Abstract
A small library of anticancer, cell-permeating, stapled peptides based on potent dual-specific antagonist of p53–MDM2/MDMX interactions, PMI-N8A, was synthesized, characterized and screened for anticancer activity against human colorectal cancer cell line, HCT-116. Employed synthetic modifications included: S-alkylation-based stapling, point mutations increasing hydrophobicity in key residues as well as improvement of cell-permeability by introduction of polycationic sequence(s) that were woven into the sequence of parental peptide. Selected analogue, ArB14Co, was also tested in vivo and exhibited potent anticancer bioactivity at the low dose (3.0 mg/kg). Collectively, our findings suggest that application of stapling in combination with rational design of polycationic short analogues may be a suitable approach in the development of physiologically active p53–MDM2/MDMX peptide inhibitors.
Keywords: Antagonists of p53, Anticancer agents, S-alkylation of peptides, Drug design, Cell permeable peptides
Introduction
In recent years lack of funds has made cutting edge research in peptide chemistry significantly more challenging. A case in point is “stapling” of peptides that emerged as a leading strategy in the development of novel peptide-based drug candidates since a pivotal Blackwell & Grubbs paper (Blackwell and Grubbs, 1998). Numerous studies used this approach to develop potent bioactive compounds even resulting in the foundation of a specialized company, Aileron Therapeutics, Inc. However, reagents necessary to carry out stapling related research are rather expensive, especially olefin functionalized non-natural amino acids which form “staple” (α-helix-stabilizing olefin bridge) during a ring-closing metathesis (RCM) reaction. This makes related SAR peptide studies particularly expensive and risky, especially if one works on difficult or long sequences where yield becomes an issue. In this “classical” approach the peptide staple is efficiently created in a two-step process between strategically positioned olefin functionalized non-natural amino acid side chains. The first step, catalyzed by Grubbs catalyst, results in olefin containing bridge that is subsequently catalytically reduced to saturated hydrocarbon (alkane), effectively locking the peptide into a stable α-helix conformation (Bernal et al., 2007; Blackwell et al., 2001; Blackwell and Grubbs, 1998; Walensky et al., 2004). Such helix stabilization had been shown to dramatically increase the helicity, potency, resistance to proteolytic degradation and cell permeability of α-helical peptides (Bautista et al., 2010; Bernal et al., 2007; Bird et al., 2010; Kim et al., 2011; Kim and Verdine, 2009; Long et al., 2013; Schafmeister et al., 2000)
Over the years novel strategies for peptide stapling have emerged as alternatives to the RCM approach (Lau et al., 2015a). These include hydrazone bridge (Cabezas and Satterthwait, 1999), oxime bridge (Haney et al., 2011), 1,4-disubstituted-[1,2,3]-triazole linkage (Holland-Nell and Meldal, 2011; Ingale and Dawson, 2011; Kawamoto et al., 2012; Lau et al., 2014c; Lau et al., 2014b; Lau et al., 2014a; Lau et al., 2015b; Scrima et al., 2010), metal chelation (Ghadiri and Choi, 1990; Ruan et al., 1990), disulfide bond formation (Almeida et al., 2012; Jackson et al., 1991; Leduc et al., 2003), lactam ring formation (Fujimoto et al., 2008; Geistlinger and Guy, 2001; Geistlinger and Guy, 2003; Houston, Jr. et al., 1995; Osapay and Taylor, 1992; Phelan et al., 1997) and S-alkylation based staples employing either α-haloacetamide alkylation of single cysteine (Brunel and Dawson, 2005; Cardoso et al., 2007; Galande et al., 2004; Woolley, 2005) or bridging two cysteines with bis-S-alkylating linker(s) (de Araujo et al., 2014; Jo et al., 2012; Muppidi et al., 2011b; Muppidi et al., 2011a; Muppidi et al., 2012; Spokoyny et al., 2013; Szewczuk et al., 1992; Timmerman et al., 2005; Wilkinson et al., 2007; Zhang et al., 2007; Zhang et al., 2008). Among these, the last seems to be most flexible approach as a wide range of inexpensive bis-thiol-reactive linkers is commercially available, including rigid aromatic derivatives (Chua et al., 2015; Jo et al., 2012; Muppidi et al., 2011b; Muppidi et al., 2011a; Muppidi et al., 2012; Timmerman et al., 2005; Zhang et al., 2007) and aliphatic counterparts (Byrne and Stites, 1995; Chua et al., 2015; Lindman et al., 2001; Wilkinson et al., 2007). In addition, the availability of various cysteine homologs: (L)Cys, (D)Cys, (L)homoCys, (D)homoCys, (L)Pen, and (D)Pen provides an additional option for “fine tuning” of pre-selected active derivatives. Moreover, those S-alkylation/stapling reactions can be carried out in water-based solutions without any protecting groups and due to use of standard amino acids (Cys and its homologs) costs are relatively low. Notably, the use of multi-thiol-reactive linkers has a remarkably long tradition, as an application for this purpose of the aromatic derivative, 1,3,5-tris(bromomethyl)benzene, was initially reported by Kemp & McNamara in 1985 (Kemp and McNamara, 1985) and use of its bis-reactive homologs, 1,2-bis(bromomethyl)benzene, and 1,3-bis(bromomethyl)benzene was described only few years later (Szewczuk et al., 1992). This methodology was also successfully applied in peptide drug development (Timmerman et al., 2007), including phage display (Angelini et al., 2012a; Baeriswyl et al., 2012; Baeriswyl et al., 2013; Baeriswyl and Heinis, 2013a; Baeriswyl and Heinis, 2013b; Bellotto et al., 2014; Chen et al., 2012; Chen et al., 2013; Chen et al., 2014b; Chen et al., 2014a; Heinis et al., 2009; Rentero-Rebollo et al., 2014; Timmerman et al., 2007) as well as peptide-albumin (Angelini et al., 2012c; Pollaro et al., 2014) and peptide-antibody drug conjugates (ADCs) (Angelini et al., 2012b).
We decided to apply this “cheap” strategy to the synthesis of p53–MDM2/MDMX bridged peptide inhibitors that are currently of great interest (Bernal et al., 2007; Bernal et al., 2010; Brown et al., 2009; Brown et al., 2013; Chang et al., 2013; Khoo et al., 2014).
The p53 pathway remains major target in anti-cancer research (Brown et al., 2009; Cheok et al., 2011). Generally, its deregulation causes malignant transformation of normal cells and approximately 50% of all cancers have a mutation in TP53 gene (Menendez et al., 2009), with varying mutations’ frequencies, ranging from ~10–70%, dependent on cancer type (Brosh and Rotter, 2009). Moreover, in all p53 pathway related cancers, only half is associated with TP53 mutations, with remaining cases being caused by overexpression of the p53-negative regulators: MDM2 (Momand et al., 1992) and MDM4, (Danovi et al., 2004; Laurie et al., 2006; Riemenschneider et al., 1999) or deletion/inactivation of ARF, the p53-positive regulator and MDM2 inhibitor (Esteller et al., 2001; Lowe and Sherr, 2003; Sherr and Weber, 2000).
The p53 stress response protein is a transcription factor with a crucial role in tumour suppression (Lowe et al., 2004; Vousden and Lane, 2007). It regulates various biological activities (Menendez et al., 2007; Menendez et al., 2009; Vousden and Prives, 2009) including, apoptosis (Bensaad et al., 2006; Vousden, 2006), cell cycle (Kastan et al., 1992; Kuerbitz et al., 1992), senescence (Garbe et al., 2007), immune response (Taura et al., 2008), cell differentiation (Molchadsky et al., 2008), motility (Qin et al., 2009) and migration (Roger et al., 2006; Singh et al., 2007), angiogenesis (Pal et al., 2001; Teodoro et al., 2006; Zhang et al., 2000), DNA (Helton and Chen, 2007) and energy (Green and Chipuk, 2006; Matoba et al., 2006; Vousden and Ryan, 2009) metabolism, microRNA processing (Suzuki et al., 2009), cell–cell communication (Yu et al., 2006) and the DNA damage response and repair (de Souza-Pinto et al., 2004; Sengupta and Harris, 2005; Sommers et al., 2005). Normal levels of p53 protein are low, due to rapid ubiquitin-dependent proteasomal degradation which is controlled largely by the E3 ubiquitin ligase MDM2 (Brooks and Gu, 2006) (known as HDM2 in human), which in turn is also a target of transcriptional regulation by p53 (Murray-Zmijewski et al., 2008). Additionally, MDM2 may regulate p53 function through direct, high affinity binding to p53’s N-terminal transactivation domain which effectively blocks its function (Momand et al., 1992), and by impairing nuclear import of the p53 protein (Haupt et al., 1997; Wade and Wahl, 2009). MDM4 (also known as HDM4, MDMX or HDMX), another negative regulator of p53, also plays an important role in its function. Although MDM4 shows a similar to MDM2 p53-binding properties, it lacks its E3 ubiquitin ligase activity and consequently cannot affect p53s stability (Marine et al., 2007; Marine, 2011; Migliorini et al., 2002a; Migliorini et al., 2002b; Toledo et al., 2006; Toledo and Wahl, 2006; Wade and Wahl, 2009). However, MDM4, a structural homologue of MDM2, can form a MDM2/MDMX heterodimers through their C-terminal RING finger domains, which potentiate the ubiquitination of p53, and subsequently its degradation (de Graaf et al., 2003; Linares et al., 2003; Pan and Chen, 2003). Notably, in contrast to MDM2, MDMX is not under transcriptional control of p53 (Riedinger and McDonnell, 2009). In addition stability of p53 is influenced by ARF (also known as p14ARF) a tumour suppressor that interacts with MDM2 and inhibits p53 degradation(Sherr and Weber, 2000).
Various stressors, including DNA damage and oncogenic stress, increase the amount of p53 by disrupting its degradation resulting in p53’s stabilization and functional activation (Lavin and Gueven, 2006; Murray-Zmijewski et al., 2008; Sherr, 2006). Once activated, p53 can elicit various cellular responses, including growth arrest, senescence and apoptosis.
Since p53 is commonly inactivated in various human cancers, its “re-activation” represents a major therapeutic strategy. Specifically, targeting the interactions of p53 with its major negative regulators, MDM2 and MDM4, recently emerged as an important therapeutic approach, with multiple examples of both, small molecules and peptides being utilized as p53-MDM2/MDM4 inhibitors (Li and Lozano, 2013; Saha et al., 2013; Wang et al., 2012; Zhao et al., 2013). p53-MDM2/MDM4 interactions are mediated primarily by three key hydrophobic residues in p53: 19Phe, 23Trp, and 26Leu (Kussie et al., 1996; Pazgier et al., 2009; Popowicz et al., 2008), and similar though not identical, hydrophobic surface grooves in MDM2 and MDM4 (Zhao et al., 2013). Over past few years, mutational studies and phage display has produced multiple peptide-based p53–MDM2/MDMX inhibitors, including all-D, stapled and bridged compounds with increased resistance to proteolysis (Bernal et al., 2007; Bernal et al., 2010; Chang et al., 2013; Guo et al., 2014; Hu et al., 2007; Li et al., 2009; Li et al., 2010b; Li et al., 2010a; Liu et al., 2010b; Liu et al., 2010a; Madden et al., 2011; Madhumalar et al., 2009; Pazgier et al., 2009; Zhan et al., 2012). Available in vitro/in vivo data (Chang et al., 2013; Hu et al., 2007; Liu et al., 2010a) suggest that the limiting factor in the inhibitory activity of these compounds is low cell-permeability since liposomal based formulations appear to be significantly more potent than “naked” peptide (Liu et al., 2010a). Even though, peptide stapling was postulated to increase cell permeability (Bernal et al., 2007) its effects seems to be limited as liposomal formulation (Liu et al., 2010a) of DPMI-α (dosage: 3 and 7.5 mg/kg, KD-MDM2=219 nM(Liu et al., 2010b)) was significantly more potent than MPEGDSPE-based micellar formulation of ATSP-7041(Chang et al., 2013) (dosage: 15–30 mg/kg, KD-MDM2=0.91 nM), despite a superior binding affinity of the latter, although this assessment may not be totally valid since different cell lines/schedules were used in both studies. Conjugates of similar peptides with cell permeating molecules (Arg3, (D)Arg9 and spermine) were also prepared, though respective in vivo studies were not reported (Lau et al., 2014c; Lau et al., 2014b; Liu et al., 2010a; Muppidi et al., 2011a). Other limited studies employing an S-alkylation of a pair of cysteines located at i,i+7 positions of analogous compounds, using 4,4’-bis-bromomethyl-biphenyl (Bph) and 6,6’-bis-bromomethyl-[3,3’]-bipyridine (Bpy) were also reported (Muppidi et al., 2011b; Muppidi et al., 2011a). Again, in vivo anticancer activity was not presented.
