Abstract
Plastid genomes of photosynthetic flowering plants are usually highly conserved in both structure and gene content. However, the plastomes of parasitic and mycoheterotrophic plants may be released from selective constraint due to the reduction or loss of photosynthetic ability. Here we present the greatly reduced and highly divergent, yet functional, plastome of the nonphotosynthetic holoparasite Hydnora visseri (Hydnoraceae, Piperales). The plastome is 27 kb in length, with 24 genes encoding ribosomal proteins, ribosomal RNAs, tRNAs, and a few nonbioenergetic genes, but no genes related to photosynthesis. The inverted repeat and the small single copy region are only approximately 1.5 kb, and intergenic regions have been drastically reduced. Despite extreme reduction, gene order and orientation are highly similar to the plastome of Piper cenocladum, a related photosynthetic plant in Piperales. Gene sequences in Hydnora are highly divergent and several complementary approaches using the highest possible sensitivity were required for identification and annotation of this plastome. Active transcription is detected for all of the protein-coding genes in the plastid genome, and one of two introns is appropriately spliced out of rps12 transcripts. The whole-genome shotgun read depth is 1,400× coverage for the plastome, whereas the mitochondrial genome is covered at 40× and the nuclear genome at 2×. Despite the extreme reduction of the genome and high sequence divergence, the presence of syntenic, long transcriptionally active open-reading frames with distant similarity to other plastid genomes and a high plastome stoichiometry relative to the mitochondrial and nuclear genomes suggests that the plastome remains functional in H. visseri. A four-stage model of gene reduction, including the potential for complete plastome loss, is proposed to account for the range of plastid genomes in nonphotosynthetic plants.
Keywords: parasitic plants, holoparasite, nonphotosynthetic, Hydnoraceae, plastome, plastid genome
Introduction
The plastids of green plants (Viridiplantae) are double membrane-bound organelles derived from cyanobacteria through endosymbiosis. The primary function of plastids in most green algae and land plants is the fixation of carbon dioxide through photosynthesis (chloroplasts); however, plastids may also function in storage of starch (amyloplasts), lipids (elaioplasts), or proteins (proteinoplasts) (Bock 2007; Wicke et al. 2011; Ruhlman and Jansen 2014). Plastids maintain a separate, circular-mapping, DNA genome that is uniparentally inherited independent of the nuclear and mitochondrial genomes. The plastid genome (plastome) of land plants is usually about 120–170 kb in size, is highly conserved in photosynthetic plants, and typically encodes about 120–130 unique genes (reviewed in Ruhlman and Jansen 2014). Plastome organization is highly conserved, containing a large single copy (LSC) region and a small single copy (SSC) region separated by two copies of an inverted repeat (IRa, IRb) (Shinozaki et al. 1986; Ohyama et al. 1996; Jansen et al. 2007).
Parasitic and mycoheterotrophic plants have repeatedly reduced or lost the need for photosynthesis in their chloroplasts by establishing a physiological connection with host plants or fungi to obtain carbohydrates, water, and other nutrients. The reduced demand for photosynthetic ability in these heterotrophic plants has relaxed or eliminated evolutionary constraints on photosynthesis and other genes related to plastid function, resulting in a divergent and greatly reduced plastid genome (dePamphilis and Palmer 1990; Wicke et al. 2013; Barrett et al. 2014). The first plastid genome of a nonphotosynthetic plant was sequenced over 20 years ago (Epifagus virginiana; Wolfe et al. 1992).
Currently, there are over 535 complete plastid genomes of land plants deposited in GenBank (retrieved June 1, 2015). Among the published parasitic flowering plant plastomes, 12 are members of the broomrape family (Orobanchaceae) (Wolfe et al. 1992; Li et al. 2013; Wicke et al. 2013; Uribe-Convers et al. 2014), four are dodders (Cuscuta) in the morning glory family (Convolvulaceae) (Funk et al. 2007; McNeal et al. 2007), and four are mistletoes from the Santalales (Petersen et al. 2015). In addition, 17 plastomes have been sequenced for mycoheterotrophic plants, including several orchids (Rhizanthellagardneri, Neottianidus-avis, Epipogium aphyllum, Epipogium roseum, and ten Corallorhiza species) (Delannoy et al. 2011; Logacheva et al. 2011; Barrett and Davis 2012; Barrett et al. 2014; Schelkunov et al. 2015), other monocots Petrosavia stellaris and Sciaphila densiflora (Logacheva et al. 2014; Lam et al. 2015), and the liverwort Aneura mirabilis (Wickett et al. 2008). All of these plastomes retain a core set of genes that support production of plastid ribosomes and one or several genes whose transcripts encode proteins that may be essential to plastid processes in nonphotosynthetic plants, including intron processing (matK), lipid synthesis (acetyl-CoA carboxylase [accD]), and protein synthesis and processing. In nonphotosynthetic species, genes related to photosynthesis, transcription, and NAD(P)H dehydrogenase subunits are often nonfunctional or lost (Wicke et al. 2011; Barrett et al. 2014).
Although the reduction in plastome gene content of nonphotosynthetic plants has been well documented, there has been a long-standing debate about the minimal plastid genome in the absence of photosynthetic constraint, and whether plastids or their genomes could be entirely lost in some heterotrophic plants (dePamphilis and Palmer 1990; Wolfe et al. 1992; Nickrent et al. 1997; Race et al. 1999; Bungard 2004; Barbrook et al. 2006; Krause 2008; Wicke et al. 2011; Molina et al. 2014; Schelkunov et al. 2015). Among nonphotosynthetic plant plastomes sequenced to date, 27–35 genes are typically retained (Wicke et al. 2011, 2013; Barrett et al. 2014; Lam et al. 2015; Schelkunov et al. 2015), including machinery for translation (rRNA and ribosomal protein genes) that may be required only for the expression of a small number of potentially indispensable protein-coding sequences (Wolfe et al. 1992; Krause and Scharff 2014, and references therein). Although each of these core nonbioenergetic, nontranslational genes, including accD, clpP, ycf1, and ycf2, has been lost from the plastid genome in at least some photosynthetic angiosperm lineages (Wicke et al. 2011), these genes are typically retained in most nonphotosynthetic plants (Braukmann and Stefanović 2012; Barrett et al. 2014). Given enough evolutionary time, the ongoing process of gene transfer from the plastome to the nuclear and mitochondrial genomes could result in the functional transfer of the last of these essential nonbioenergetic, nontranslational genes, and at that point, genes involved only in translational function would be unnecessary, allowing the continued deletion and potential complete loss of the plastome. Alternatively, some core sequences may not be transferrable out of the plastome because of redox balance requirements (Race et al. 1999) or other still unknown processes that require certain genes, or even nongenic sequences, to remain plastid encoded. The plastid trnE gene has been discussed as a compelling candidate out of all the plastid genes that has to be retained due to a dual function (Barbrook et al. 2006). Ancient holoparasitic lineages provide evolutionary test cases for the minimal plastid genome, and whether complete loss of the plastid genome has ever occurred.
In one recent study, the plastid genome appears to be missing from whole-genome shotgun data from the holoparasitic flowering plant Rafflesia lagascae (Molina et al. 2014). Different search strategies failed to identify a plastid genome in the genomic assembly. Reference-based mapping, a BLASTn approach, and profile Hidden Markov Models of plastid gene alignments identified only short and low coverage fragments of plastid genes at less than 2× depth of coverage, whereas assembled portions of the mitochondrial genome were readily detected at much higher depth of coverage (350×). At the same time, another study reported the putative loss of the plastid genome from the nonphotosynthetic unicellular alga Polytomella (Smith and Lee 2014). Using related chlorophyte organellar genomes as queries, both Basic Local Alignment Search Tool (BLAST) and reference mapping-based approaches from whole-genomic Illumina data of four Polytomella species did not recover any reads corresponding to the plastid genome. Compared with Rafflesia, where microscopy shows only a plastid-like organelle without a known function, Polytomella possesses starch-storing plastids (Moore et al. 1970; Brown et al.1976).
In photosynthetic plants, there are typically dozens of copies of mitochondrial DNA and hundreds to thousands of copies of plastid DNA per cell. Therefore, in a genomic sequence sample where no attempt has been made to enrich for DNA from one genomic compartment or another, a plastid genome would be expected to have a relative depth of read coverage that is 1 or even 2 orders of magnitude greater than the mitochondrial genome and 3–4 orders of magnitude greater than the nuclear genome (Straub, Fishbein, et al. 2011; Straub, Parks, et al. 2011; Wolf et al. 2015). In a nonphotosynthetic plant, however, where the plastome has lost part of its functionality, the normal stoichiometric relationships may be altered. Nevertheless, stoichiometry is important evidence for the detection of organellar genomes in genomic sequence data (Wolf et al. 2015; Wu et al. 2015), and differences in coverage depth can help diagnose the genomic location of particular sequences, including intragenomic and horizontal transfers. Sequenced plant genomes often display significant quantities of plastome DNA translocated to the mitochondrion (Rice et al. 2013; Park et al. 2014). The rate of sequence evolution is typically much lower in the plant mitochondrial genome compared with the plastid and nuclear genomes (Wolfe et al. 1987; Palmer and Herbon 1988; Drouin et al. 2008). Hence, in a plant lineage that has been nonphotosynthetic for many millions of years and may have lost its plastid genome, the mitochondrial genome is the most likely place to find persistent, “fossilized” genes or gene fragments transferred from the plastome.
