, Curley DP, Cartlidge RA, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41(5):544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng W, Gopal YN, Scott A, Chen G, Woodman SE, Davies MA. Role and therapeutic potential of PI3K-mTOR signaling in de novo resistance to BRAF inhibition. Pigment Cell Melanoma Res. 2012;25(2):248–258. doi: 10.1111/j.1755-148X.2011.00950.x. [DOI] [PubMed] [Google Scholar]
- 31.Halaban R, Zhang W, Bacchiocchi A, et al. PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells. Pigment Cell Melanoma Res. 2010;23(2):190–200. doi: 10.1111/j.1755-148X.2010.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao M, Tian F, Mariadason JM, et al. Resistance to BRAF inhibition in BRAF-mutant colon cancer can be overcome with PI3K inhibition or demethylating agents. Clin Cancer Res. 2013;19(3):657–667. doi: 10.1158/1078-0432.CCR-11-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paraiso KH, Xiang Y, Rebecca VW, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Res. 2011;71(7):2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smalley KS, Lioni M, Dalla Palma M, et al. Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E-mutated melanomas. Mol Cancer Ther. 2008;7(9):2876–2883. doi: 10.1158/1535-7163.MCT-08-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 36.Joseph EW, Pratilas CA, Poulikakos PI, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc Natl Acad Sci U S A. 2010;107(33):14903–14908. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montero-Conde C, Ruiz-Llorente S, Dominguez JM, et al. Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer Discov. 2013;3(5):520–533. doi: 10.1158/2159-8290.CD-12-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mittapalli RK, Vaidhyanathan S, Sane R, Elmquist WF. Impact of P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2) on the brain distribution of a novel BRAF inhibitor: vemurafenib (PLX4032) J Pharmacol Exp Ther. 2012;342(1):33–40. doi: 10.1124/jpet.112.192195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durmus S, Sparidans RW, Wagenaar E, Beijnen JH, Schinkel AH. Oral availability and brain penetration of the B-RAFV600E inhibitor vemurafenib can be enhanced by the P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2) inhibitor elacridar. Mol Pharm. 2012;9(11):3236–3245. doi: 10.1021/mp3003144. [DOI] [PubMed] [Google Scholar]
- 40.Wu CP, Sim HM, Huang YH, et al. Overexpression of ATP-binding cassette transporter ABCG2 as a potential mechanism of acquired resistance to vemurafenib in BRAF(V600E) mutant cancer cells. Biochem Pharmacol. 2013;85(3):325–334. doi: 10.1016/j.bcp.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung SS, Kim E, Park JH, et al. Hematopoietic stem cell origin of BRAFV600E mutations in hairy cell leukemia. Sci Transl Med. 2014;6(238):238ra271. doi: 10.1126/scitranslmed.3008004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mercer K, Giblett S, Green S, et al. Expression of endogenous oncogenic V600EB-raf induces proliferation and developmental defects in mice and transformation of primary fibroblasts. Cancer Res. 2005;65(24):11493–11500. doi: 10.1158/0008-5472.CAN-05-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamata T, Dankort D, Kang J, et al. Hematopoietic expression of oncogenic BRAF promotes aberrant growth of monocyte-lineage cells resistant to PLX4720. Mol Cancer Res. 2013;11(12):1530–1541. doi: 10.1158/1541-7786.MCR-13-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Badalian-Very G, Vergilio JA, Degar BA, et al. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 2010;116(11):1919–1923. doi: 10.1182/blood-2010-04-279083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haroche J, Charlotte F, Arnaud L, et al. High prevalence of BRAF V600E mutations in Erdheim-Chester disease but not in other non-Langerhans cell histiocytoses. Blood. 2012;120(13):2700–2703. doi: 10.1182/blood-2012-05-430140. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.