Materials and Methods
Peptide Synthesis
All peptides were synthesized as C-terminal amides (except for ArB14Ao) by the solid phase method using CEM Liberty automatic microwave peptide synthesizer (CEM Corporation Inc., Matthews, NC), applying 9-fluorenylmethyloxycarbonyl (Fmoc) chemistry and commercially available amino acid derivatives and reagents (EMD Biosciences, San Diego, CA and Chem-Impex International, Inc., Wood Dale, IL). Rink Amide MBHA Resin (Piperazine-2-Chlorotrityl for ArB14Ao) (EMD Biosciences, San Diego, CA) was used as a solid support. Peptides were cleaved from resin using modified reagent K (TFA 94% (v/v); phenol, 2% (w/v); water, 2% (v/v); TIS, 1% (v/v); EDT, 1% (v/v); 2 hours) and precipitated by addition of ice-cold diethyl ether. Linear peptides were purified by preparative reverse-phase high performance liquid chromatography (RP-HPLC) and their purity evaluated by matrix-assisted laser desorption ionization spectrometry (MALDI-MS) as well as analytical RP-HPLC.
Stapling using the bis(halogenomethyl)aryl linkers
1,2-bis(bromomethyl)benzene,
1,3-bis(bromomethyl)benzene,
1,4-bis(bromomethyl)benzene,
1-(bromomethyl)-3-[3-(bromomethyl)benzyl]benzene,
1-(chloromethyl)-4-[4-(chloromethyl)phenoxy]benzene, may be performed either on the solid support or in solution.
In solution
Peptide (1 eq) containing 2 properly placed cysteines was mixed in the solution of DMF (N,N-dimethylformamide) and DMSO (dimethylsulfoxide) (1:1 ratio, concentration of peptide: 0.5–1 mg/mL) with the bis(halogenomethyl)aryl compound (XCH2-Ar-CH2X, 1 eq.) in the presence of cesium carbonate (Cs2CO3, 10 eq.) and tetrabutylammonium iodide (TBAI, 1 eq.). Reaction was monitored by HPLC/mass spectrometry. Alternatively stapled compounds were obtained using modified Timmerman protocol (Timmerman et al., 2005). Briefly, linear peptides were solubilized in 50% acetonitrile in water containing 50 mM NH4HCO3 (~0.5 mg/mL) and then appropriate bis-functional S-alkylating reagent was added. Reaction(s) were stirred for 18 h and solutions subsequently lyophilized. Obtained crude compounds were purified by preparative RP-HPLC and their purity evaluated by MALDI-MS as well as analytical RP-HPLC
On the solid support
After assembly of the peptide, final Fmoc group was removed and side chains of Cys residues deprotected (2% trifluoroacetic acid (TFA), 2% triisopropyl silane (TIS) in dichloromethane (DCM), 3×10 min). Then peptide was washed with DMF and bis(thioether)arylbridging performed on resin using appropriate bis-functional S-alkylating reagent (1 eq.) in the presence of Cs2CO3, (10 eq) and TBAI (1 eq) in DMF (overnight). Subsequently, peptides were cleaved and processed as described above.
Stapling with 2-chloro-N-(2-(2-chloro-acetyloamino)-ethyl)-acetamide linker: was performed using Timmerman protocol (Timmerman et al., 2005) described above.
Stapling with divinylsulfone (DVS): was performed using previously described protocol (Wilkinson et al., 2007) from corresponding purified linear peptides in reaction with 1 eq. (mole:mole ratio) of DVS in solution of 10 mM NH4HCO3 in 50% ACN in water (overnight). Subsequently, solution was lyophilized, and obtained crude solid residues purified by preparative RP-HPLC.
Stapling with 1,4-dibromobutane and 1,7-dibromoheptane: was performed in solution using modified Włostowski S-alkylation protocol (Wlostowski et al., 2010) that we adapted to peptides (Chua et al., 2015). Briefly, linear peptides were solubilized in anhydrous methanol (~0.5 mg/mL) containing, if necessary up 25% of DMSO. Subsequently 1 equivalent of either 1,4-dibromobutane and 1,7-dibromoheptane was added followed by 1,1,3,3-tetramethylguanidine (final conc.=0.35%, vol/vol). Reaction mixture was stirred 15 min at room temperature and then 90 min at 50 °C. Subsequently, solution was lyophilized, and obtained crude solid residues purified by preparative RP-HPLC.
Synthesis of analogues ArB14k1÷k4 and ArB18Ek: Peptides ArB14k1÷k4 and ArB18Ek were assembled using SPPS (Fmoc-(D)Lys(Dde)-OH was used). Then side chain of (D)Lys was selectively deprotected using 2% hydrazine in DMF (3×10 min) and subsequently chloroacetylated using chloroacetic anhydride (5 eq./30 min). Then peptides were cleaved from the resin and cyclized in solution of 65 mM NH4HCO3 in 70% ACN in water (overnight) giving thioether bridge. Subsequently, solution was lyophilized, and obtained crude solid residues purified by preparative RP-HPLC.
Oxidation of thioethers to corresponding sulfones: (ArB14Dmx and ArB14Emx): Oxidation of thioethers to sulfones was performed using OXONE® (Duran et al., 2006) (Sigma-Aldrich, KHSO5×0.5 KHSO4×0.5 K2SO4) in methyl alcohol:water mix (3:1) using 3 eq. of OXONE® per each thioether group. Reaction was carried out in room temperature for 3 h. Subsequently solutions were freeze-dried and purified using preparative RP-HPLC.
Synthesis of ArB conjugates with
Azidothymidine (AZT): was conjugated using Cu(I)-catalyzed azide-alkyne 1,3-dipolar Huisgen's cycloaddition (CuAAC, “click chemistry”). Alkyne component was synthesized from ArB14Co in one step synthesis: 5-Hexynoic acid in dry DMSO (1 eq., Sigma-Aldrich Corp., St. Louis, MO) was pre-activated with N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide (EDC) in the presence of 3 eq. of N-hydroxysuccinimide (NHS) for 3 h. Subsequently, 1 eq. of ArB14Co in dry DMSO was added followed by 5 eq. of NMM. The solution was agitated overnight and subsequently evaporated under reduced pressure on SpeedVac. Remaining residue was purified on RP-HPLC. Obtained ArB-alkyne derivative was then “clicked” with AZT using published protocol (Lau et al., 2015b) using CuSO4/sodium ascorbate catalyst in 50% tert-buthanol in water. Subsequently, reaction mixture was freeze-dried and purified using RP-HPLC.
Bleomycin A5 (BLM): was conjugated with ArB analogue(s) using bis-amine-reactive linker, dithiobis(succinimidyl propionate) (DSP, Pierce Biotechnology, Rockford, lL). Briefly, specific ArB peptide was synthesized with an additional 6-aminohexanoic acid (Ahx) residue at N-terminus. Then chosen stapling procedure was performed and stapled peptide was purified using RP-HPLC. Subsequently, equimolar amounts of Ahx-ArB peptide and BLM (EMD Millipore, Temecula, CA) were mixed in dry DMSO (5 mg/ml). Then 10 eq. of N-methylmorpholine (NMM) was added followed by 1 eq. of DSP. The reaction was mixed for 18 h and subsequently evaporated under reduced pressure on SpeedVac. Remaining residue was purified using RP- HPLC.
Chlorambucil (CLB): was introduced using N-terminal amine group employing N,N’-diisopropylcarbodiimide (DIC, Sigma-Aldrich Corp., St. Louis, MO) as a coupling reagent. Reactions were carried out on resin using N,N-dimethylformamide as a solvent (2h).
Cholesterol (Chol): was introduced using N-terminal amine group employing cholesterol chloroformate (Sigma-Aldrich Corp., St. Louis, MO) resulting in urethane type connectivity. Reactions were carried out on resin in the presence of DIPEA using dichloromethane as a solvent (2h).
Docetaxel (DTX): was conjugated with ArB analogue(s) using the same strategy that was employed for BLM conjugates. Suitable DTX derivative, 2’-O-glycyl-docetaxel, was synthesized using previously described method (Zhigaltsev et al., 2010). Briefly, Trt-Gly-OH (1 eq.) and DTX (1 eq.) were dissolved in dry dichloromethane and cooled in ice bath under the inert-gas blanket. Subsequently, 2 eq. of 2-fluoro-1-ethyl pyridinium tetrafluoroborate (FEP) dissolved in minimal amount of dry dichloromethane were added followed by 3 eq. of DIPEA. The reaction was agitated for 18 h with gradual warming to room temperature and then evaporated. Solid residue was re-dissolved in solution of 2% TFA in dry dichloromethane to remove Trt protecting group (30 min) and evaporated again. Obtained residue was purified using RP-HPLC giving 2’-O-glycyl-docetaxel that was subsequently used for conjugation(s).
Methotrexate (MTX): was introduced using N-terminal amine group employing N,N’-diisopropylcarbodiimide (DIC, Sigma-Aldrich Corp., St. Louis, MO) as a coupling reagent. Reactions were carried out on resin in the presence of DIPEA using N,N-dimethylformamide as a solvent (2h).
Analytical RP-HPLC
Experiments were performed on a Varian ProStar 210 HPLC system equipped with ProStar 325 Dual Wavelength UV-Vis detector with the wavelengths set at 220 nm and 280 nm (Varian Inc., Palo Alto, CA). Mobile phases consisted of solvent A, 0.1% TFA in water, and solvent B, 0.1% TFA in acetonitrile. Analyses of peptides were performed with an analytical reversed-phase C18 SymmetryShield™ column, 4.6×250 mm, 5 µm (Waters, Milford, MA) or (*) an analytical reversed-phase C18 Vydac 218TP54 column, 4.6×250 mm, 5 µm (Grace, Deerfield, IL) applying linear gradient of solvent B from 0 to 100% over 100 min (flow rate: 1 ml/min).