Hydnora visseri (Hydnoraceae), the focal holoparasitic plant of this study, represents one of the 11 independent lineages of parasitic plants (Barkman et al. 2007). The small and entirely heterotrophic family consists of only two genera, Hydnora and Prosopanche. Given a stem group age of 100 Ma, and a crown group age of 54 Ma (the split between the two genera Hydnora and Prosopanche), Hydnoraceae are among the oldest parasitic lineages (Naumann et al. 2013). Hydnoraceae are different from other parasitic flowering plants in many ways (Visser and Musselman 1986; Bolin et al. 2011) and have been described as the “strangest plants in the world” (Visser and Musselman 1986). The fleshy trap flower and a massive, horizontally growing underground stem whose haustoria connect to the host (fig. 1), are the only remaining plant organs (Bolin et al. 2011; Wagner et al. 2014). The highly modified flowers of Hydnoraceae have three large, sometimes very brightly colored tepals that emit volatiles reminiscent of rotting flesh and attract and temporarily imprison huge numbers of carrion beetles for their pollination services (Bolin et al. 2009). Although the extraordinary flowers of Hydnora triceps are strictly subterranean, flowers of most species break through the surface to reproduce; the emerging flowers grow with so much force that they can crack asphalt or concrete (Maass and Musselman 2001). Hydnora visseri grows in desert habitats of Namibia and feeds exclusively on Euphorbia gregaria and Euphorbia gummifera (Bolin et al. 2011), whereas other members of the genus Hydnora feed upon a wider range of host plants in the spurge (Euphorbiaceae), legume (Fabaceae), and torchwood (Burseraceae) families (Musselman and Visser 1989; Beentje and Luke 2002; Bolin et al. 2010). In addition to Fabaceae and Euphorbiaceae, Prosopanche has a much wider host range, including Anacardiaceae, Apiaceae, Aquifoliaceae, Asteraceae, Amaranthaceae, Malvaceae, Rhamnaceae, and Solanaceae (Cocucci AE and Cocucci AA 1996). Hydnoraceae are placed in the order Piperales (Nickrent et al. 2002; Naumann et al. 2013), with their closest relatives among the first successive branches of living angiosperms, commonly referred to as the “basal angiosperms” (Jansen et al. 2007). Given the age of Hydnoraceae and the highly modified morphology following the ancient loss of photosynthesis, this lineage is a potential candidate, along with Rafflesia, to have lost the plastid genome entirely (Nickrent et al. 1997). Here we 1) describe the challenges of identifying and annotating the full plastome from genomic sequence data of H. visseri, 2) describe the extreme reduction in both size and gene content that goes far beyond the loss of genes related to photosynthesis, and 3) discuss the relevance of the Hydnora plastome in the context of extreme genome reduction and sequence divergence.
Fig. 1.—
Hydnora visseri (A). Flower (B). Excavated underground stem (dark) connected to host plant Euphorbia gregaria (light) and a close-up. (C) Flower bud of H. visseri in foreground next to shovel and E. gregaria stems in background (D). Cross section of H. visseri underground stem.
Materials and Methods
Plant Material, DNA Extraction, and Genome Sequencing
Plant material of H. visseri and Hydnora longicollis was collected on private property (Gondwana Cañon Preserve) (Namibian MET Permit No. 1350/2009). The tissue was snap frozen after collection, shipped and stored at −80 °C. Genomic DNA (gDNA) was extracted using a DNeasy Plant Mini Kit (Qiagen) and used for library preparation from H. visseri (insert size of 300 bp) for the Illumina HiSeq 2000 in the laboratory of Stephan C. Schuster (Penn State University). One lane of 100 bp × 100 bp paired-end sequence was obtained, yielding 162,683,243 trimmed reads and comprising over 16 GB total DNA sequence.
Data Processing, Genome Assembly, and Read Mapping
The genomic raw data were processed using CLC assembly cell beta (Version 4.0.6 for Linux). This program was also used to remove duplicate reads created during the polymerase chain reaction (PCR) amplification step of library preparation and to trim adapter sequences and low quality bases (<Q20) from the read data. The genomic reads were de novo assembled in CLC, with the scaffolding mode accounting for the precomputed paired-end insert size, producing 135 Mb of assembled genomic sequence data.
To identify any organellar scaffolds, “gene feature” data extracted from 33 plastid and 14 mitochondrial genomes were aligned with BLASTn (e value 1e-10) against the Hydnora genomic assembly. This search included the closest available plastome to Hydnora, that of Piper (Piperaceae; Cai et al. 2006), a nonparasitic relative of Hydnoraceae also from the order Piperales (Naumann et al. 2013). This search returned 78 putative organellar scaffolds that were further assembled in Geneious (Version 7.1.2, Biomatters Limited, Kearse et al. 2012) using the “De novo assemble” tool, reducing the number of scaffolds to 58.
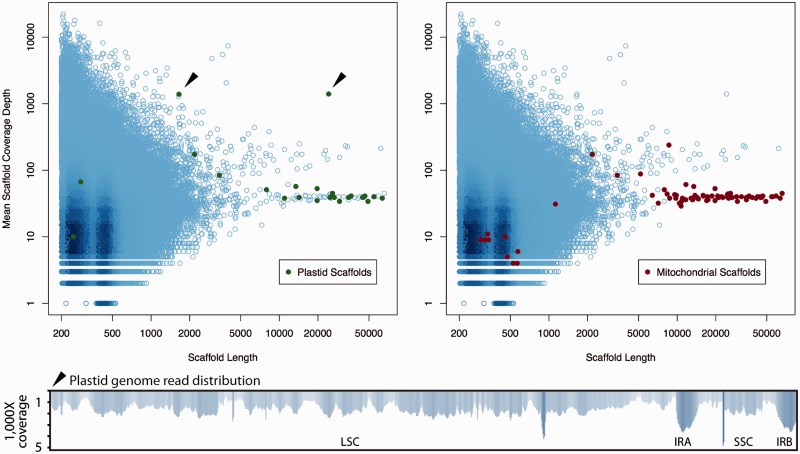
Having identified a total of 58 scaffolds with BLAST alignments to organelle genes (plastid or mitochondrial) we next sought to characterize the relative sequence depths (stoichiometries) of each contig with and without detected organelle sequences. The read mapping was performed with CLC Cell (version beta 4.0.6 for Linux) using “ref-assemble”, and read densities were then visualized using R (R 3.2.0 GUI 1.65 Mavericks build [6931]) and the “RColorBrewer” package. Contigs containing positive BLAST hits to mitochondrial or plastid genes are indicated in red and green, respectively (fig. 2).
Fig. 2.—
Plot of read depths relative to scaffold length. Cyan circles are all scaffolds from the genomic assembly creating darker spots at greater read depths. Scaffolds containing BLAST hits to plastid genes are overplotted by green filled circles (left). Scaffolds containing BLAST hits to mitochondrial genes are overplotted by red filled circles (right). All scaffolds at around 40× coverage that contain plastid gene fragments also contain mitochondrial genes. This indicates plastid sequences that have migrated into the mitochondrial genome. The two green filled circles at around 1,400× coverage (filled black arrows) contain only plastid genes and comprise the Hydnora plastid genome. The remaining cyan circles correspond to the nuclear genome. The remaining red and green filled circles are presumably mitochondrial and plastid sequences that are located in the nucleus.
One plastid scaffold of length 24,268 bp was identified with very high (∼1,400×) average read depth. To see whether this scaffold connects to any additional sequences in the assembly, it was used as a query in another BLASTn search. A second scaffold of length 1,650 bp was observed at a similar sequence depth (1,389×). A 50-bp overlap allowed the two high depth fragments to be merged, and closed to form a circle with a short inverted repeat (IR). PCR primers were designed to amplify across all four SC to IR junctions and the 50-bp scaffold joins, confirming a circular structure with an IR. This circular-mapping DNA molecule represents the complete plastid genome of Hydnora visseri (GenBank accession number KT970098). In contrast to the plastome, most mitochondrial genes were present on scaffolds of much lower (∼40×) depth of coverage. However, a few more plastid and mitochondrial gene fragments were identified on scaffolds at around 2× coverage; these are presumably part of the nuclear genome (supplementary fig. S1, Supplementary Material online).
Annotation
Just two genes were identified on the plastome by the initial BLASTn search (rrn16 and rrn23). To further complete the annotation of the plastid genome, DOGMA (http://dogma.ccbb.utexas.edu/, last accessed January 11, 2016; Wyman et al. 2004) was used at different stringencies. Settings less stringent than the default settings (50% sequence identity in protein-coding genes and 60% in RNA genes) and an e value of 1e−5 identified 13 additional genes, including the three tRNAs (supplementary table S1, Supplementary Material online).
Furthermore, four additional alignment tools (1) Geneious tool “Map to Reference” [Version 7.1.2, Biomatters Limited, Kearse et al. 2012]; 2) LASTZ “Align Whole Genomes” [Harris 2007] implemented in Geneious; 3) BWA-MEM version 0.7.8 [Li and Durbin 2009]; and 4) GMAP version 2014-02-28 [Wu and Watanabe 2005]) were applied to all scaffolds identified in the initial BLASTn search. All of these programs were set up to align angiosperm organellar genes (the same that were used as a query in the initial BLASTn search) to the Hydnora organellar scaffolds as a reference sequence. All of those approaches returned different results with respect to genes identified, and the results had to be compared carefully and in some cases adjusted manually to obtain the longest alignments with the fewest gaps. A summary of all identified plastid and mitochondrial genes and gene fragments found with each method is provided in supplementary table S1, Supplementary Material online. With respect to the Hydnora plastome, four additional genes were identified with this approach (rps4, rps7, ycf1, and rrn4.5). Next, we identified all open-reading frames (ORFs) larger than 100 bp using Geneious (Version 7.1.2, Biomatters Limited, Kearse et al. 2012), and used tBLASTx and PSI-BLAST in National Center for Biotechnology Information to assign unannotated ORFs, which identified rps2, rps3, rps11, rps18, and ycf2. Also, unannotated sections of the plastome were used to query the database using BLASTn, but did not recover any new genes.
To verify and complete the annotation of the plastid genome, DOGMA (http://dogma.ccbb.utexas.edu/, last accessed January 11, 2016; Wyman et al. 2004) was used at very low stringencies (25% sequence identity in protein-coding genes and 30% in RNA genes) and an e value of 1e−5. These settings identified 23 of 24 genes (not including rrn4.5). Final annotation (gene boundaries) was based on the identified ORFs for all of the protein-coding genes. Short exons for rpl16 and rps12 were identified manually by aligning the corresponding Piper sequence to Hydnora. The resulting annotation was submitted to OrganellarGenomeDRAW (http://ogdraw.mpimp-golm.mpg.de/, last accessed January 11, 2016; Lohse et al. 2007, 2013).