In vitro cells’ growth inhibition assay
Experiments were carried out using PrestoBlue™ Cell Viability Reagent (Invitrogen, Carlsbad, CA) according to manufacturer's protocol. Briefly, HCT-116 cells were plated in a 96-well plate at a density of 5×103 cells/well in a total volume of 50 µl of culture media and treated with various concentrations of tested peptides (50 µl of 0–200 µM peptides in culture media). The cells' viability was assessed after 48 h by fluorescence measurement (Ex/Em:560/590, incubation time 30 min) employing the SpectraMAX M2 microplate reader (Molecular Devices, Sunnyvale, CA). All experiments were carried out in triplicate.
Animal experiments
All animal experiments were approved by the UCLA Animal Care and Use Committee and conformed to local and national guidelines. For subcutaneous engraftment model experiments BALB/SCID gnotobiotic mice (8 weeks old) were obtained from the UCLA AALAC-accredited Department of Radiation Oncology Facility and subcutaneously injected with 2.0×106 cells of human colorectal cancer cell line (HCT-116, leg). After 3 weeks, palpable tumours of approximately 5 mm diameter appeared and treatment was initiated. In general, each animal received intraperitoneally a total of 10 doses of ArB14Co at 3.0 mg/kg or 7.5 mg/kg on days 1–5 and 8–12 with 8 mice per group. The peptide was formulated in 2% Cremophor EL (Sigma-Aldrich, St Louis, MO) in phosphate-buffered saline (PBS, vol/vol). Control animals were injected with vehicle alone. Tumour(s) size was assessed with calliper and its volume calculated using formula: V=L*W2/2 (V=tumour volume, L=length, W=width, L>W) and animals sacrificed as necessary according to the UCLA Animal Care guidelines.
Results and Discussion
Based on the analysis of literature data, we decided to synthesize library of p53–MDM2/MDMX inhibitors with enhanced cell permeability afforded by multiple Arg residues “woven” into the sequence of the parent peptide. Such modifications provide desired permeability effect and simultaneously limit the size/costs of making the potential drug candidate. Physiological stability/enzymatic resistance of these peptides was enhanced by the use of unusual- and (D)-amino acids as well as by stapling based on bis-cysteine bridges obtained by means of S-alkylation with selected bis-functional compounds. Employed modifications are schematically represented in Fig. 1.
Fig. 1.
Schematic representation of modifications in ArB analogues
As a starting point for our modifications we decided to use PMI-N8A peptide, the potent dual-specific antagonist of p53–MDM2/MDMX interactions with respective Kd values of 490 pM and 2.4 nM. PMI and its derivative PMI-N8A were originally developed in Lu’s group using phage display (Li et al., 2010a; Pazgier et al., 2009). Analogous all-D-peptide inhibitors called DPMI-α, DPMI-γ and DPMI-δ were also discovered by the same group using mirror-image phage display (Liu et al., 2010b; Liu et al., 2010a; Zhan et al., 2012) and subsequently tested in an animal model. However even proteolysis resistant DPMI-α required a liposomal delivery system to be active in vivo.
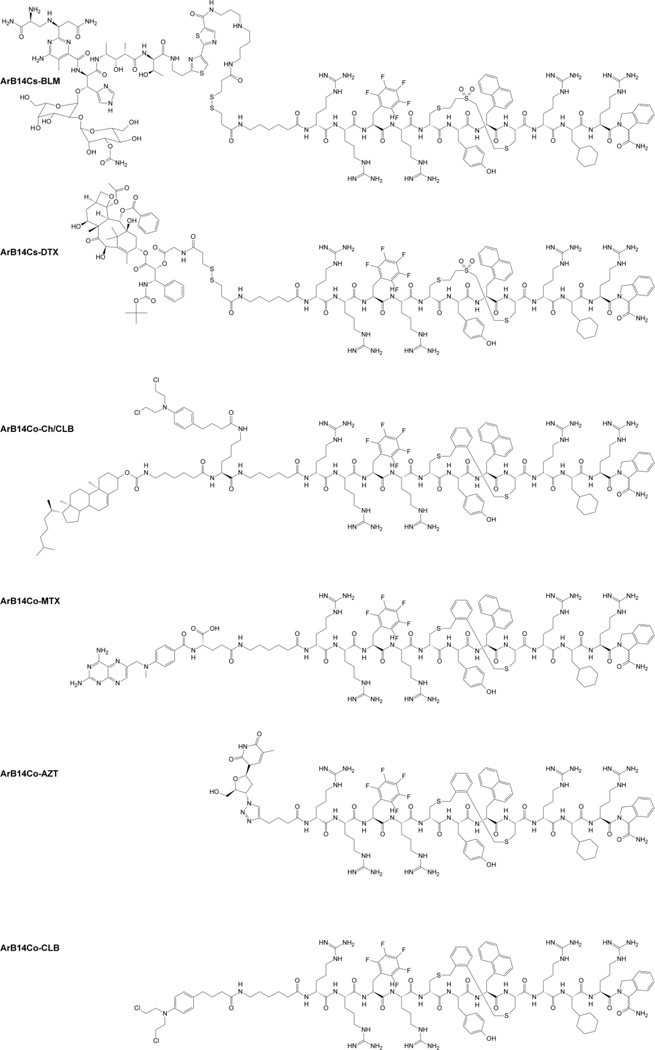
Based on analysis of p53/MDM2 and PMI-N8A/MDM2 crystal structures we placed various combinations of (L)- and (D)Cys residues in specific positions of the PMI sequence taking into account the importance of particular amino acids for peptide bioactivity (Table 1). These peptides were in turn stapled using selected bis-thiol-reactive compounds (Fig. 2) that fitted into the available spatial arrangements/dimensions, based on computer-aided molecular modeling. Pairs of cysteines were placed in positions: i,i+3; i,i+4; i,i+6; and i,i+7 of the peptides. Since DPMI-α presumably lacks cell permeating properties, in addition, we engineered several analogues with multiple (L)/(D)Arg residues to increase cell permeability, in combination with (D)- and non-proteinaceous amino acids to enhance stability and binding properties of the peptides. Selected analogues were also conjugated with cholesterol (Chol), bleomycin A5 (BLM), chlorambucil (CLB), docetaxel (DTX) and methotrexate (MTX), using the thiol-cleavable linker, dithiobis(succinimidyl propionate) (DSP, Pierce Biotechnology, Rockford, lL).
Table 1.
Analytical data for ArB analogues synthesized for these studies
| Name | Sequence | Composition | MW Calc / Found |
RT (min) | EC50 [µM] HCT-116 |
|---|---|---|---|---|---|
| PMI-N8A | Thr-Ser-Phe-Ala-Glu-Tyr-Trp-Ala-Leu-Leu-Ser-Pro | C67H94N14O18 | 1383.56 / 1383.59 | 38.17* | 178.8 |
| (15–29)p53 | Ser-Gln-Glu-Thr-Phe-Ser-Asp-Leu-Trp-Lys-Leu-Leu-Pro-Glu-Asn | C82H124N20O26 | 1806.00 / 1806.88 | 37.32* | NA |
| ArB14o | Thr-Ser-Phe-Ala-Cys-Tyr-Trp-Cys-Ala-Leu-Ser-Pro | C70H92N14O16S2 | 1449.71 / 1449.86 | 39.38* | 248.6 |
| ArB14Ao | Thr-Ser-PF5-Ala-Cys-Tyr-Na1-Cys-Ala-Cha-Ser-Pipz | C74H92N13O15F5S2 | 1562.74 / 1562.46 | 45.71* | 83.3 |
| ArB14Bo | Arg-Arg-Na2-Arg-Cys-Tyr-Na1-Cys-Arg-Cha-Arg-DISC | C97H132N28O13S2 | 1962.41 / 1962.36 | 37.05 | 10.7±0.7 |
| ArB14Co | Arg-Arg-PF5-Arg-Cys-Tyr-Na1-Cys-Arg-Cha-Arg-DISC | C93H125N28O13F5S2 | 2002.30 / 2003.02 | 36.94 | 11.2±1.2 |
| ArB14Cs | Arg-Arg-PF5-Arg-Cys-Tyr-Na1-Cys-Arg-Cha-Arg-DISC | C89H125N28O15F5S3 | 2018.32 / 2018.63 | 35.61 | 5.6±2.0 |
| ArB14C2s | Arg-Arg-PF3-Arg-Cys-Tyr-Na1-Cys-Arg-Cha-Arg-DISC | C90H129N28O15F3S3 | 1996.36 / 1997.12 | 37.13 | 14.3± 2.5 |
| ArB14Ck1 | Ac-Aib-Arg-PF3-Arg-Lys-Tyr-Trp-Cys-Arg-Cha-Arg | C80H119N26O14F3S | 1758.04 / 1758.35 | 36.21 | 82.14 |
| ArB14Ck2 | Ac-Arg-Arg-PF3-Arg-Lys-Tyr-Trp-Cys-Arg-Cha-Arg | C82H124N29O14F3S | 1829.12 / 1829.45 | 34.16 | 66.32 |
| ArB14Ck3 | Ac-Aib-Arg-PF5-Arg-Lys-Tyr-Trp-Cys-Arg-Cha-Arg | C79H115N26O14F5S | 1779.99 / 1780.75 | 34.79 | >250 |
| ArB14Ck4 | Ac-Arg-Arg-PF5-Arg-Lys-Tyr-Trp-Cys-Arg-Cha-Arg | C81H120N29O14F5S | 1851.08 / 1851.56 | 32.92 | 69.59 |
| ArB14Co-AZT | AZT-Arg-Arg-PF5-Arg-Cys-Tyr-Na1-Cys-Arg-Cha-Arg-DISC | C110H145N32O18F5S2 | 2362.65 / 2363.66 | 37.96 | 30.2±2.5 |
| ArB14Co-CLB | CLB-Ahx-Arg-Arg-PF5-Arg-Cys-Tyr-Na1-Cys-Arg-Cha-Arg-DISC | C113H153N30O15Cl2F5S2 | 2401.66 / 2402.78 | 56.41 | 7.5±1.1 |
| ArB14Co-Ch | Chol-Ahx-Lys-Ahx-Arg-Arg-PF5-Arg-Cys-Tyr-Na1-Cys-Arg-Cha-Arg-DISC | C139H203N32O18F5S2 | 2769.45 / 2769.14 | 57.99 | NT |
| ArB14Co-Ch/CLB | Chol-Ahx-LysCLB-Ahx-Arg-Arg-PF5-Arg-Cys-Tyr-Na1-Cys-Arg-Cha-Arg-DISC | C153H220N33O19Cl2F5S2 | 3055.65 / 3057.04 | 58.30 | 7.9±1.3 |
| ArB14Co-MTX | MTX-Ahx-Arg-Arg-PF5-Arg-Cys-Tyr-Na1-Cys-Arg-Cha-Arg-DISC | C119H156N37O18F5S2 | 2551.89 / 2553.86 | 46.51 | 6.9±0.4 |
| ArB14Co-TFAo | Tfa-Arg-Arg-PF5-Arg-Cys-Tyr-Na1-Cys-Arg-Cha-Arg-DISC | C95H124N28O14F8S2 | 2098.31 / 2098.91 | 38.00 | 18.7±3.4 |
| ArB14Co-BLM | BLM-SS-Ahx-Arg-Arg-PF5-Arg-Cys-Tyr-Na1-Cys-Arg-Cha-Arg-DISC | C162H231N48O37F5S6 | 3730.26 / 3731.46 | 38.96 | 5.5±1.1 |
| ArB14Co-DTX | DTX-SS-Ahx-Arg-Arg-PF5-Arg-Cys-Tyr-Na1-Cys-Arg-Cha-Arg-DISC | C150H198N31O31F5S4 | 3154.64 / 3154.61 | 46.89 | 21.6±1,4 |
| ArB14Cs-BLM | BLM-SS-Ahx-Arg-Arg-PF5-Arg-Cys-Tyr-Na1-Cys-Arg-Cha-Arg-DISC | C158H231N48O39F5S7 | 3746.28 / 3746.15 | 34.19 | 1.3±0.5 |
| ArB14Cs-DTX | DTX-SS-Ahx-Arg-Arg-PF5-Arg-Cys-Tyr-Na1-Cys-Arg-Cha-Arg-DISC | C146H198N31O33F5S5 | 3170.65 / 3171.19 | 45.90 | 8.4±1.5 |
| ArB14Dm | Thr-Ser-Phe-Arg-Arg-Tyr-Cys-Arg-Arg-Cys-Ser-Pro | C71H109N25O16S2 | 1632.92 / 1632.89 | 28.59* | 118.3 |
| ArB14Em | Thr-Ser-Phe-Arg-Arg-Tyr-Cys-Arg-Arg-Cys-Ser-Pro | C71H109N25O16S2 | 1632.92 / 1633.52 | 28.17* | 113.9 |
| ArB14Dmx | Thr-Ser-Phe-Arg-Arg-Tyr-Cys-Arg-Arg-Cys-Ser-Pro | C71H109N25O20S2 | 1696.92 / 1696.72 | 24.00* | 141.5 |
| ArB14Emx | Thr-Ser-Phe-Arg-Arg-Tyr-Cys-Arg-Arg-Cys-Ser-Pro | C71H109N25O20S2 | 1696.92 / 1697.21 | 24.79* | NA |
| ArB15Am | Thr-Ser-Phe-Ala-Cys-Tyr-Trp-Gly-Cys-Leu-Ser-Pro | C69H90N14O16S2 | 1435.68 / 1436.82 | 40.10 | NA |
| ArB15Bm | Thr-Ser-Phe-Ala-Cys-Tyr-Trp-Gly-Cys-Leu-Ser-Pro | C69H90N14O16S2 | 1435.68 / 1436.59 | 41.01 | NA |
| ArB15Cp | Thr-Ser-Phe-Ala-Cys-Tyr-Trp-Gly-Cys-Leu-Ser-Pro | C69H90N14O16S2 | 1435.68 / 1436.04 | 40.07 | >250 |
| ArB15Dp | Thr-Ser-Phe-Ala-Cys-Tyr-Trp-Gly-Cys-Leu-Ser-Pro | C69H90N14O16S2 | 1435.68 / 1436.67 | 39.99 | NA |
| ArB15Fo | Arg-Phe-Cys-Arg-Tyr-Trp-Cys-Arg-Leu-Arg-Pro | C78H113N25O12S2 | 1657.03 / 1657.86 | 32.64 | 49.64 |
| ArB15Gm | Arg-Phe-Cys-Arg-Tyr-Trp-Cys-Arg-Leu-Arg-Pro | C78H113N25O12S2 | 1657.03 / 1657.46 | 31.92 | 52.19 |
| ArB15FNico | Nic-Arg-Phe-Cys-Arg-Tyr-Trp-Cys-Arg-Leu-Arg-Pro | C84H116N26O13S2 | 1762.13 / 1763.63 | 35.97 | 189.0 |
| ArB15GNicm | Nic-Arg-Phe-Cys-Arg-Tyr-Trp-Cys-Arg-Leu-Arg-Pro | C84H116N26O13S2 | 1762.13 / 1762.51 | 35.36 | 126.4 |
| ArB15F/Gr | Arg-Phe-Cys-Arg-Tyr-Trp-Cys-Arg-Leu-Arg-Pro | C70H107N25O12S2 | 1554.89 / 1555.39 | 25.06* | >250 |
| ArB15F/GNicr | Nic-Arg-Phe-Cys-Arg-Tyr-Trp-Cys-Arg-Leu-Arg-Pro | C76H110N26O13S2 | 1659.99 / 1660.56 | 28.24* | 64.5 |
| ArB15F/Gs | Arg-Phe-Cys-Arg-Tyr-Trp-Cys-Arg-Leu-Arg-Pro | C74H113N25O14S3 | 1673.05 / 1674.05 | 27.06 | 49.9 |
| ArB15F/Gs-BLM | BLM-SS-Arg-Phe-Cys-Arg-Tyr-Trp-Cys-Arg-Leu-Arg-Pro | C137H208N44O37S7 | 3287.85 / 3288.42 | 29.49 | 1.1±0.7 |
| ArB15F/Gs-DTX | DTX-SS-Arg-Phe-Cys-Arg-Tyr-Trp-Cys-Arg-Leu-Arg-Pro | C125H175N27O31S5 | 2712.22 / 2712.60 | 43.77 | 62.2 |
| ArB15F/GNics | Nic-Arg-Phe-Cys-Arg-Tyr-Trp-Cys-Arg-Leu-Arg-Pro | C80H116N26O15S3 | 1778.14 / 1779.20 | 30.63 | 34.2±5.9 |
| ArB15F/Gt | [Arg-Phe-Cys-Arg-Tyr-Trp-Cys-Arg-Leu-Arg-Pro]2Ph(CH2)4 | C150H220N50O24S4 | 3235.95 / 3235.80 | 30.72 | NT |
| ArB15F/GNict | [Nic-Arg-Phe-Cys-Arg-Tyr-Trp-Cys-Arg-Leu-Arg-Pro]2Ph(CH2)4 | C162H226N52O26S4 | 3446.14 / 3447.36 | 33.62 | NT |
| ArB15F/G0/Nic | [Nic-Arg-Phe-Cys-Arg-Tyr-Trp-Cys-Arg-Leu-Arg-Pro]Ph(CH2)4 | C156H223N51O25S4 | 3341.05 / 3341.51 | 34.42* | NT |
| Mix | [Arg-Phe-Cys-Arg-Tyr-Trp-Cys-Arg-Leu-Arg-Pro] | 34.86* | |||
| ArB15Eb | Thr-Ser-Phe-Ala-Cys-Tyr-Trp-Gly-Cys-Leu-Ser-Pro | C76H96N14O16S2 | 1525.81 / 1526.03 | 46.95 | 162.9 |
| ArB15Hr | Ac-Thr-Ser-Phe-Cys-Glu-Tyr-Trp-Cys-Leu-Leu-Ser | C64H89N13O18S2 | 1392.60 / 1393.25 | 46.46 | NA |
| ArB15Ho | Ac-Thr-Ser-Phe-Cys-Glu-Tyr-Trp-Cys-Leu-Leu-Ser | C72H95N13O18S2 | 1494.74 / 1494.80 | 49.23 | NA |
| ArB15Hm | Ac-Thr-Ser-Phe-Cys-Glu-Tyr-Trp-Cys-Leu-Leu-Ser | C72H95N13O18S2 | 1494.74 / 1494.83 | 48.12 | NA |
| ArB15Hg | Ac-Thr-Ser-Phe-Cys-Glu-Tyr-Trp-Cys-Leu-Leu-Ser | C68H95N13O18S2 | 1446.69 / 1446.79 | 46.31 | NA |
| ArB15Hs | Ac-Thr-Ser-Phe-Cys-Glu-Tyr-Trp-Cys-Leu-Leu-Ser | C68H95N13O20S3 | 1510.75 / 1510.90 | 39.17 | NA |
| ArB17Ac | Thr-Cys-Phe-Ala-Gly-Tyr-Trp-Cys-Ala-Leu-Ser-Pro | C75H94N14O16S2 | 1511.78 / 1511.01 | 44.24 | NA |
| ArB17Bc | Thr-Cys-Phe-Ala-Gly-Tyr-Trp-Cys-Ala-Leu-Ser-Pro | C75H94N14O16S2 | 1511.78 / 1511.26 | 45.53 | NA |
| ArB18Ac | Thr-Cys-Phe-Ala-Gly-Tyr-Trp-Gly-Cys-Leu-Ser-Pro | C74H92N14O16S2 | 1497.75 / 1499.28 | 42.66 | NA |
| ArB18Bc | Thr-Cys-Phe-Ala-Gly-Tyr-Trp-Gly-Cys-Leu-Ser-Pro | C74H92N14O16S2 | 1497.75 / 1498.49 | 45.85 | NA |
| ArB18Cr | Arg-Arg-Phe-Cys-Arg-Tyr-Trp-Arg-Arg-Leu-Cys | C71H112N28O12S2 | 1613.97 / 1614.23 | 28.47* | >250 |
| ArB18Dr | Aib-Arg-Phe-Cys-Arg-Tyr-Trp-Arg-Arg-Leu-Cys | C69H107N25O12S2 | 1542.88 / 1543.26 | 29.75* | >250 |
| ArB18Cd | Arg-Arg-Phe-Cys-Arg-Tyr-Trp-Arg-Arg-Leu-Cys | C77H120N30O14S2 | 1754.11 / 1754.28 | 24.89 | NA |
| ArB18Dd | Aib-Arg-Phe-Cys-Arg-Tyr-Trp-Arg-Arg-Leu-Cys | C75H115N27O14S2 | 1683.03 / 1683.35 | 26.30 | NA |
| ArB18Ch | Arg-Arg-Phe-Cys-Arg-Tyr-Trp-Arg-Arg-Leu-Cys | C78H124N28O12S2 | 1710.14 / 1710.72 | 23.74 | NA |
| ArB18Dh | Aib-Arg-Phe-Cys-Arg-Tyr-Trp-Arg-Arg-Leu-Cys | C76H119N25O12S2 | 1639.06 / 1639.25 | 25.31 | NA |
| ArB18Er | Ac-Thr-Ser-Phe-Cys-Glu-Tyr-Trp-Ala-Leu-Leu-Cys | C64H89N13O17S2 | 1376.60 / 1377.82 | 49.03 | NA |
| ArB18Ed | Ac-Thr-Ser-Phe-Cys-Glu-Tyr-Trp-Ala-Leu-Leu-Cys | C70H97N15O19S2 | 1516.74 / 1516.20 | 42.72 | >250 |
| ArB18Ec | Ac-Thr-Ser-Phe-Cys-Glu-Tyr-Trp-Ala-Leu-Leu-Cys | C78H99N13O18S2 | 1570.84 / 1571.21 | 55.34 | NA |
| ArB18Es | Ac-Thr-Ser-Phe-Cys-Glu-Tyr-Trp-Ala-Leu-Leu-Cys | C68H95N13O19S3 | 1494.75 / 1494.67 | 47.06 | >250 |
| ArB18Ek | Ac-Thr-Ser-Phe-Cys-Glu-Tyr-Trp-Ala-Leu-Leu-Lys | C69H96N14O18S | 1441.66 / 1442.86 | 43.50 | 74.8 |
| ArB18Eh | Ac-Thr-Ser-Phe-Cys-Glu-Tyr-Trp-Ala-Leu-Leu-Cys | C71H101N13O17S2 | 1472.77 / 1472.99 | 53.28 | NA |
| ArBMim | [-Bmpa-bhPro-Arg-Ala-Trp-Arg-Lys-Inp-CH2CH2CH2CH2-S-]2 | C114H172N32O16S2 | 2310.94 / 2311.61 | 33.70* | 23.4±2.1 |
| BLM | 0.3±0.6 | ||||
| DTX | 74.5±3.7 |
Abbreviations: Aib-Aminoisobutyric acid; AZT-azidothymidine; BLM-Bleomycin A5; Bmpa-4-(Bromomethyl)phenylacetic acid; Cha-Cyclohexyl-(L)-alanine; CLB-Chlorambucil; Chol-Cholesterol; DISC -(R,S)-1,3-dihydro-2H-isoindole carboxylic acid; DTX-Docetaxel; Inp-isonipecotic acid; MTX-Methotrexate; Nic-Nicotinic acid; Na1-1-Naphthyl-(L)-alanine; Na2-2-Naphthyl-(L)-alanine; PF5-Pentafluoro-(L)-phenylalanine; bhPro –β-homo-(L)-proline; Pipz-Piperazine monoamide; PF3-p-Trifluoromethyl-(L)-phenylalanine; Tfa-Tfrifluoroacetic acid. D-amino acids are underlined cursive e.g. Cys.