Amplification of gDNA and cDNA
The structure of the plastid genome of H. visseri was validated using PCR of gDNA. All genes found on the H. visseri plastid genome, as well as the IR boundaries, were amplified and resequenced from gDNA of H. visseri and H. longicollis using custom primers designed from the H. visseri plastome sequence.
Transcription of 19 plastid genes was confirmed using reverse transcription (RT)-PCR (not including the three short tRNAs, rps18, and rrn4.5). Experimental design for RT-PCR confirmation of rps12 splicing was modeled after Ems et al. (1995) using RNA and DNA inputs and multiple experimental controls. All primers used here are listed in supplementary table S2, Supplementary Material online. Total RNA was extracted from H. visseri tepal and H. longicollis floral bud tissue using a cetyltrimethylammonium bromide (CTAB) RNA isolation protocol (Chang et al. 1993). Total nucleic acids were divided equally for serial DNase I (Qiagen) and RNase A (Qiagen) treatments. RNA digestions were performed in solution with 300 µg RNase A at 37 °C for 1 h. DNA digestions were performed following Appendix C of the RNeasy MineElute Clean-up Handbook (Qiagen). DNAs and RNAs were then purified using the DNeasy and RNeasy Mini Kit, respectively. Nucleic acid concentrations were estimated using Qubit High Sensitivity DNA and RNA assays. One microgram RNA from each of the extracted RNA treatments was reverse transcribed using Maxima First Stand Synthesis Kit (Thermo Scientific).
RT-PCR amplifications were performed using DreamTaq (Thermo Scientific) in an Eppendorf Thermocycler using the following parameters: 5 min initial melt (95 °C) followed by 35 cylcles of 30 s melt (95 °C), 30 s annealing (50 °C), 30 s extension (72 °C), and a final extension of 10 min (72 °C). Three nanograms of gDNA and cDNA, as estimated by RNA mass added to cDNA synthesis reactions, and added to each reaction mix. PCR products were run on a 2% agarose gel containing 0.5× SyberSafe Dye (Life Technologies) at 125 V for 1 h. Images were taken on a Molecular Imager Gel Doc XR system (Bio Rad) and Quantity One (Bio Rad) used for estimation of PCR product sizes with respect to the 1 kb Plus ladder (Life Technologies). PCR product for both Hydnora species was purified using MinElute PCR Purification Kit (Qiagen): Purified product was sequenced at GeneWiz.
Phylogenetic Analyses
Nineteen plastid genes derived from the plastid genome were added to the respective angiosperm-wide alignments published by Jansen et al. (2007). Phylogenetic trees for a concatenated alignment of all 20 genes were calculated in RAxML v7.2.6 (Stamatakis 2006) applying the GTR+G model for the rapid Bootstrap (BS) algorithm that is combined with the search for the best scoring maximum-likelihood (ML) tree. In total, 1,000 BS replicates were applied for all analyses. Due to the high sequence divergence of the Hydnora sequences, a starting tree for the nonparasitic taxa (Jansen et al. 2007) was used. Using the “-t” function allowed to add the Hydnora sequences to the existing tree, that is then optimized under ML (Stamatakis 2006). The phylogenetic trees were formatted with TreeGraph2 (Stöver and Müller 2010).
Test for Relaxed Selection of Plastid Genes
To test for relaxed selection of the Hydnora plastid genes, different hypotheses were tested for 14 genes and the concatenated data set using CodeML implemented in PAML (Yang 2007; pamlX v1.2, Xu and Yang 2013). As the individual plastid genes do not provide enough phylogenetic information to obtain a correct species tree, the input trees for CodeML were calculated in RAxML (Stamatakis 2006) using a starting tree (“-t” function) that comprises the full sampling (including the two Hydnora species this time). The basic model was used to calculate the dN/dS ratio of the background, whereas the branch model was used to calculate the dN/dS ratio of the Hydnora branches and the background separately. Significance was tested using the difference of likelihood ratios of both models (background vs. branch model) in a simple chi-square test and with 1 degree of freedom (http://www.socscistatistics.com/pvalues/chidistribution.aspx, last accessed January 11, 2016). For the genes that were tested to be significant for relaxed selection, a second branch model (selection), which allows several dN/dS ratios for branches, was used to identify codons that are under positive selection.
Results
Plastids of Hydnora Produce Starch Granules
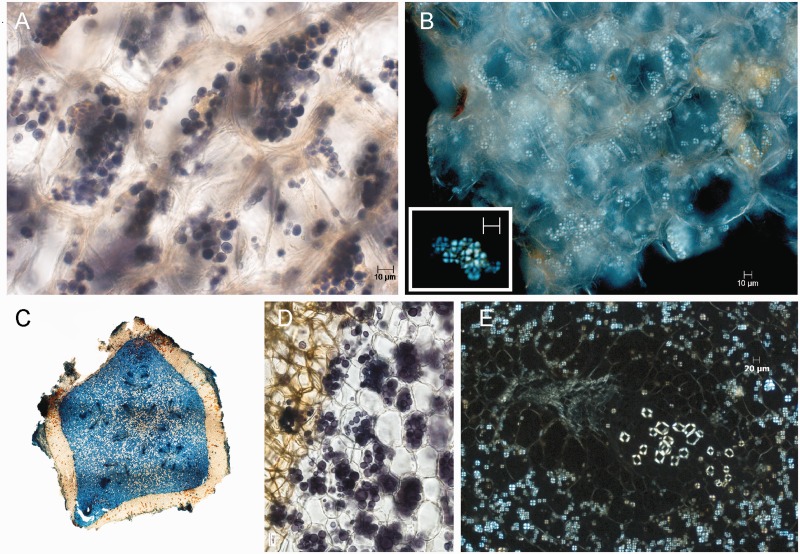
In parasitic plants lacking photosynthesis, there are often questions related to plastid function and the state of decay of the plastid genome. Light microscopic images of tepal and underground stem transverse sections of H. visseri and H. longicollis stained with iodine–potassium iodide clearly show several starch grains per cell (fig. 3A and D). Using polarized light, typically a single starch grain per plastid is observed (fig. 3B and E). As plastids are the exclusive location for building and storing starch (amyloplasts) in a plant cell, this is clear evidence for the presence of plastids in these extreme heterotrophs.
Fig. 3.—
Light microscopy of two different starch-containing tissues of Hydnora visseri. (A) Section of tepal, starch grains in cells stained with iodine–potassium iodide. (B) Section of tepal, showing starch grains under polarized light; inset: enlarged starch grains. (C) Transverse section of underground organ; stained with Astrablue-safranin; 5-merous organization of vascular system is visible. (D) Starch grains in the underground organ; stained with iodine-potassium iodide. (E) Starch grains and vascular bundle in underground organ under polarized light.
Size and Structure of the H. visseri Plastome
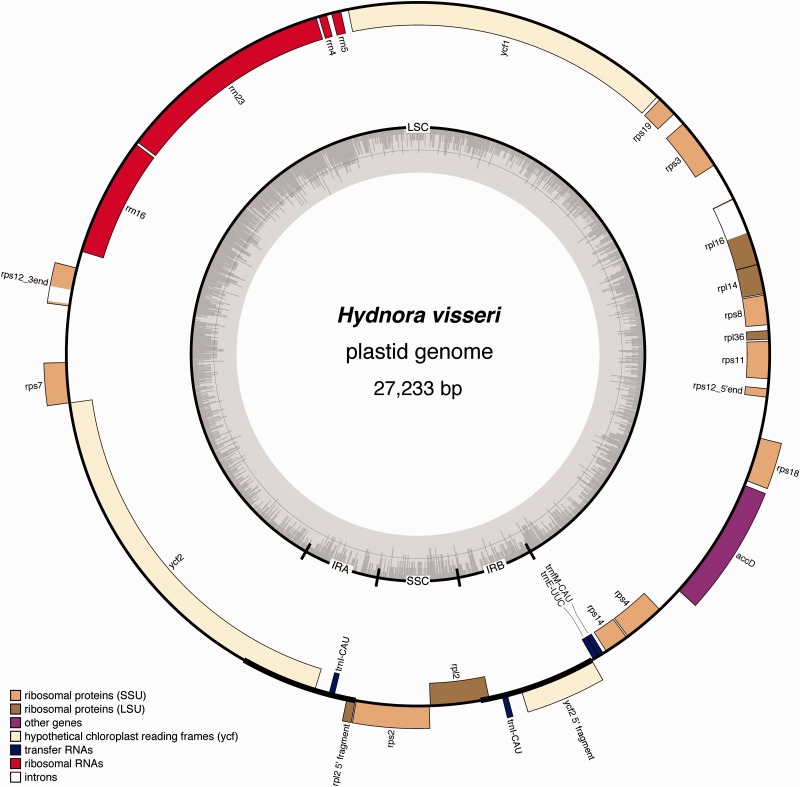
The 27,233 bp plastid genome of H. visseri is only one-sixth the size of the plastome of Piper cenocladum (160 kb; Cai et al. 2006), a close photosynthetic relative, and nearly half the size of the plastome of Conopholis americana (46 kb), a holoparasitic Orobanchaceae with the smallest potentially functional plastome yet known in parasitic plants (Wicke et al. 2013). The circular plastome of H. visseri retains the quadripartite structure typical of most characterized plastomes (Wicke et al. 2011; Jansen and Ruhlman 2012), but with much reduced size (fig. 4, table 1). The LSC region of 22,751 bp and a very short SSC region of 1,550 bp are separated by two short IRs, each 1,466 bp in length. Structurally, however, the IR-boundaries have shifted drastically in Hydnora. The genes ycf1, rps7, as well as the four rRNAs are located in the IR in Piper, but in Hydnora they are part of the LSC. The only two genes in the Hydnora SSC are rps2 and rpl2, which are found in the LSC in Piper. The IR contains only trnI-CAU plus parts of ycf2 and rpl2. As expected, read mapping clearly shows twice the sequencing depth in the IR region (fig. 3).
Fig. 4.—
Map of the plastid genome of Hydnora visseri. Genes are color coded according to the legend, which indicates functional groups. The inner ring illustrates the boundaries of the LSC and SSC regions, separated by the two copies of the inverted repeat (IRa, IRb). The innermost ring shows the GC content across the plastid genome.
Table 1.
Comparison of the Plastid Genomes of Hydnora visseri and Piper cenocladum
| Piper | Hydnora | |
|---|---|---|
| Size (bp) | 160,624 | 27,233 |
| Genic (bp) | 100,645 | 21,617 |
| Intergenic (bp) | 59,979 | 5,616 |
| Percentage genic | 62.66% | 79.38% |
| Percentage intergenic | 37.34% | 20.62% |
| LSC length (bp) | 87,668 | 24,114 |
| SSC length (bp) | 18,878 | 1,550 |
| IR length (bp) | 27,039 | 1,466 |
| Number of genes (unique genes) | 130 (113) | 25 (24) |
| Number of genes duplicated in IR | 17 | 1 |
| Number of genes with introns | 18 | 3 |
| GC content | 38.3% | 23.4% |
A direct comparison of the nucleotide sequence of Piper and Hydnora shows very few colinear regions visible in the dotplot relative the background noise (supplementary fig. S2, Supplementary Material online, word size 12 and 100 percent identity, implemented in Geneious [Version 7.1.2, Biomatters Limited, Kearse et al. 2012]). Only a LASTZ alignment graph shows a few more clear short lines of identity. That the dissimilarity is due to a very high sequence divergence of Hydnora plastome sequences is illustrated by a similar dotplot comparison of Piper versus Arabidopsis plastomes (supplementary fig. S2, Supplementary Material online). At the same stringency (word size 12, percent identity 100), Piper and Arabidopsis alignments are easily seen, despite Hydnora and Piper being members of the Piperales and Arabidopsis being a distantly related eudicot. The GC content of the Hydnora plastome is 23.7%, which is extremely low compared with 38.3% in Piper and 33.2% in Conopholis, and is consistent with Hydnora’s high sequence divergence (table 1).
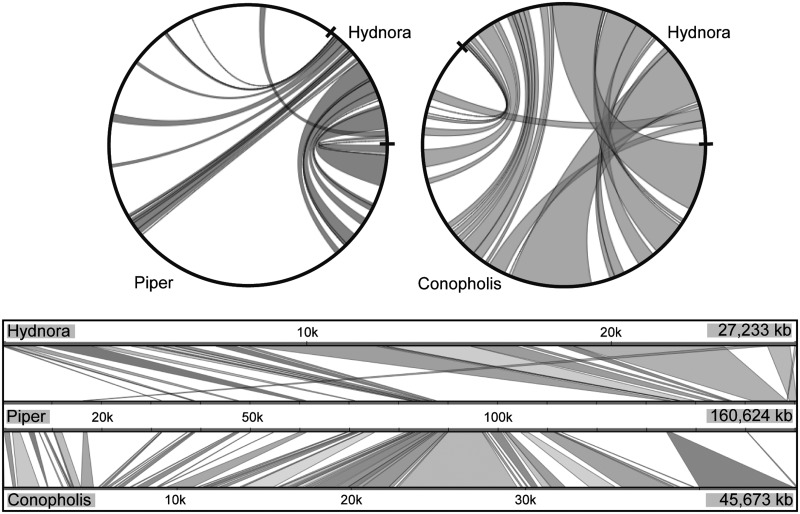
To assess any rearrangements in the Hydnora plastome, it was compared with P. cenocladum and C. americana. As sequence divergence is very high in Hydnora compared with these two plants, colinearity was visualized based on annotation, using Multi-Genome Synteny viewer (http://cas-bioinfo.cas.unt.edu/mgsv/index.php, last accessed January 11, 2016; fig. 5). Both the gene order and the gene orientation are nearly identical compared with Piper and Conopholis (fig. 5). The only exception being the gene block of ycf1-rrn5-rrn4.5-rrn23-rrn16-rps12-rps7 that is part of the IR in more typical plastomes, but is found in reverse complement orientation in Hydnora compared with Conopholis (fig. 5). Although Conopholis has lost one copy of the IR (Wicke et al. 2013; supplementary fig. S2, Supplementary Material online), Hydnora has retained a very short IR, but this gene block, originally part of the IR, is not in the IR anymore in Hydnora. Hence, this looks like an inversion, but retention of this gene from one side of a once-larger IR is more likely to explain this pattern.
Fig. 5.—
Maps of plastid genomes showing colinearity. The two circles show annotation-based syntenic regions between Hydnora and Piper and, respectively, Hydnora and Conopholis (generated in mGSV, http://cas-bioinfo.cas.unt.edu/mgsv/, last accessed January 11, 2016). At the bottom, there is a comparison between all three species. Gene order is colinear with the exception of an inversion of a large gene block between Hydnora and Conopholis that originally stems from opposite IRs.
Furthermore, the extreme downsizing of the Hydnora plastome has led to a high gene density: 85% of the total length (23,159 of 27,233 bp) is occupied by genes, leaving only 5,616 bp of intergenic DNA. In Piper the ratio of genic:intergenic DNA is 1.6:1, and in Hydnora 3.8:1, which indicates that the density of genes is much greater in the Hydnora plastome. The intergenic DNA includes an approximately 600 bp highly repetitive region between the rps12 3′-end and rps7 (supplementary fig. S2, Supplementary Material online).
The H. visseri Plastome Encodes Just 24 Genes
The gene content of the 27-kb plastid genome of H. visseri has been greatly reduced to just 24 potentially functional genes: 14 ribosomal protein genes (rps2, rps3, rps4, rps7, rps8, rps11, rps12, rps14, rps18, rps19, rpl2, rpl14, rpl16, and rpl36), four rRNAs (rrn4.5, rrn5, rrn16, and rrn23), three tRNAs (trnICAU, trnEUUC, and trnfMCAU), a single biosynthetic protein-coding gene (accD), and two protein-coding genes of unknown function (ycf1 and ycf2) (fig. 4, table 2; supplementary table S1, Supplementary Material online). The function of ycf1 is still under debate (de Vries et al. 2015), although recent experimental evidence in model plants suggested a function of ycf1 in the TOC/TIC machinery (Kikuchi et al. 2013) and it has been proposed to rename ycf1 as tic214 (Nakai et al. 2015). Although ycf2 is the largest plastid coding sequence (if it is not lost, e.g., Epipogium or Sciaphila; Lam et al. 2015; Schelkunov et al. 2015), its function has remained unknown for decades. However, recent evidence shows that the protein encoded by ycf2 might be important for water regulation (Ruiz-Nieto et al. 2015). Due to high sequence divergence, successful annotation of the Hydnora plastome required multiple approaches unlike the relatively straightforward annotation of typical chloroplast genomes. An initial BLASTn search provided only two genes (rrn16 and rrn23). Default settings in DOGMA (Wyman et al. 2004; http://dogma.ccbb.utexas.edu/, last accessed January 11, 2016) provided only five protein-coding genes (accD, rps12, rpl14, rpl16, and rpl36) and the three tRNAs (60% identity cutoff for protein coding genes, 80% identity cutoff for tRNAs, and e-value of 1e−5). At very low stringency (25% identity cutoff for protein coding genes, 30% identity cutoff for tRNAs, and e-value cutoff of 1e−5), nearly all genes are identified (except the short rrn4.5 gene). All of the protein-coding genes predict ORFs that are full length or almost full length compared with the Piper plastome, and the longest ORF (ycf2) is nearly 5,000 bp long.
Table 2.
Summary of plastid genes present and absent in Hydnora
| Present on Hydnora plastome | Deleted from Hydnora platome |
|
|---|---|---|
| Ribosomal RNA genes | ||
| rrn4.5 | ||
| rrn5 | ||
| rrn16 | ||
| rrn23 | ||
| Transfer RNA genes | ||
| trnE-UUC | trnA-UGC | trnP-UGG |
| trnfM-CAU | trnC-GCA | trnQ-UUG |
| trnI-CAU | trnD-GUC | trnR-ACG |
| trnF-GAA | trnR-UCU | |
| trnG-GCC | trnS-GCU | |
| trnH-GUG | trnS-GGA | |
| trnI-GAU | trnS-UGA | |
| trnK-UUU | trnT-GGU | |
| trnL-CAA | trnT-UGU | |
| trnL-UAA | trnV-GAC | |
| trnL-UAG | trnV-UAC | |
| trnM-CAU | trnW-CCA | |
| trnN-GUU | trnY-GUA | |
| Ribosomal protein genes | ||
| rpl2 | rpl20 | |
| rpl14 | rpl22 | |
| rpl16 | rpl23 | |
| rpl36 | rpl32 | |
| rps2 | rpl33 | |
| rps3 | rps15 | |
| rps4 | rps16 | |
| rps7 | ||
| rps8 | ||
| rps11 | ||
| rps12 | ||
| rps14 | ||
| rps18 | ||
| rps19 | ||
| Other genes | ||
| accD | clpP | |
| ycf1 | matK | |
| ycf2 | ycf15 | |
| ycf3 | ||
| ycf4 | ||
| RNA polymerase | ||
| rpoA | ||
| rpoB | ||
| rpoC1 | ||
| rpoC2 | ||
| Photosynthetic and chlororespiratory genes | ||
| atpB | infA | |
| atpE | ccsA | |
| atpF | cemA | |
| atpH | psaA | |
| atpI | psaB | |
| petA | psaC | |
| petB | psaI | |
| petD | psaJ | |
| petG | psbA | |
| petL | psbB | |
| petN | psbC | |
| ndhA | psbD | |
| ndhB | psbE | |
| ndhC | psbF | |
| ndhD | psbH | |
| ndhE | psbI | |
| ndhF | psbJ | |
| ndhG | psbK | |
| ndhH | psbL | |
| ndhI | psbM | |
| ndhJ | psbN | |
| ndhK | psbT | |
| rbcL | psbZ | |
Plastid genes residing on the mitochondrial genome as pseudogenes are indicated in bold.