All peptides were synthesized as C-terminal amides except for ArB14Ao – that was synthesized as C-terminal piperazine monoamide (Pipz). An analytical RP-HPLC was performed with reversed-phase C18 SymmetryShield™ column, 4.6×250 mm, 5 µm (Waters, Milford, MA) or (*) an reversed-phase C18 Vydac 218TP54 column, 4.6×250 mm, 5 µm (Grace, Deerfield, IL). Small letter at the end of peptide’s name indicates that peptide was bridged using: (b) 1-(bromomethyl)-3-[3-(bromomethyl)benzyl]benzene; (c) 1-(chloromethyl)-4-[4-(chloromethyl)phenoxy]benzene; (d) 2-chloro-N-(2-(2-chloro-acetylamino)-ethyl)-acetamide; (g) 1,4-dibromobutane; (h) 1,7-dibromoheptane; (m) 1,3-bis(bromomethyl)benzene; (o) 1,2-bis(bromomethyl)benzene; (p) 1,4-bis(bromomethyl)benzene; (r) reduced/not stapled peptide; (s) divinylsulfone (DVS); (t) peptide was stapled and dimerized using 1,2,4,5-tetrakis(bromomethyl)benzene; (x) thioether groups were oxidized to sulfones with OXONE®. For peptides ArB14k1÷k4 and ArB18Ek intramolecular bridge was achieved via side chain of Lys that was chloroacetylated and subsequently reacted with side chain of Cys forming thioether linker.
Fig. 2.
Cysteine(s) based staples used in the synthesis of ArB-analogues
Generally, all analogues were synthesized as C-terminal amides (except ArB14Ao) by the solid phase method using CEM Liberty automatic microwave peptide synthesizer (CEM Corporation Inc., Matthews, NC), applying 9-fluorenylmethyl-oxycarbonyl (Fmoc) chemistry (Fields and Noble, 1990) and standard, commercially available amino acid derivatives and reagents (EMD Biosciences, San Diego, CA and Chem-Impex International, Inc., Wood Dale, IL). Non-stapled peptides were purified by preparative reverse-phase high performance liquid chromatography (RP-HPLC) and their purity evaluated by matrix-assisted laser desorption ionization spectrometry (MALDI-MS) as well as analytical RP-HPLC (see Table 1 and Fig. 3).
Fig. 3.
The representative analytical RP-HPLC profile and corresponding MALDI-MS spectra obtained for analogue ArB14Cs
The stapling/S-alkylation reactions were carried out in solution using various protocols (for details see Experimental Section) using both, water-based solutions and “exclusively” organic solvents. Generally, for S-alkylation of synthetic linear peptides using bis-halogenomethyl-aryl derivatives and 2-chloro-N-(2-(2-chloro-acetylamino)-ethyl)-acetamide, we employed the protocol described by Timmerman and co-workers (Timmerman et al., 2005). Those reactions were carried out at ambient temperature in a solution of 50 mM ammonium bicarbonate (NH4HCO3) dissolved in a mixture of acetonitrile (ACN) and water. The synthesis of mono-S-alkylated compounds (ArB14Ck1-k4) was achieved using the same protocol. The stapling with divinylsulfone (DVS) was performed using a previously described protocol (Wilkinson et al., 2007) with conditions very similar to NH4HCO3/ACN method, with the only difference being the NH4HCO3 concentration (10 mM). The highest efficiency of stapling reaction(s) was observed for divinylsulfone (DVS), 2-chloro-N-(2-(2-chloro-acetylamino)-ethyl)-acetamide and 1,2- and 1,3-bis(bromomethyl)benzene bridged analogues. Simultaneously, 1-(bromomethyl)-3-[3-(bromomethyl)benzyl]benzene and 1-(chloromethyl)-4-[4-(chloromethyl)phenoxy]benzene staples were created with particularly low efficiency (yield <3%). Therefore in these cases (ArB15Eb, ArB17Ac and ArB18Bc) we also attempted to synthesize desired analogues using DMF/DMSO/Cs2CO3/TBAI system that we recently described (Micewicz et al., 2014). Unfortunately, this method did not improve synthetic efficiency. Notably, stapling with ortho- and meta-bis(bromomethyl)benzene can be also performed on resin using analogous protocol (see Experimental Section for details). The poor synthetic yields were also attributed to the stapling performed with 1,4-dibromobutane and 1,7-dibromoheptane that was carried out using a previously described 1,1,3,3-tetramethylguanidine (TMG) driven alkylation of thiol(s) in organic solvents that we adapted to peptides (Chua et al., 2015). Synthesis of oxidized analogues ArB14Dmx and ArB14Emx was performed using OXONE® (Duran et al., 2006) (Sigma-Aldrich, KHSO5×0.5 KHSO4×0.5 K2SO4) in a 75% solution of methyl alcohol in water employing 3 eq. of OXONE® per each thioether group. The reaction was carried out in room temperature for 3 h giving desired sulfones‘ containing products. For the analogues ArB14Co and ArB14Cs we additionally synthesized conjugates with known anticancer compounds, namely azidothymidine (AZT), bleomycin A5 (BLM), chlorambucil (CLB), docetaxel (DTX) and methotrexate (MTX) (see Table 1 and Fig. 4) to ascertain possible synergy effects. AZT conjugate was synthesized using Cu(I)-catalyzed azide-alkyne 1,3-dipolar Huisgen's cycloaddition (CuAAC, “click chemistry”). CLB and MTX were attached as N-terminal moieties directly on resin during SPPS. In the case of BLM and DTX conjugates, we employed direct conjugation of proper unprotected ArB peptides with the desired anticancer entity using bis-amine reactive crosslinking reagent, dithiobis(succinimidyl propionate) (DSP, Pierce Biotechnology, Rockford, lL) in DMSO solution (5 mg/ml, 18 h).
Fig. 4.
Structures of selected conjugates of ArBs prepared for this study
An initial evaluation of bioactivity of our compounds was carried out in vitro using growth inhibition assay (PrestoBlue™, Invitrogen, Carlsbad, CA) and HCT-116 colorectal cancer cell line that is sensitive to MDM2 inhibitor(s) treatment (Hu et al., 2007; Tovar et al., 2006; Vassilev et al., 2004). In our view, this simple approach provides more reliable data which take into account many factors like the compounds’ cell permeability, stability, binding potency, etc., and in this particular case is better suited for the screening than pure biophysical method(s) e.g. measurements of binding affinity, especially in the light of reported discrepancies between physicochemical/in vitro data and in vivo activity (Chang et al., 2013; Liu et al., 2010b; Liu et al., 2010a). Notably, we had previously employed this approach in the synthesis of drug candidates targeting other intracellular targets (Smac) with positive results (Micewicz et al., 2014). Examples of cell growth curves are presented in Fig. 5 and the in vitro screening data are summarized in Table 1. The most active compounds were found to be i,i+3 stapled analogues (ArB14Cs, ArB14Co, and ArB14Bo) containing a polyarginine motif (Fig. 6) with staple created either by 1,2-bis(bromomethyl)benzene (ArB14Co and ArB14Bo) or DVS (ArB14Cs) between (D)Cys in position 5 and (L)Cys in position 8. Notably, ArB14Cs seems to be ~2× more active than ArB14Co/ArB14Bo counterparts suggesting that the DVS-staple produces a sterically less constrained compound than the ortho-substituted-thio-methyl-benzene-linker. Moreover, the DVS staple is structurally/electrostatically similar to the Glu side chain that is present in this particular position in the parental compound (PMI-N8A), which has previously been reported important for overall activity (Li et al., 2010a). Position 3 seems to be also quite permissive as the activity observed for the pentafluoro-phenylalanine-containing analogue (ArB14Co) is almost identical to that with a 2-naphthyl-alanine residue in the same position (ArB14Bo). The polyArg motif consists of 5 arginine residues in positions 1, 2, 4, 9 and 11, with positions 1 and 9 being occupied by (D)Arg residues. Introduction of (D)Arg enantiomers in these particular positions allowed for formation of a polycationic face for the molecule, which was reported as necessary feature for efficient cell entry (Futaki et al., 2002; Wender et al., 2008), while simultaneously providing additional resistance to proteolytic cleavage afforded by unnatural/(D) amino acids. Notably, all ArB analogues lacking the polyArg motif showed poor anticancer properties. Modifications in the crucial hydrophobic triad: 19Phe→PheF5/2-Nal, 23Trp→1-Nal, and 26Leu→Cha were prompted by reports that certain modifications enhancing hydrophobicity may result in analogues with enhanced properties (Liu et al., 2010b; Zhan et al., 2012), with an additional benefit being enhanced resistance to oxidative degradation afforded by elimination from the molecule of the Trp residue which is prone to such process (Simat and Steinhart, 1998). Interestingly, in the case of 19Phe→PheCF3 substitution effects seem to be less pronounced than for 19Phe→PheF5 analogue, suggesting that the additional para-trifluoromethyl-group may be associated with limited steric hindrance, which is contrary to previously reported results (Zhan et al., 2012), although such conclusion may be partially incorrect since compared peptide sequences are not identical. The observed in vitro anticancer effects were sequence specific and seem to be independent on polArg toxicity as various other polycationic sequences synthesized for this study showed limited potency (e.g. ArB14Dm, ArB15Em, ArB15Fo, ArB15FNico, etc.). In certain cases, conjugation with known anticancer entities seems to be beneficial as well. Specifically, BLM-conjugates had improved anticancer activity (ArB14Cs-BLM versus ArB14Cs; ArB14Co-BLM versus ArB14Co). A similar, but less pronounced, trend was also observed for MTX- and CLB-conjugated counterparts.
Fig. 5.
Cell viability data for selected compounds. Compounds possess following EC50 values (µM): ArB14Co:11.2±1.2; ArB14Bo:10.7±0.7; ArB14Co-CLB:7.5±1.1; ArB14Co-MTX:6.9±0.4; ArB14Co-AZT:30.2±2.5
Fig. 6.