The 24 plastid genes present in the Hydnora plastome are a perfect subset of those found in Conopholis (fig. 6). Genes missing from Hydnora that are present and potentially functional in the already drastically reduced Conopholis plastome are clpP, matK, rpl20, and rpl33 plus 15 tRNAs (trnSUGA, trnSGGA, trnSGCU, trnYGUA, trnLUAG, trnHGUG, trnDGUC, trnPUGG, trnFGAA, trnMCAU, trnNGUU, trnQUUG, trnRUCU, trnWCCA, and trnGUCC). Comparing the H. visseri plastome with the extremely reduced mycoheterotrophs E. roseum, E. aphyllum (Schelkunov et al. 2015), and S. densiflora (Lam et al. 2015), Hydnora has the fewest genes (fig. 6 and supplementary fig. S3, Supplementary Material online). The difference in length of those plastomes is mostly due to the retention of ycf1 and ycf2, as well as the retention of one versus two copies of rrn16 and rrn23 in Hydnora.
Fig. 6.—
Gene content of highly reduced plastid genomes of seven nonphotosynthetic plants. The gene set shown here excludes gene categories that are missing in most of these extremely reduced plastomes. Black boxes indicate a present gene, white boxes indicate absent gene, and gray boxes indicate a pseudogene. A more comprehensive version of this figure including the full plastome gene set and including the available complete plastomes of hemiparasitic or nonphotosynthetic plants, as well as close photosynthetic relatives, is shown in supplementary figure S4, Supplementary Material online.
The Native Origin of the Hydnora Plastome Is Phylogenetically Verified
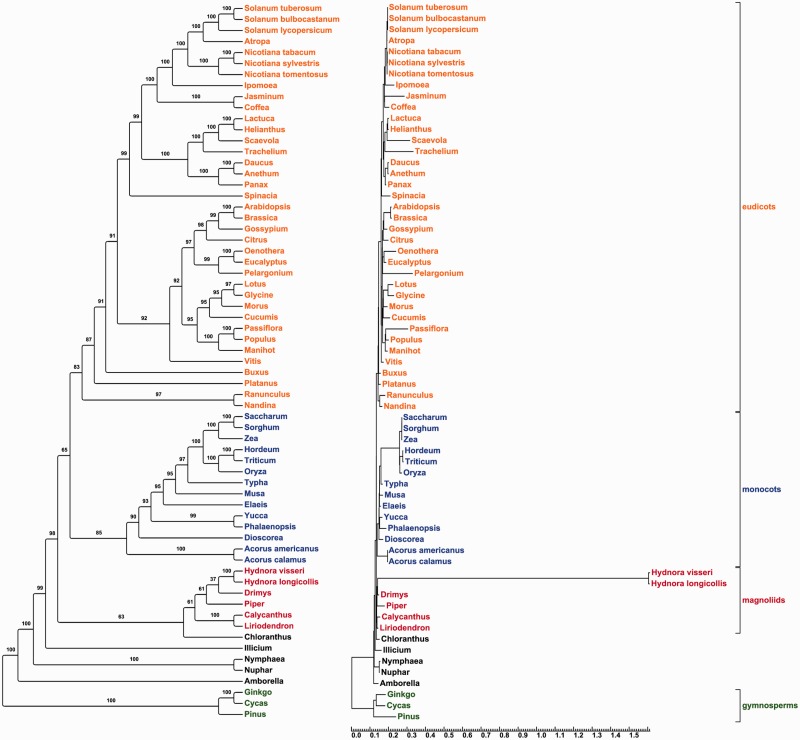
Phylogenetic evidence was collected for plastid genes of H. visseri and H. longicollis using a concatenated data set of 20 genes (not including the three tRNAs and ycf1). Because the Hydnora sequences of some genes are extremely divergent compared with the sequences from other angiosperms, a starting plastome tree (Jansen et al. [2007] with the addition of unplaced Hydnora sequences) was used for all of the calculations. This approach seemed most reasonable because we were only interested in the placement and apparent divergence of the Hydnora sequences from those of photosynthetic relatives. In the phylogeny, Hydnora is placed sister to Drimys (Canellales), in the magnoliids. A high sequence divergence for Hydnora is revealed by an extremely long branch in the phylogram (fig. 7).
Fig. 7.—
ML phylogenetic trees of the Hydnora visseri plastid genes. A phylogenetic tree has been estimated for a concatenated data set of 20 plastid genes, based on alignments published by Jansen et al. (2007). The tree was estimated with RAxML (Stamatakis 2006), applying the rapid bootstrapping algorithm (1,000 bootstrap replicates). The topology of Jansen et al. (2007) was used as a starting tree (-t function in RAxML). The cladogram (left) verifies a magnoliid origin of the Hydnora plastid genes. Bootstrap values are plotted above nodes. The scale of the phylogram is in substitutions per site. The phylogram (right) shows a much higher number of substitutions for the Hydnora sequences compared with the other taxa.
Evidence for Functionality of the Hydnora Plastome
Because ORFs have been retained for each of the protein-coding sequences in H. visseri, while at the same time being highly divergent compared with plastid genes of related flowering plants, we posit that all the genes found on the plastid genome are potentially functional.
To obtain additional evidence for the potential functionality of the plastid genome, we next amplified and sequenced 16 protein-coding genes and 3 ribosomal RNA genes on the plastid genome from both gDNA and cDNA of two Hydnora species (H. visseri and H. longicollis; supplementary fig. S2, Supplementary Material online, GenBank accession numbers KT922054–KT922083) using primers derived from the H. visseri plastome (supplementary table S2, Supplementary Material online). The gene sequences were nearly identical between H. visseri and H. longicollis, resulting in 5,109 bp of alignable nonambiguous gene sequence in the two species. Rpl2 is very likely a pseudogene in H. longicollis (supplementary fig. S4, Supplementary Material online) and was excluded from the alignment. The remaining 4,647 bp alignment shows 97.2% identity (4,515 bp identical sites). The ratio of nonsynonymous and synonymous sites (dN/dS ratio) between the two species is 0.093 (gene-wise ranging between 0 and 0.5599), indicating that the plastid proteome as a whole has been subject to purifying selection in these two closely related holoparasites (supplementary table S3, Supplementary Material online). The dS estimates are extremely high, and likely saturated with substitutions for the individual genes on the long branch leading to the two Hydnora species (dS between 2 and 28, not including the two very short gene alignments of rpl36 and rps2) and also of the concatenated data set (dS = 3.9); leading to a lower dN/dS ratio compared with the background. The lower dN/dS ratio is likely a result of the saturated synonymous divergence plus continued nonsynonymous divergence and may not indicate an increase in the intensity of purifying selection. We found four codons that are significant for positive selection (accD, codon 9, Q–V; rps12, codon 56, R–E; rps12 codon 60, F–P; rpl2, codon 5, L–N), indicating that the pattern we see here might in part be adaptive evolution. In the future, sequences from a less closely related member of Hydnoraceae, such as Prosopanche, will provide additional insights into the selective constraints of the Hydnoraceae plastome genes.
Positive amplification from cDNA verified active transcription of each of the 16 protein-coding and 3 ribosomal RNA genes in both Hydnora species, including the likely pseudogene of rpl2 in H. longicollis (supplementary fig. S5, Supplementary Material online). Genomic sequences and their corresponding cDNA sequences were identical (8,231 bp of corresponding sequence), meaning that there was no evidence of RNA editing in the Hydnora-coding regions.
Rps12 and rpl16 are the only intron-containing genes in the H. visseri plastome. In Piper, rps12 has three exons (e.g., Cai et al. 2006), where, generally in angiosperms, the first intron is a trans-spliced group IIb intron, and the second intron is a cis-spliced group IIa intron (Kroeger et al. 2009). Relative locations of the exons on the plastome are similar in Hydnora. However, comparison of the gDNA and cDNA sequences in Hydnora indicates trans-splicing of the first intron, but no splicing of the 230-bp second intron, whose homolog is cis-spliced in Piper (supplementary fig. S6, Supplementary Material online). This is surprising since the exons potentially encode a full-length rps12 ORF. If it is true that the second and third exons of this gene are not brought together in mature transcripts, the third exon would be out of reading frame due to the length of the intron. Then, the rps12 sequence may be a very recent pseudogene. For rpl16 it was not possible to obtain splicing evidence as the 5′-exon of this gene is only 9 bp long, which was too short for placing a primer. Although rpl2 has an intron in most flowering plant lineages, particularly in the “basal angiosperms,” including Piper (Cai et al 2006), this intron is absent from Hydnora.
Only three predicted tRNA genes were detected in the Hydnora plastome. All three can be folded into characteristic stem-loop structures essentially identical to their homologs in Piper and more distantly related angiosperm species (supplementary fig. S7, Supplementary Material online). The structures of the three tRNAs on the plastid genome were calculated using tRNA-Scan (http://lowelab.ucsc.edu/tRNAscan-SE/, last accessed January 11, 2016) and redrawn in RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi, last accessed January 11, 2016) to obtain higher resolution figures. The predicted anticodons (trnICAU, trnEUUC, and trnfMCAU) are unaltered in Hydnora. Compared with Piper, Hydnora shows one compensatory mutation in the stem of trnICAU, and three in trnfMCAU, all of which retain the stem-loop structures. Two of the three identified tRNAs have a single base pair indel compared with their homologs in Piper, but in both cases, the indel is located in the variable loop and may not affect functionality (supplementary fig. S7, Supplementary Material online). TrnICAU and trnfMCAU have the same anticodon, but trnICAU has been shown to be posttranscriptionally modified to AUA (Alkatib et al. 2012).
Discussion
The Minimal Plastid Genome of H. visseri
The plastid genome of H. visseri, a plant belonging to one of the most ancient parasitic angiosperm lineages (Naumann et al. 2013), shows extreme reduction in both size and gene content. The retention of only 24 genes encoded in the plastome and the loss of 89 genes compared with the close photosynthetic relative P. cenocladum (Cai et al. 2006) makes Hydnora the most minimal plastome sequenced to date, with respect to gene number, yet multiple lines of evidence suggest that it remains functional. The very long branch of Hydnora in the phylogram based on 20 plastid genes indicates a very high base substitution rate that is apparently even greater than in the also highly divergent plastomes of Sciaphila (Lam et al. 2015) or the Corsiaceae (Mennes et al. 2015). Although extremely divergent, all 17 protein-coding sequences encode potentially full-length ORFs, the sequences are highly similar and have experienced purifying selection between two related Hydnora species, and transcripts are detected for all 19 tested genes. Compensatory mutations help maintain stem–loop structures in conserved tRNA genes. Although the retained genes are dramatically divergent, the gene order is remarkably colinear with Piper, indicating that deletions have clearly occurred at a much higher rate than inversions in the plastome of Hydnora and its ancestors. This strong bias of deletions being much more numerous than inversions or other changes in gene order has also been observed in the highly reduced plastomes of holoparasitic Orobanchaceae (Wolfe et al. 1992; Wicke et al. 2013) and Cuscuta (Funk et al. 2007; McNeal et al. 2007).
Over the past decade, the number of sequenced plastomes of parasitic and mycoheterotrophic plants has increased significantly. Plastomes representing various evolutionary stages leading to and following complete heterotrophy show that similar patterns of gene loss and size reduction have occurred in both parasitic plants and mycoheterotrophs (Wolfe et al. 1992; Funk et al. 2007; McNeal et al. 2007; Wickett et al. 2008; Delannoy et al. 2011; Logacheva et al. 2011; Barrett and Davis 2012; Li et al. 2013; Wicke et al. 2013; Barrett et al. 2014; Logacheva et al. 2014; Uribe-Convers et al. 2014; Lam et al. 2015; Schelkunov et al. 2015). Independent evidence from multiple taxonomic lineages suggests that the evolution of plastid decay seems to follow a general pattern (Barrett and Davis 2012; Wicke et al. 2013; Barrett et al. 2014) associated with the reduction and eventual loss of photosynthetic constraints occurring in both groups with an increase of heterotrophic dependence (Lemaire et al. 2011; Barrett et al. 2014). The high degree of plastome reduction found in the ancient holoparasite Hydnora fits this pattern to an extreme, as the total coding capacity of Hydnora is the smallest yet observed in a potentially functional plastome.
The typical quadripartite structure of plastid genomes (a large IR separating two single copy regions) is conserved in most seed plants (Palmer 1985). Exceptions have been reported in a few plant lineages, including Geraniaceae (Guisinger et al. 2011), Poaceae (Guisinger et al. 2010), Vaccinium (Fajardo et al. 2013), Arbutus unedo (Martinez-Alberola et al. 2013), Fabaceae (Cai et al. 2008), Pinaceae and cupressophytes (Wu et al. 2011). However, nonphotosynthetic plants, which represent a small fraction of angiosperm species, possess remarkably varied plastomes, with structures including large IRs and only one single copy region (E. aphyllum; Schelkunov et al. 2015), or a very small IR (E. roseum; Schelkunov et al. 2015), to the complete loss of one IR copy (C. americana; Wicke et al. 2013). In the H. visseri plastid genome, all three plastome regions are retained, but each has been drastically reduced in size. The extreme contraction in size of the IR of Hydnora (to approximately 1.5 kb compared with 27 kb in Piper) has led to relocation of the genes that are located in the IR in Piperales (and possibly also in the immediate ancestors of Hydnora) to mainly the LSC. The gene order has remained mostly unaltered. The retention of the IR in Hydnora, although small, supports the hypothesis that the IR might be important for stabilizing and retaining the plastome over tens of millions of years (Palmer and Thompson 1982; Perry and Wolfe 2002; Maréchal and Brisson 2010).
Entire classes of genes that are commonly pseudogenized or lost during or soon after the transition to the heterotrophic lifestyle are entirely missing from the plastome of Hydnora: NADH dehydrogenase (ndh genes), ATP synthase (atp genes), RNA polymerase (rpo genes), photosystem (psa and psb genes), and cytochrome-related genes (pet genes). In Hydnora, the plastid genome reduction has gone far beyond that seen in assembled plastomes of most other heterotrophic plants. Only ribosomal proteins, ribosomal RNAs, some housekeeping genes, and three tRNAs genes are retained in the Hydnora plastome. The four ribosomal RNAs retained in Hydnora have also been found in all other plastomes and each is expected to be functionally essential (Wicke et al. 2013; Barrett et al. 2014; Lam et al. 2015; Schelkunov et al. 2015).
The plastomes of photosynthetic plants typically contain 21 ribosomal protein-coding genes, and at most, a handful are missing from nonphotosynthetic plants (Wicke et al. 2013; Barrett et al. 2014). The retention of 14 ribosomal protein genes in the Hydnora plastome is similar to the very reduced plastomes of other parasitic or mycotrophic plants where 15 (Sciaphila), 16 (Conopholis, Epipogium), and 19 (Epifagus) are retained. The ribosomal protein genes from Hydnora are a perfect subset of those from Conopholis, where rpl20 and rpl33 are retained in addition to those retained in Hydnora. To date, rpl20 has previously been identified as retained in all functional plastomes, and rpl33 is only reported missing from the plastomes of the mycoheterotrophic orchids (Delannoy et al. 2011; Schelkunov et al. 2015). Unless functional ribosomes can be assembled from a slightly smaller number of ribosomal proteins, the absence of seven ribosomal protein genes from the Hydnora plastome suggests that these proteins may be imported into the Hydnora plastid for ribosome assembly. This would imply either that these genes have been functionally transferred to another genomic compartment or that the missing components have been replaced by proteins normally functioning in the mitochondrial or nuclear ribosomes.
The retention of only 3 of 30 plastid-encoded tRNAs (trnICAU, trnEUUC, and trnfMCAU) is many fewer than what is expected for a minimal functional plastome (Lohan and Wolfe 1998) and is the smallest set of plastid tRNAs that has ever been observed. In comparison, six are retained in the extremely reduced plastomes of E. aphyllum and S. densiflora (Lam et al. 2015; Schelkunov et al. 2015) and 14 in Conopholis (Wicke et al. 2013). It has been discussed previously, and demonstrated with computer simulations, that some tRNAs could escape deletion by chance because of their small size and only moderate sequence divergence from an autotrophic ancestor (Lohan and Wolfe 1998). Their characteristic cloverleaf structure of tRNAs is highly conserved and is sensitive to mutations, especially in the stem regions. This is possibly the case in Orobanchaceae, as it is a rather young parasitic plant family (20 Myr old, including numerous photosynthetic members; Naumann et al. 2013). Hydnoraceae, however, is an ancient parasitic family, with at least 54 Myr to over 90 Myr of evolution as a holoparasite (Naumann et al. 2013); retention of a plastome by chance becomes more and more unlikely over time, especially considering the extreme downsizing and condensation of the plastome that Hydnora has experienced. Additionally, plastomes sequenced to date retain the same set of three tRNAs found in Hydnora (trnICAU, trnEUUC, and trnfMCAU), suggesting an essential function for all three tRNAs. As proposed by Howe and Smith (1991), trnE has a dual function in plastid biology (tetrapyrrole biosynthesis and protein biosynthesis), and this could be the reason why it cannot be replaced by its cytosolic counterpart making it an essential plastid-encoded gene. Isoleucine and Methionine, both encoded by two tRNAs (trnICAU and trnIGAU, as well as trnfMCAU and trnMCAU), seem to be essential for any plastome. It has been shown that for each of these two aminoacids, at least one tRNA has to be retained (Alkatib et al. 2012).
As many as four plastid protein-coding genes were proposed to be essential for a minimal plastome (ycf1, ycf2, accD, and clpP) (based on Epifagus virginiana; Wolfe et al. 1992). Three of these (ycf1, ycf2, and accD) are retained in Hydnora and in most other plant plastomes (supplementary fig. S4, Supplementary Material online) though one or more of these genes has been lost on occasion from nonphotosynthetic or even photosynthetic plastomes (Straub, Fishbein, et al. 2011; Wicke et al. 2011; Barrett et al. 2014). The caseinolytic protease encoded by clpP, which is part of the stromal proteolytic machinery (Adam and Clarke 2002), has been lost from Hydnora. ClpP is retained even in the most reduced plastomes of nonphotosynthetic plants sequenced to date (Delannoy et al. 2011; Wicke et al. 2013; Lam et al. 2015; Schelkunov et al. 2015). In photosynthetic plants, clpP has not been found in Scaevola and Passiflora (Jansen et al. 2007); it is a pseudogene in Asclepias (Straub, Fishbein, et al. 2011), Monsonia and Geranium (Geraniaceae, Guisinger et al. 2011), Trachelium (Haberle et al. 2008), Arbutus (Martínez-Alberola et al. 2013), and may encode a nonfunctional protein in Acacia (Williams et al. 2015). In those cases, it is possible that a nuclear-encoded homolog is transported into the plastid and functionally compensates for the missing or nonfunctional protease (Williams et al. 2015). Making imported proteins functional seems essential for any plastid, especially in holoparasitic plants where gene products might be retrieved from the host and act as substitutes for plastid-encoded proteins. A functional transfer of clpP to the nucleus in Hydnora is unlikely, as both the genome (albeit only 2× coverage) and a thorough transcriptome sequence (Naumann J, unpublished data) have been screened for plastid genes at different stringencies. As there is no evidence for clpP in Hydnora at all, some other protein may have adapted to serve the essential functions of clpP in the plastid.
The beta-carboxyl transferase subunit of accD as well as ycf1 and ycf2 is present and potentially functional in Hydnora. Ycf2, in particular, is the longest plastid gene in most plant plastomes (6,945 bp in Piper). Although it has an extremely high sequence divergence in Hydnora, it encodes a long ORF of 4,920 bp, which would be virtually impossible to be retained by chance during tens of millions of years of heterotrophic evolution and considerable sequence divergence (Leebens-Mack and dePamphilis 2002). The shorter total plastome sequence of E. roseum and S. densiflora (Lam et al. 2015; Schelkunov et al. 2015) as compared with Hydnora is mostly due to the loss of ycf1, and ycf2. The absence of these two genes appears to be a common pattern in the extremely reduced mycoheterotrophs (Lam et al. 2015; Schelkunov et al. 2015), but not in the sequenced parasitic plants (Wicke et al. 2013), including Hydnora.