The most active ArB analogues
To ascertain whether our ArB compounds possess therapeutic potential in vivo, we performed studies in a murine subcutaneous tumour engraftment model (HCT-116/SCID). Based on the in vitro results and ease of synthesis/availability, we selected stapled peptide ArB14Co as the drug candidate for in vivo experiments. The treatment of cancer-bearing animals with this analogue resulted in potent anticancer effects (Fig. 7). Animals treated with 10 doses of the compound at a concentration of 3.0 mg/kg showed ~14.6 days delay in tumour growth. Interestingly, 2.5× higher dose (7.5 mg/kg) showed similar efficacy. Since all reported to date in vivo data for p53–MDM2/MDMX peptide inhibitors utilized different murine models, direct comparison between them should be rather cautious due to inherent differences in tumour growth rates, physiology, etc. Nonetheless, comparison with recently reported results for ATSP-7041 (Chang et al., 2013) (Aileron Therapeutics, Inc.) seems to be very favorable as therapeutic dosage for ATSP-7041 was in much higher range (15–30 mg/kg). Interestingly, in an MDM2-amplified osteosarcoma xenograft model (SJSA-1) anticancer activity of ATSP-7041, like ArB14Co, showed no dose response as the in vivo activitiy for 15 mg/kg and 30 mg/kg doses was basically the same, providing roughly ~4–5 days of tumour growth delay, which is ~3× less than for ArB14Co administered at significantly lower dose (3.0 mg/kg). Activity of ATSP-7041 in the MDMX overexpressing MCF-7 human breast cancer xenograft model is more promising, however the dosage utilized is still high (20–30 mg/kg). Comparison of ArB14Co with targeted liposomal formulation of DPMI-α (Liu et al., 2010a) (human glioma cell line U87/nude BALB/c model) is somewhat difficult as presented tumour volumes are reported in relative scale and tumour growth delay values are not reported, Nonetheless, presented results suggest significant activity, at least at 7.5 mg/kg dose. However, in this case ArB14Co may possess an advantage as well since it does not require liposomal delivery system. Notably, naked DPMI-α was virtually inactive in reported in vitro experimets in concentrations as high as 100 µM.
Fig. 7.
In vivo results from subcutaneous engraftment mouse model (HCT-116/SCID). Administration of ArB14Co results in ~14.6 days of tumour growth delay
Conclusions
A new family of anticancer, cell-permeating, stapled peptides was synthesized, characterized and screened for anticancer activity against human colorectal cancer cell line, HCT-116. Selected analogue, ArB14Co was tested in vivo showing potent anticancer activity at the low dose (3.0 mg/kg). Utilized stapling/modification strategy yielding short, cell permeating analogues, containing polycationic motif woven into parental sequence of peptide(s) seems to be a suitable approach in the development of p53–MDM2/MDMX bridged peptide inhibitors.
Acknowledgements
This project was partially supported by funds from the Adams and Burnham endowments provided by the Dean’s Office of the David Geffen School of Medicine at UCLA (PR) and the NIH/NIAID award 5U19AI067769 (EDM and WHM).
Footnotes
Conflict of interest and Ethical Standards Authors declare that this article content has no conflicts of interest.
Statement of Informed Consent The article does not contain any studies in patients by any of the authors.
Statement of Human and Animal Rights This article does not contain studies involving human subjects. All animal experiments were approved by the UCLA Animal Care and Use Committee and conformed to local and national guidelines.
Contributor Information
Ewa D. Micewicz, Department of Radiation Oncology, University of California at Los Angeles, 10833 Le Conte Avenue, Los Angeles, CA 90095, USA
Shantanu Sharma, Materials and Process Simulation Center, California Institute of Technology, 1200 East California Boulevard, Pasadena, CA 91125, USA.
Alan J. Waring, Department of Medicine, Los Angeles Biomedical Research Institute at Harbor-UCLA, Medical Center, 1000 West Carson Street, Torrance, CA 90502, USA
Hai T. Luong, Department of Analytical Operations, Gilead Sciences, Inc., 4049 Avenida de la Plata, Oceanside CA, 92056, USA
William H. McBride, Department of Radiation Oncology, University of California at Los Angeles, 10833 Le Conte Avenue, Los Angeles, CA 90095, USA
Piotr Ruchala, Email: pruchala@mednet.ucla.edu, Department of Psychiatry and Biobehavioral Sciences, University of California at Los Angeles, 760 Westwood Plaza, Los Angeles, CA 90024, USA.
References
- Almeida AM, Li R, Gellman SH. Parallel beta-sheet secondary structure is stabilized and terminated by interstrand disulfide cross-linking. J. Am. Chem. Soc. 2012;134:75–78. doi: 10.1021/ja208856c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini A, Cendron L, Chen S, et al. Bicyclic peptide inhibitor reveals large contact interface with a protease target. ACS Chem. Biol. 2012a;7:817–821. doi: 10.1021/cb200478t. [DOI] [PubMed] [Google Scholar]
- Angelini A, Diderich P, Morales-Sanfrutos J, et al. Chemical macrocyclization of peptides fused to antibody Fc fragments. Bioconjug. Chem. 2012b;23:1856–1863. doi: 10.1021/bc300184m. [DOI] [PubMed] [Google Scholar]
- Angelini A, Morales-Sanfrutos J, Diderich P, Chen S, Heinis C. Bicyclization and tethering to albumin yields long-acting peptide antagonists. J. Med. Chem. 2012c;55:10187–10197. doi: 10.1021/jm301276e. [DOI] [PubMed] [Google Scholar]
- Baeriswyl V, Calzavarini S, Gerschheimer C, Diderich P, Angelillo-Scherrer A, Heinis C. Development of a selective peptide macrocycle inhibitor of coagulation factor XII toward the generation of a safe antithrombotic therapy. J. Med. Chem. 2013;56:3742–3746. doi: 10.1021/jm400236j. [DOI] [PubMed] [Google Scholar]
- Baeriswyl V, Heinis C. Phage selection of cyclic peptide antagonists with increased stability toward intestinal proteases. Protein Eng. Des. Sel. 2013a;26:81–89. doi: 10.1093/protein/gzs085. [DOI] [PubMed] [Google Scholar]
- Baeriswyl V, Heinis C. Polycyclic peptide therapeutics. Chem Med Chem. 2013b;8:377–384. doi: 10.1002/cmdc.201200513. [DOI] [PubMed] [Google Scholar]
- Baeriswyl V, Rapley H, Pollaro L, et al. Bicyclic peptides with optimized ring size inhibit human plasma kallikrein and its orthologues while sparing paralogous proteases. Chem Med Chem. 2012;7:1173–1176. doi: 10.1002/cmdc.201200071. [DOI] [PubMed] [Google Scholar]
- Bautista AD, Appelbaum JS, Craig CJ, Michel J, Schepartz A. Bridged beta(3)-peptide inhibitors of p53-hDM2 complexation: correlation between affinity and cell permeability. J. Am. Chem. Soc. 2010;132:2904–2906. doi: 10.1021/ja910715u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellotto S, Chen S, Rentero RI, Wegner HA, Heinis C. Phage selection of photoswitchable peptide ligands. J. Am. Chem. Soc. 2014;136:5880–5883. doi: 10.1021/ja501861m. [DOI] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J. Am. Chem. Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal F, Wade M, Godes M, et al. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 2010;18:411–422. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird GH, Madani N, Perry AF, et al. Hydrocarbon double-stapling remedies the proteolytic instability of a lengthy peptide therapeutic. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14093–14098. doi: 10.1073/pnas.1002713107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell HE, Grubbs RH. Highly efficient synthesis of covalently cross-linked peptide helices by ring-closing metathesis. Angew. Chem. Int. Ed Engl. 1998;37:3281–3284. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Blackwell HE, Sadowsky JD, Howard RJ, et al. Ring-closing metathesis of olefinic peptides: design, synthesis, and structural characterization of macrocyclic helical peptides. J. Org. Chem. 2001;66:5291–5302. doi: 10.1021/jo015533k. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol. Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat. Rev. Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Quah ST, Jong J, et al. Stapled peptides with improved potency and specificity that activate p53. ACS Chem. Biol. 2013;8:506–512. doi: 10.1021/cb3005148. [DOI] [PubMed] [Google Scholar]
- Brunel FM, Dawson PE. Synthesis of constrained helical peptides by thioether ligation: application to analogs of gp41. Chem. Commun. (Camb.) 2005:2552–2554. doi: 10.1039/b419015g. [DOI] [PubMed] [Google Scholar]
- Byrne MP, Stites WE. Chemically crosslinked protein dimers: stability and denaturation effects. Protein Sci. 1995;4:2545–2558. doi: 10.1002/pro.5560041211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezas E, Satterthwait AC. The hydrogen bond mimic approach: solid-phase synthesis of a peptide stabilized as an a-helix with a hydrazone link. J. Am. Chem. Soc. 1999;121:3862–3875. [Google Scholar]
- Cardoso RM, Brunel FM, Ferguson S, et al. Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10. J. Mol. Biol. 2007;365:1533–1544. doi: 10.1016/j.jmb.2006.10.088. [DOI] [PubMed] [Google Scholar]
- Chang YS, Graves B, Guerlavais V, et al. Stapled alpha-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E3445–E3454. doi: 10.1073/pnas.1303002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Bertoldo D, Angelini A, Pojer F, Heinis C. Peptide ligands stabilized by small molecules. Angew. Chem. Int. Ed Engl. 2014a;53:1602–1606. doi: 10.1002/anie.201309459. [DOI] [PubMed] [Google Scholar]
- Chen S, Gopalakrishnan R, Schaer T, et al. Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides. Nat. Chem. 2014b;6:1009–1016. doi: 10.1038/nchem.2043. [DOI] [PubMed] [Google Scholar]
- Chen S, Morales-Sanfrutos J, Angelini A, Cutting B, Heinis C. Structurally diverse cyclisation linkers impose different backbone conformations in bicyclic peptides. Chembiochem. 2012;13:1032–1038. doi: 10.1002/cbic.201200049. [DOI] [PubMed] [Google Scholar]
- Chen S, Rentero Rebollo I, Buth SA, et al. Bicyclic peptide ligands pulled out of cysteine-rich peptide libraries. J. Am. Chem. Soc. 2013;135:6562–6569. doi: 10.1021/ja400461h. [DOI] [PubMed] [Google Scholar]
- Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nat. Rev. Clin. Oncol. 2011;8:25–37. doi: 10.1038/nrclinonc.2010.174. [DOI] [PubMed] [Google Scholar]
- Chua K, Fung E, Micewicz ED, Ganz T, Nemeth E, Ruchala P. Small cyclic agonists of iron regulatory hormone hepcidin. Bioorg. Med. Chem. Lett. 2015 doi: 10.1016/j.bmcl.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danovi D, Meulmeester E, Pasini D, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol. Cell Biol. 2004;24:5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Araujo AD, Hoang HN, Kok WM, et al. Comparative alpha-helicity of cyclic pentapeptides in water. Angew. Chem. Int. Ed Engl. 2014;53:6965–6969. doi: 10.1002/anie.201310245. [DOI] [PubMed] [Google Scholar]
- de Graaf P, Little NA, Ramos YF, Meulmeester E, Letteboer SJ, Jochemsen AG. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J. Biol. Chem. 2003;278:38315–38324. doi: 10.1074/jbc.M213034200. [DOI] [PubMed] [Google Scholar]
- de Souza-Pinto NC, Harris CC, Bohr VA. p53 functions in the incorporation step in DNA base excision repair in mouse liver mitochondria. Oncogene. 2004;23:6559–6568. doi: 10.1038/sj.onc.1207874. [DOI] [PubMed] [Google Scholar]
- Duran FJ, Ghini AA, Coirini H, Burton G. Synthesis of 6-thia analogs of the natural neurosteroid allopregnanolone. Tetrahedron. 2006;62:4762–4768. [Google Scholar]
- Esteller M, Cordon-Cardo C, Corn PG, et al. p14ARF silencing by promoter hypermethylation mediates abnormal intracellular localization of MDM2. Cancer Res. 2001;61:2816–2821. [PubMed] [Google Scholar]
- Fields GB, Noble RL. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 1990;35:161–214. doi: 10.1111/j.1399-3011.1990.tb00939.x. [DOI] [PubMed] [Google Scholar]
- Fujimoto K, Kajino M, Inouye M. Development of a series of cross-linking agents that effectively stabilize alpha-helical structures in various short peptides. Chem. Eur. J. 2008;14:857–863. doi: 10.1002/chem.200700843. [DOI] [PubMed] [Google Scholar]
- Futaki S, Nakase I, Suzuki T, Youjun Z, Sugiura Y. Translocation of branched-chain arginine peptides through cell membranes: flexibility in the spatial disposition of positive charges in membrane-permeable peptides. Biochemistry. 2002;41:7925–7930. doi: 10.1021/bi0256173. [DOI] [PubMed] [Google Scholar]
- Galande AK, Bramlett KS, Burris TP, Wittliff JL, Spatola AF. Thioether side chain cyclization for helical peptide formation: inhibitors of estrogen receptor-coactivator interactions. J. Pept. Res. 2004;63:297–302. doi: 10.1111/j.1399-3011.2004.00152.x. [DOI] [PubMed] [Google Scholar]
- Garbe JC, Holst CR, Bassett E, Tlsty T, Stampfer MR. Inactivation of p53 function in cultured human mammary epithelial cells turns the telomere-length dependent senescence barrier from agonescence into crisis. Cell Cycle. 2007;6:1927–1936. doi: 10.4161/cc.6.15.4519. [DOI] [PubMed] [Google Scholar]
- Geistlinger TR, Guy RK. An inhibitor of the interaction of thyroid hormone receptor beta and glucocorticoid interacting protein 1. J. Am. Chem. Soc. 2001;123:1525–1526. doi: 10.1021/ja005549c. [DOI] [PubMed] [Google Scholar]
- Geistlinger TR, Guy RK. Novel selective inhibitors of the interaction of individual nuclear hormone receptors with a mutually shared steroid receptor coactivator 2. J. Am. Chem. Soc. 2003;125:6852–6853. doi: 10.1021/ja0348391. [DOI] [PubMed] [Google Scholar]
- Ghadiri MR, Choi C. Secondary structure nucleation in peptides. Transition metal ion stabilized alpha-helices. J. Am. Chem. Soc. 1990;112:1630–1632. [Google Scholar]
- Green DR, Chipuk JE. p53 and metabolism: Inside the TIGAR. Cell. 2006;126:30–32. doi: 10.1016/j.cell.2006.06.032. [DOI] [PubMed] [Google Scholar]
- Guo Z, Streu K, Krilov G, Mohanty U. Probing the origin of structural stability of single and double stapled p53 peptide analogs bound to MDM2. Chem. Biol. Drug Des. 2014;83:631–642. doi: 10.1111/cbdd.12284. [DOI] [PubMed] [Google Scholar]
- Haney CM, Loch MT, Horne WS. Promoting peptide alpha-helix formation with dynamic covalent oxime side-chain cross-links. Chem. Commun. (Camb.) 2011;47:10915–10917. doi: 10.1039/c1cc12010g. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Heinis C, Rutherford T, Freund S, Winter G. Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat. Chem. Biol. 2009;5:502–507. doi: 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]
- Helton ES, Chen X. p53 modulation of the DNA damage response. J. Cell Biochem. 2007;100:883–896. doi: 10.1002/jcb.21091. [DOI] [PubMed] [Google Scholar]
- Holland-Nell K, Meldal M. Maintaining biological activity by using triazoles as disulfide bond mimetics. Angew. Chem. Int. Ed Engl. 2011;50:5204–5206. doi: 10.1002/anie.201005846. [DOI] [PubMed] [Google Scholar]
- Houston ME, Jr, Gannon CL, Kay CM, Hodges RS. Lactam bridge stabilization of alpha-helical peptides: ring size, orientation and positional effects. J. Pept. Sci. 1995;1:274–282. doi: 10.1002/psc.310010408. [DOI] [PubMed] [Google Scholar]
- Hu B, Gilkes DM, Chen J. Efficient p53 activation and apoptosis by simultaneous disruption of binding to MDM2 and MDMX. Cancer Res. 2007;67:8810–8817. doi: 10.1158/0008-5472.CAN-07-1140. [DOI] [PubMed] [Google Scholar]
- Ingale S, Dawson PE. On resin side-chain cyclization of complex peptides using CuAAC. Org. Lett. 2011;13:2822–2825. doi: 10.1021/ol200775h. [DOI] [PubMed] [Google Scholar]
- Jackson DY, King DS, Chmielewski J, Singh S, Schultz PG. General approach to the synthesis of short a-helical peptides. J. Am. Chem. Soc. 1991;113:9391–9392. [Google Scholar]
- Jo H, Meinhardt N, Wu Y, et al. Development of alpha-helical calpain probes by mimicking a natural protein-protein interaction. J. Am. Chem. Soc. 2012;134:17704–17713. doi: 10.1021/ja307599z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Zhan Q, el-Deiry WS, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Kawamoto SA, Coleska A, Ran X, Yi H, Yang CY, Wang S. Design of triazole-stapled BCL9 alpha-helical peptides to target the beta-catenin/B-cell CLL/lymphoma 9 (BCL9) protein-protein interaction. J. Med. Chem. 2012;55:1137–1146. doi: 10.1021/jm201125d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp DS, McNamara M. Conformationally restricted cyclic nonapeptides derived from L-cysteine and LL-3-Amino-2-piperidone-6-carboxylic acid (LL-Acp), a potent b-turn-inducing dipeptide analogue. J. Org. Chem. 1985;50:5834–5838. [Google Scholar]
- Khoo KH, Verma CS, Lane DP. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 2014;13:217–236. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- Kim YW, Grossmann TN, Verdine GL. Synthesis of all-hydrocarbon stapled alpha-helical peptides by ring-closing olefin metathesis. Nat. Protoc. 2011;6:761–771. doi: 10.1038/nprot.2011.324. [DOI] [PubMed] [Google Scholar]
- Kim YW, Verdine GL. Stereochemical effects of all-hydrocarbon tethers in i,i+4 stapled peptides. Bioorg. Med. Chem. Lett. 2009;19:2533–2536. doi: 10.1016/j.bmcl.2009.03.022. [DOI] [PubMed] [Google Scholar]
- Kuerbitz SJ, Plunkett BS, Walsh WV, Kastan MB. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Lau YH, de Andrade P, McKenzie GJ, Venkitaraman AR, Spring DR. Linear aliphatic dialkynes as alternative linkers for double-click stapling of p53-derived peptides. Chembiochem. 2014a;15:2680–2683. doi: 10.1002/cbic.201402374. [DOI] [PubMed] [Google Scholar]
- Lau YH, de Andrade P, Skold N, et al. Investigating peptide sequence variations for 'double-click' stapled p53 peptides. Org. Biomol. Chem. 2014b;12:4074–4077. doi: 10.1039/c4ob00742e. [DOI] [PubMed] [Google Scholar]
- Lau YH, de Andrade P, Wu Y, Spring DR. Peptide stapling techniques based on different macrocyclisation chemistries. Chem. Soc. Rev. 2015a;44:91–102. doi: 10.1039/c4cs00246f. [DOI] [PubMed] [Google Scholar]
- Lau YH, de Andrade P, Quah ST, et al. Functionalised staple linkages for modulating the cellular activity of stapled peptides. Chem. Sci. 2014c;5:1804–1809. [Google Scholar]
- Lau YH, Wu Y, de Andrade P, Galloway WR, Spring DR. A two-component 'double-click' approach to peptide stapling. Nat. Protoc. 2015b;10:585–594. doi: 10.1038/nprot.2015.033. [DOI] [PubMed] [Google Scholar]
- Laurie NA, Donovan SL, Shih CS, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death. Differ. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- Leduc AM, Trent JO, Wittliff JL, et al. Helix-stabilized cyclic peptides as selective inhibitors of steroid receptor-coactivator interactions. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11273–11278. doi: 10.1073/pnas.1934759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Pazgier M, Li C, et al. Systematic mutational analysis of peptide inhibition of the p53-MDM2/MDMX interactions. J. Mol. Biol. 2010a;398:200–213. doi: 10.1016/j.jmb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Pazgier M, Li J, et al. Limitations of peptide retro-inverso isomerization in molecular mimicry. J. Biol. Chem. 2010b;285:19572–19581. doi: 10.1074/jbc.M110.116814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Pazgier M, Liu M, Lu WY, Lu W. Apamin as a template for structure-based rational design of potent peptide activators of p53. Angew. Chem. Int. Ed Engl. 2009;48:8712–8715. doi: 10.1002/anie.200904550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lozano G. Molecular pathways: targeting Mdm2 and Mdm4 in cancer therapy. Clin. Cancer Res. 2013;19:34–41. doi: 10.1158/1078-0432.CCR-12-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindman S, Lindeberg G, Gogoll A, Nyberg F, Karlen A, Hallberg A. Synthesis, receptor binding affinities and conformational properties of cyclic methylenedithioether analogues of angiotensin II. Bioorg. Med. Chem. 2001;9:763–772. doi: 10.1016/s0968-0896(00)00294-7. [DOI] [PubMed] [Google Scholar]
- Liu M, Li C, Pazgier M, et al. D-peptide inhibitors of the p53-MDM2 interaction for targeted molecular therapy of malignant neoplasms. Proc. Natl. Acad. Sci. U. S. A. 2010a;107:14321–14326. doi: 10.1073/pnas.1008930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Pazgier M, Li C, Yuan W, Li C, Lu W. A left-handed solution to peptide inhibition of the p53-MDM2 interaction. Angew. Chem. Int. Ed Engl. 2010b;49:3649–3652. doi: 10.1002/anie.201000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long YQ, Huang SX, Zawahir Z, et al. Design of cell-permeable stapled peptides as HIV-1 integrase inhibitors. J. Med. Chem. 2013;56:5601–5612. doi: 10.1021/jm4006516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Sherr CJ. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Madden MM, Muppidi A, Li Z, Li X, Chen J, Lin Q. Synthesis of cell-permeable stapled peptide dual inhibitors of the p53-Mdm2/Mdmx interactions via photoinduced cycloaddition. Bioorg. Med. Chem. Lett. 2011;21:1472–1475. doi: 10.1016/j.bmcl.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhumalar A, Lee HJ, Brown CJ, Lane D, Verma C. Design of a novel MDM2 binding peptide based on the p53 family. Cell Cycle. 2009;8:2828–2836. doi: 10.4161/cc.8.17.9516. [DOI] [PubMed] [Google Scholar]
- Marine JC. MDM2 and MDMX in cancer and development. Curr. Top. Dev. Biol. 2011;94:45–75. doi: 10.1016/B978-0-12-380916-2.00003-6. [DOI] [PubMed] [Google Scholar]
- Marine JC, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. J. Cell Sci. 2007;120:371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Menendez D, Inga A, Jordan JJ, Resnick MA. Changing the p53 master regulatory network: ELEMENTary, my dear Mr Watson. Oncogene. 2007;26:2191–2201. doi: 10.1038/sj.onc.1210277. [DOI] [PubMed] [Google Scholar]
- Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat. Rev. Cancer. 2009;9:724–737. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- Micewicz ED, Luong HT, Jung CL, Waring AJ, McBride WH, Ruchala P. Novel dimeric Smac analogs as prospective anticancer agents. Bioorg. Med. Chem. Lett. 2014;24:1452–1457. doi: 10.1016/j.bmcl.2014.02.024. [DOI] [PubMed] [Google Scholar]