A comparison of ten plastid genomes from the Orobanchaceae additionally suggests an essential function of matK in that parasite lineage, because genes containing group IIA introns are retained in the plastomes of that family of parasites (Wicke et al. 2013). On the other hand, matK has been lost in Cuscuta obtusifolia, Cuscuta campestris, and Cuscuta obtusifolia (Funk et al. 2007; McNeal et al. 2007; Braukmann et al. 2013) following the progressive and eventually complete loss of all group IIA introns from Cuscuta (McNeal et al. 2007). Parallel loss of matK and group IIA intron-containing genes has occurred in Rhizanthella gardneri (Delannoy et al. 2011). Maturase K (MATK) is an enzyme that is required for group IIA intron splicing of several plastid genes. It is retained in most reduced plastomes, although especially in the genus Cuscuta many cases of a pseudogenized matK have been reported (Braukmann et al. 2013). If all seven plastid genes containing group IIA introns (trnVUAC, trnIGAU, trnAUGC, trnKUUU, rpl2, rps12, and atpF; Zoschke et al. 2010) or the introns themselves were lost, matK would no longer be required (McNeal et al. 2007). In rps12, the first intron is a group IIB-intron, and the second is a group IIA-intron, being dependent on MATK for splicing. Amplification evidence of rps12 shows that the group IIA-intron is not being spliced out from Hydnora transcripts, which is in accordance with the absence of matK from the Hydnora plastome. This implies that this gene is being misspliced and is potentially nonfunctional (i.e., pseudogene), or it is spliced by another enzyme that is not plastid encoded. Although all genes on the plastome of the white albostrians barley mutants are transcribed, their translation is deficient due to lack of chloroplast ribosomes. It has been shown that the lack of MATK results in an immature mRNA of rps12, where the group IIA intron is not being spliced out (Hübschmann et al. 1996). In a green plant, where many genes require MATK to produce mature mRNAs, the loss of this protein would likely be lethal. In Hydnora, the second rps12 intron is the only MATK-dependent intron left, and thus proper splicing is disrupted in only one gene as opposed to seven. A similar pattern is found in plastomes of both Epipogium species (Lam et al. 2015). Both plastomes have lost matK, but have retained group IIA introns: The rpl2 intron (both species) and the second intron of clpP (E. roseum) are still retained. Loss of matK prior to the loss of one or more genes with intron sequences that depend on splicing by MATK is an alternative scenario from that proposed by McNeal et al. (2009) to explain the eventual loss of both matK and group IIA intron containing genes. Whether matK is lost prior to the final groupIIA intron loss (as suggested in Hydnora) or lost simultaneously to or after the loss of all group IIA introns (McNeal et al. 2009) could depend on the particular lineage in question, but the ultimate outcome—loss of both introns and maturase—would be identical.
Rpl2 is another gene that contains a group IIA intron in most angiosperms, but one that has also lost its intron several times in different angiosperm lineages independently (Downie et al. 1991). Although rpl2 seems functional in H. visseri, it appears to be a pseudogene in H. longicollis because of several indels throughout the gene that lead to frameshifts and numerous inferred stop codons. Both rps12 (both species) and rpl2 (only H. longicollis) may be in early stages of pseudogenization. They are transcribed in both H. visseri and H. longicollis, but it is unlikely that they could be translated into functional proteins. In some Orobanchaceae, rbcL is in early stages of pseudogenization (specifically in Hyobanche). The rbcL pseudogene was shown to be transcribed, but active RuBisCo enzyme was detected in some tissues and was hypothesized to be parasitized host enzyme (Randle and Wolfe 2005).
When the Epifagus plastome was first mapped, sequenced, and discussed (dePamphilis and Palmer 1990; Wolfe et al. 1992), it was hypothesized that a minimal plastome would require the functional retention of at least one gene required for a retained plastid-specific process, plus any nonexpendable machinery for its expression (Wolfe et al. 1992). This hypothesis has persisted through many sequenced plastomes of nonphotosynthetic plants and still holds in light of the Hydnora plastome. In addition to the dual function of trnE (Howe and Smith 1991), at least some ribosomal protein-coding genes and plastid ribosomal RNAs may be retained because they cannot be transferred to the nucleus due to interference with their cytosolic equivalents (Howe and Smith 1991; Barbrook et al. 2006). A third hypothesis to explain retention of some plastid genes (and therefore the plastid genome) is that plastid encoding is required for correct regulation of plastid gene expression based on redox balance (Allen 2003). As opposed to earlier stages in the evolution toward holoparasitism, other than retention of rDNAs and some ribosomal protein and tRNA genes in all of the reduced plastomes, there seems to be no universal pattern to the loss or retention of the very last few genes such as accD, clpP, ycf1, and ycf2. In different plant lineages, losses of one or more of these “potentially essential” genes have been reported repeatedly (reviewed in Wicke et al. 2011) and may depend upon whether nuclear or mitochondrial homologs can substitute for the loss of function of plastid copies in specific lineages of plants.
The Possible Loss of Plastid Genomes Revisited
The holoparasitic R. lagascae is a recently reported case of a potentially lost plastid genome in a flowering plant (Molina et al. 2014). This remarkable claim reopens the debate as to whether or not a plastid genome could be lost in plants (dePamphilis and Palmer 1990; Wolfe et al. 1992; Nickrent et al. 1997; Race et al. 1999; Bungard 2004; Barbrook et al. 2006; Krause 2008; Wicke et al. 2011; Janouškovec et al. 2015). Whenever the question of the loss or retention of a plastid genome is raised in a specific plant, first and foremost, the presence or absence of any particular type of plastid should be investigated. In H. visseri we find amyloplasts, plastids that are specialized for starch synthesis and storage. The storage of large amounts of starch might be important for Hydnora for floral thermogenesis (Seymour et al 2009). An additional possibility is that the starch might help Hydnora outlast recurrent periods of extreme drought in the desert habitat where it lives, because when the stored starch is degraded into monomers, water can be derived by oxidation of glucose in a subsequent reaction (Spoehr 1919).
The unicellular algae Polytomella appear to have also retained amyloplasts, but strong evidence was presented that the plastome has been lost from Polytomella (Smith and Lee 2014). In Rafflesia, the situation is less clear. Although plastid-like structures have been observed, they are not reported to contain starch (Molina et al. 2014). On a cellular level, whether or not the plastid-like structures in Rafflesia are derived plastids could be further explored with fluorescence labeling of known mitochondrial and nuclear-encoded plastid markers or fluorescence staining of the organellar membranes. The endophytic life style of Rafflesia could possibly reduce the spectrum of required plastid types, as well as enable the drastic evolutionary step of a complete plastome loss. If it is true that Rafflesia has lost its plastid genome, but retained its plastids, it has apparently retained function(s) other than starch storage.
However, it remains arguable whether or not Rafflesia has a lost its plastid genome. If Rafflesia’s gene sequences were highly divergent from available plastid genome sequences, as we have shown to be the case for Hydnora, there is a chance it could have escaped detection by all of the approaches used in Molina et al. (2014) (mapping scaffolds to a photosynthetic reference, BLASTn to plastid genomes, and Hidden Markov Models of plastid gene alignments). Due to the reduced size and gene content, as well as the high sequence divergence and compositional bias of coding genes, finding plastid sequences in the H. visseri assembly was extremely challenging. Basic similarity-based approaches did not serve to identify plastid genes as they would for the vast majority of plants, where the plastid genome is very straightforward to extract from genomic sequence assemblies (Straub, Parks, et al. 2011). The identification and annotation of the Hydnora plastid genome was a long process that required multiple approaches, where especially the relative read depths for the three genomic compartments were found to be valuable evidence for identifying organellar genomes.
Even in low coverage genomic data (Straub, Fishbein, et al. 2011; Wolf et al. 2015), the plastid and the mitochondrial genomes are expected to be captured at distinct stoichiometries in the sample (Bock 2007; Straub et al. 2011; Wolf et al. 2015). Usually, there are tens to hundreds of mitochondrial genomes per cell, but thousands of plastomes, which will typically show a higher stoichiometry for a functional plastome (Straub et al. 2011). Nonphotosynthetic or senescent tissues with reduced photosynthetic activity, however, can show a decreased plastome copy number (Fulgosi et al. 2012) and thus, show a reduced read depth of the plastome in the genomic sample (Bowman and Simon 2013). If the plastome cannot be identified by its stoichiometry in the genomic assembly, the mitochondrial genome is a potentially important place to carefully look for old plastid gene “fossils.” As gene transfer from the plastid to the mitochondrion is commonly discovered in flowering plants, and plant mitochondrial genes generally show a very low substitution rate, the origin of integrated genes, and sometimes even ancient transfers, can be tracked back to their plastid origins (Wolfe et al. 1987; Mower et al. 2007; Rice et al. 2013).