- Migliorini D, Danovi D, Colombo E, Carbone R, Pelicci PG, Marine JC. Hdmx recruitment into the nucleus by Hdm2 is essential for its ability to regulate p53 stability and transactivation. J. Biol. Chem. 2002a;277:7318–7323. doi: 10.1074/jbc.M108795200. [DOI] [PubMed] [Google Scholar]
- Migliorini D, Lazzerini DE, Danovi D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol. Cell Biol. 2002b;22:5527–5538. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molchadsky A, Shats I, Goldfinger N, et al. p53 plays a role in mesenchymal differentiation programs, in a cell fate dependent manner. PLoS. One. 2008;3:e3707. doi: 10.1371/journal.pone.0003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- Muppidi A, Doi K, Edwardraja S, et al. Rational design of proteolytically stable, cell-permeable peptide-based selective Mcl-1 inhibitors. J. Am. Chem. Soc. 2012;134:14734–14737. doi: 10.1021/ja306864v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muppidi A, Li X, Chen J, Lin Q. Conjugation of spermine enhances cellular uptake of the stapled peptide-based inhibitors of p53-Mdm2 interaction. Bioorg. Med. Chem. Lett. 2011a;21:7412–7415. doi: 10.1016/j.bmcl.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muppidi A, Wang Z, Li X, Chen J, Lin Q. Achieving cell penetration with distance-matching cysteine cross-linkers: a facile route to cell-permeable peptide dual inhibitors of Mdm2/Mdmx. Chem. Commun. (Camb.) 2011b;47:9396–9398. doi: 10.1039/c1cc13320a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat. Rev. Mol. Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- Osapay G, Taylor JW. Multicyclic polypeptide model compounds. 2. Synthesis and conformational properties of a highly .alpha.-helical uncosapeptide constrained by three side-chain to side-chain lactam bridges. J. Am. Chem. Soc. 1992;114:6966–6973. [Google Scholar]
- Pal S, Datta K, Mukhopadhyay D. Central role of p53 on regulation of vascular permeability factor/vascular endothelial growth factor (VPF/VEGF) expression in mammary carcinoma. Cancer Res. 2001;61:6952–6957. [PubMed] [Google Scholar]
- Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol. Cell Biol. 2003;23:5113–5121. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazgier M, Liu M, Zou G, et al. Structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4665–4670. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phelan JC, Skelton NJ, Braisted AC, McDowell RS. A general method for constraining short peptides to an a-helical conformation. J. Am. Chem. Soc. 1997;119:455–460. [Google Scholar]
- Pollaro L, Raghunathan S, Morales-Sanfrutos J, Angelini A, Kontos S, Heinis C. Bicyclic peptides conjugated to an albumin-binding tag diffuse efficiently into solid tumors. Mol. Cancer Ther. 2014 doi: 10.1158/1535-7163.MCT-14-0534. [DOI] [PubMed] [Google Scholar]
- Popowicz GM, Czarna A, Holak TA. Structure of the human Mdmx protein bound to the p53 tumor suppressor transactivation domain. Cell Cycle. 2008;7:2441–2443. doi: 10.4161/cc.6365. [DOI] [PubMed] [Google Scholar]
- Qin Q, Baudry M, Liao G, Noniyev A, Galeano J, Bi X. A novel function for p53: regulation of growth cone motility through interaction with Rho kinase. J. Neurosci. 2009;29:5183–5192. doi: 10.1523/JNEUROSCI.0420-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentero-Rebollo I, Sabisz M, Baeriswyl V, Heinis C. Identification of target-binding peptide motifs by high-throughput sequencing of phage-selected peptides. Nucleic Acids Res. 2014;42:e169. doi: 10.1093/nar/gku940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedinger C, McDonnell JM. Inhibitors of MDM2 and MDMX: a structural perspective. Future Med. Chem. 2009;1:1075–1094. doi: 10.4155/fmc.09.75. [DOI] [PubMed] [Google Scholar]
- Riemenschneider MJ, Buschges R, Wolter M, et al. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res. 1999;59:6091–6096. [PubMed] [Google Scholar]
- Roger L, Gadea G, Roux P. Control of cell migration: a tumour suppressor function for p53? Biol. Cell. 2006;98:141–152. doi: 10.1042/BC20050058. [DOI] [PubMed] [Google Scholar]
- Ruan FQ, Chen YQ, Hopkins PB. Metal ion-enhanced helicity in synthetic peptides containing unnatural, metal-ligating residues. J. Am. Chem. Soc. 1990;112:9403–9404. [Google Scholar]
- Saha MN, Qiu L, Chang H. Targeting p53 by small molecules in hematological malignancies. J. Hematol. Oncol. 2013;6:23. doi: 10.1186/1756-8722-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafmeister CE, Po J, Verdine GL. An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J. Am. Chem. Soc. 2000;122:5891–5892. [Google Scholar]
- Scrima M, Le Chevalier-Isaad A, Rovero P, Papini AM, Chorev M, D'Ursi AM. CuI-catalyzed azide-alkyne intramolecular i-to-(i+4) side-chain-to-side-chain cyclization promotes the formation of helix-like secondary structures. Eur. J. Org. Chem. 2010;2010:446–457. [Google Scholar]
- Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nat. Rev. Mol. Cell Biol. 2005;6:44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat. Rev. Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Weber JD. The ARF/p53 pathway. Curr. Opin. Genet. Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- Simat TJ, Steinhart H. Oxidation of Free Tryptophan and Tryptophan Residues in Peptides and Proteins. J. Agric. Food Chem. 1998;46:490–498. doi: 10.1021/jf970818c. [DOI] [PubMed] [Google Scholar]
- Singh K, Mogare D, Giridharagopalan RO, Gogiraju R, Pande G, Chattopadhyay S. p53 target gene SMAR1 is dysregulated in breast cancer: its role in cancer cell migration and invasion. PLoS. One. 2007;2:e660. doi: 10.1371/journal.pone.0000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommers JA, Sharma S, Doherty KM, et al. p53 modulates RPA-dependent and RPA-independent WRN helicase activity. Cancer Res. 2005;65:1223–1233. doi: 10.1158/0008-5472.CAN-03-0231. [DOI] [PubMed] [Google Scholar]
- Spokoyny AM, Zou Y, Ling JJ, Yu H, Lin YS, Pentelute BL. A perfluoroaryl-cysteine S(N)Ar chemistry approach to unprotected peptide stapling. J. Am. Chem. Soc. 2013;135:5946–5949. doi: 10.1021/ja400119t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- Szewczuk Z, Rebholz KL, Rich DH. Synthesis and biological activity of new conformationally restricted analogues of pepstatin. Int. J. Pept. Protein Res. 1992;40:233–242. doi: 10.1111/j.1399-3011.1992.tb00296.x. [DOI] [PubMed] [Google Scholar]
- Taura M, Eguma A, Suico MA, et al. p53 regulates Toll-like receptor 3 expression and function in human epithelial cell lines. Mol. Cell Biol. 2008;28:6557–6567. doi: 10.1128/MCB.01202-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoro JG, Parker AE, Zhu X, Green MR. p53-mediated inhibition of angiogenesis through up-regulation of a collagen prolyl hydroxylase. Science. 2006;313:968–971. doi: 10.1126/science.1126391. [DOI] [PubMed] [Google Scholar]
- Timmerman P, Beld J, Puijk WC, Meloen RH. Rapid and quantitative cyclization of multiple peptide loops onto synthetic scaffolds for structural mimicry of protein surfaces. Chembiochem. 2005;6:821–824. doi: 10.1002/cbic.200400374. [DOI] [PubMed] [Google Scholar]
- Timmerman P, Puijk WC, Meloen RH. Functional reconstruction and synthetic mimicry of a conformational epitope using CLIPS technology. J. Mol. Recognit. 2007;20:283–299. doi: 10.1002/jmr.846. [DOI] [PubMed] [Google Scholar]
- Toledo F, Krummel KA, Lee CJ, et al. A mouse p53 mutant lacking the proline-rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell. 2006;9:273–285. doi: 10.1016/j.ccr.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Tovar C, Rosinski J, Filipovic Z, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Vousden KH. Outcomes of p53 activation--spoilt for choice. J. Cell Sci. 2006;119:5015–5020. doi: 10.1242/jcs.03293. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Ryan KM. p53 and metabolism. Nat. Rev. Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- Wade M, Wahl GM. Targeting Mdm2 and Mdmx in cancer therapy: better living through medicinal chemistry? Mol. Cancer Res. 2009;7:1–11. doi: 10.1158/1541-7786.MCR-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Kung AL, Escher I, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhao Y, Bernard D, Aguilar A, Kumar S. In: Targeting the MDM2-p53 protein-protein interaction for new cancer therapeutics, Vol. 8, Protein-protein interactions. Wendt MD, editor. Berlin-Heidelberg: Springer-Verlag; 2012. pp. 57–80. [Google Scholar]
- Wender PA, Galliher WC, Goun EA, Jones LR, Pillow TH. The design of guanidinium-rich transporters and their internalization mechanisms. Adv. Drug Deliv. Rev. 2008;60:452–472. doi: 10.1016/j.addr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson RA, Evans JR, Jacobs JM, et al. Peptides selected from a phage display library with an HIV-neutralizing antibody elicit antibodies to HIV gp120 in rabbits, but not to the same epitope. AIDS Res. Hum. Retroviruses. 2007;23:1416–1427. doi: 10.1089/aid.2007.0027. [DOI] [PubMed] [Google Scholar]
- Wlostowski M, Czarnocka S, Maciejewski P. Efficient S-alkylation of cysteine in the presence of 1,1,3,3-tetramethylguanidine. Tetrahedron Lett. 2010;51:5977–5979. [Google Scholar]
- Woolley GA. Photocontrolling peptide a helices. Acc. Chem. Res. 2005;38:486–493. doi: 10.1021/ar040091v. [DOI] [PubMed] [Google Scholar]
- Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66:4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- Zhan C, Zhao L, Wei X, et al. An ultrahigh affinity d-peptide antagonist Of MDM2. J. Med. Chem. 2012;55:6237–6241. doi: 10.1021/jm3005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Sadovski O, Woolley GA. Synthesis and characterization of a long, rigid photoswitchable cross-linker for promoting peptide and protein conformational change. Chembiochem. 2008;9:2147–2154. doi: 10.1002/cbic.200800196. [DOI] [PubMed] [Google Scholar]
- Zhang F, Sadovski O, Xin SJ, Woolley GA. Stabilization of folded peptide and protein structures via distance matching with a long, rigid cross-linker. J. Am. Chem. Soc. 2007;129:14154–14155. doi: 10.1021/ja075829t. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yu D, Hu M, et al. Wild-type p53 suppresses angiogenesis in human leiomyosarcoma and synovial sarcoma by transcriptional suppression of vascular endothelial growth factor expression. Cancer Res. 2000;60:3655–3661. [PubMed] [Google Scholar]
- Zhao Y, Bernard D, Wang S. Small molecule inhibitors of MDM2-p53 and MDMX-p53 interactions as new cancer therapeutics. Biodiscovery. 2013;4:1–15. [Google Scholar]
- Zhigaltsev IV, Winters G, Srinivasulu M, et al. Development of a weak-base docetaxel derivative that can be loaded into lipid nanoparticles. J. Control. Release. 2010;144:332–340. doi: 10.1016/j.jconrel.2010.02.029. [DOI] [PubMed] [Google Scholar]