Conclusion
The plastid genome of H. visseri shows a unique combination of features: An extreme downsizing and gene reduction, especially of the tRNAs, and an extreme sequence divergence and base compositional bias, whereas the retained genes show multiple indications of probable functionality. This suggests the following evolutionary scenario for the plastid genome in nonphotosynthetic plants. First, in the “degradation stage I,” nonessential and photosynthesis-related genes are pseudogenized successively, followed by complete loss of those genes. The order of gene loss follows a recurring pattern in the different lineages of hemiparasitic and nonphotosynthetic plants, particularly observable in the Orobanchaceae and the Orchidaceae, where plastomes with various degrees of reduction have been examined (e.g., Barrett and Davis 2012; Wicke et al. 2013; Barrett et al. 2014; Cusimano and Wicke 2015). Second, in the “stationary stage” only genes required for nonphotosynthetic functions are retained, the rate of gene loss is much slower, and pseudogenes are expected to be rarely produced. At this stage, further gene loss is likely to be dependent upon the ability of imported or substitute proteins to serve any continuing required function in the nonphotosynthetic plastid. Alternatively, successful functional transfers of genes into the nucleus or mitochondrion, with a transit peptide to direct the protein back into the plastid, could allow additional genes to be lost from the plastome. Such events of functional gene transfer in green plant lineages are rare (Baldauf and Palmer 1990; Martin et al. 1998). Thus, this stage can potentially last much longer than the degradation stages, although the duration of the stationary stage may be lineage specific and depend on many factors. The retained plastid genes, however, continue to evolve, sometimes with a relatively high rate of net mutation after repair; hence they show a high level of sequence divergence after tens of millions of years. If the last essential plastid gene is lost or functionally replaced with a nuclear or mitochondrial copy, the plastome would become unnecessary; this would mark the third, or “degradation stage II.” If this occurs, then any remaining genes that have been maintained only to facilitate the expression of an essential gene will be pseudogenized and eventually lost; this would make a remnant plastome in this stage even harder to detect. Finally, in the “absent” stage, the plastome is completely lost, except for small fragments that may reside in other parts of the genome, and those genes functionally transferred to another genomic location whose gene products still function in the plastid. In the case of Rafflesiaceae, they either have an extremely reduced, even more divergent plastome than Hydnora (late stage 2 or stage 3), or this lineage has entirely lost its plastome (stage 4). The retained short and low coverage plastid fragments of Rafflesia and Polytomella (Molina et al. 2014; Smith and Lee 2014, respectively) remain to be characterized in more detail. The minimal, but functional Hydnora plastome, being well into stage 2, helps us understand how diminished a plastome can be while still retaining functionality.
Supplementary Material
Supplementary figures S1–S7 and tables S1–S3 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the University for Technology Dresden, and in part by the Parasitic Plant Genome Project Grant (P.P.G.P., U.S. N.S.F IOS #0701748) to James H. Westwood, C.W.D., Michael P. Timko, and John I. Yoder. The authors are also grateful for additional funding provided by the TU Dresden “starting grant” to S.W. and by the DFG Piperales project to S.W., C.N., and Nick Rowe (NE 681/11-1). They also thank Daniela Drautz, Lynn P. Tomsho, and Stephan C. Schuster (Penn State University) for generating the genomic sequence data. Personnel exchange between the TU Dresden and Penn State University was supported by a DAAD PPP USA grant to S.W. The collection of plant tissue of Hydnora visseri was conducted under Namibian MET Permit No. 1360/2009. They are also grateful for the support of Gondwana Cañon Preserve, Lytton J. Musselman, and the Namibian National Botanical Research Institute. They also thank Susann Wicke for valuable advice and many helpful suggestions that improved the manuscript, Zhenzhen Yang for help setting up CodeML (test for significance of relaxed dN/dS ratio), and Wen-Bin Yu for helpng to revise the comparison of plastome genes (supplementary fig. S4, Supplementary Material online).
Literature Cited
- Adam Z, Clarke AK. 2002. Cutting edge of chloroplast proteolysis. Trends Plant Sci. 7(10):451–456. [DOI] [PubMed] [Google Scholar]
- Alkatib S, Fleischmann TT, Scharff LB, Bock R. 2012. Evolutionary constraints on the plastid tRNA set decoding methionine and isoleucine. Nucleic Acids Res. 40(14):6713–6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF. 2003. The function of genomes in bioenergetic organelles. Philos Trans R Soc Lond B Biol Sci. 358:19–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf SL, Palmer JD. 1990. Evolutionary transfer of the chloroplast tufA gene to the nucleus. Nature 344:262–265. [DOI] [PubMed] [Google Scholar]
- Barbrook AC, Howe CJ, Purton S. 2006. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci.. 11:101–108. [DOI] [PubMed] [Google Scholar]
- Barkman TJ, et al. 2007. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol Biol. 7:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CF, Davis JI. 2012. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am J Bot. 99:1513–1523. [DOI] [PubMed] [Google Scholar]
- Barrett CF, et al. 2014. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 31(12):3095–3112. [DOI] [PubMed] [Google Scholar]
- Beentje H, Luke WQ. 2002. Hydnoraceae. In: Beentje H, Ghazanfar SA, editors. Flora of tropical east Africa. Rotterdam (The Netherlands): CRC Press; p. 1–7. [Google Scholar]
- Bock R, editor. 2007. Structure, function, and inheritance of plastid genomes In: Cell and molecular biology of plastids Springer: Berlin Heidelberg: p. 29–63. [Google Scholar]
- Bolin JF, Maass E, Musselman LJ. 2009. Pollination biology of Hydnora africana Thunb. (Hydnoraceae) in Namibia: brood-site mimicry with insect imprisonment. Int J Plant Sci. 170:157–163. [Google Scholar]
- Bolin JF, Maass E, Musselman LJ. 2011. A new species of Hydnora (Hydnoraceae) from Southern Africa. Syst Bot. 36:255–260. [Google Scholar]
- Bolin JF, Tennakoon KU, Maass E. 2010. Mineral nutrition and heterotrophy in the water conservative holoparasite Hydnora Thunb.(Hydnoraceae). Flora 205(12):802–810. [Google Scholar]
- Bowman MJ, Simon PW. 2013. Quantification of the relative abundance of plastome to nuclear genome in leaf and root tissues of carrot (Daucus carota L.) using quantitative PCR. Plant Mol Biol. 31(4):1040–1047. [Google Scholar]
- Braukmann T, Kuzmina M, Stefanović S. 2013. Plastid genome evolution across the genus Cuscuta (Convolvulaceae): two clades within subgenus Grammica exhibit extensive gene loss. J Exp Bot. 64:977–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braukmann T, Stefanović S. 2012. Plastid genome evolution in mycoheterotrophic Ericaceae. Plant Mol Biol. 79(1–2):5–20. [DOI] [PubMed] [Google Scholar]
- Brown DL, Massalski A, Patenaude R. 1976. Organization of the flagellar apparatus and associate cytoplasmic microtubules in the quadriflagellate alga Polytomella agilis. J Cell Biol. 69:106–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungard RA. 2004. Photosynthetic evolution in parasitic plants: insight from the chloroplast genome. Bioessays 26:235–247. [DOI] [PubMed] [Google Scholar]
- Cai Z, et al. 2006. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evol Biol. 6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, et al. 2008. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J Mol Evol. 67:696–704. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. 1993. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 11(2):113–116. [Google Scholar]
- Cocucci AE, Cocucci AA. 1996. Prosopanche (Hydnoraceae): somatic and reproductive structures, biology, systematics, phylogeny and potentialities as a parasitic weed In: Moreno MT, Cubero JI, Berner D, Joel D, Musselman LJ, Parker C, editors. Advances in Parasitic Plant Research: Junta de Andalucia. Cordoba (Spain): Direccion General de Investigacion Agraria p. 179–193. [Google Scholar]
- Cusimano N, Wicke S. 2015. Massive intracellular gene transfer during plastid genome reduction in nongreen Orobanchaceae. New Phytol. doi:10.1111/nph.13784 [DOI] [PubMed] [Google Scholar]
- Delannoy E, Fujii S, Colas des Francs-Small C, Brundrett M, Small I. 2011. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol. 28:2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dePamphilis CW, Palmer JD. 1990. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature 348:337–339. [DOI] [PubMed] [Google Scholar]
- de Vries J, Sousa FL, Bölter B, Soll J, Gould SB. 2015. YCF1: a green TIC? Plant Cell . tcp-114:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie SR, et al. 1991. Six independent losses of the chloroplast DNA rpl2 intron in dicotyledons: molecular and phylogenetic implications. Evolution 45(5):1245–1259. [DOI] [PubMed] [Google Scholar]
- Drouin G, Daoud H, Xia J. 2008. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol Phylogenet Evol. 49:827–831. [DOI] [PubMed] [Google Scholar]
- Ems SC, et al. 1995. Transcription, splicing and editing of plastid RNAs in the nonphotosynthetic plant Epifagus virginiana. Plant Mol Biol. 29(4):721–733. [DOI] [PubMed] [Google Scholar]
- Fajardo D, et al. 2013. Complete plastid genome sequence of Vaccinium macrocarpon: structure, gene content, and rearrangements revealed by next generation sequencing. Tree Genet Genomes 9(2):489–498. [Google Scholar]
- Fulgosi H, et al. 2012. Degradation of chloroplast DNA during natural senescence of maple leaves. Tree Physiol. 32:346–354. [DOI] [PubMed] [Google Scholar]
- Funk H, Berg S, Krupinska K, Maier U, Krause K. 2007. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK. 2010. Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J Mol Evol. 70(2):149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. 2011. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 28(1):583–600. [DOI] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK. 2008. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol. 66(4):350–361. [DOI] [PubMed] [Google Scholar]
- Harris RS. 2007. Improved pairwise alignment of genomic DNA. [PhD thesis]. The Pennsylvania State University, PA, USA.
- Howe CJ, Smith AG. 1991. Plants without chlorophyll. Nature 349:109.1986302 [Google Scholar]
- Hübschmann T, Hess WR, Börner T. 1996. Impaired splicing of the rps12 transcript in ribosome-deficient plastids. Plant Mol Biol. 30(1):109–123. [DOI] [PubMed] [Google Scholar]
- Janouškovec J, et al. 2015. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc Natl Acad Sci U S A. 112(33):10200–10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 104:19369–19374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Ruhlman TA. 2012. Plastid genomes of seed plants In: Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria, advances in photosynthesis and respiration 35. Dordrecht (The Netherlands): Springer; p. 103–126. [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, et al. 2013. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339:571–574. [DOI] [PubMed] [Google Scholar]
- Krause K. 2008. From chloroplasts to “cryptic” plastids: evolution of plastid genomes in parasitic plants. Curr Genet. 54:111–121. [DOI] [PubMed] [Google Scholar]
- Krause K, Scharff LB. 2014. Reduced genomes from parasitic plant plastids: templates for minimal plastomes? In: Lüttge U, Beyschlag W, Cushman J, editors. Progress in botany. Heidelberg (Germany): Springer Berlin Verlag Heidelberg; p. 97–115. [Google Scholar]
- Kroeger TS, Watkins KP, Friso G, van Wijk KJ, Barkan A. 2009. A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc Natl Acad Sci U S A. 106(11):4537–4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam VKY, Soto Gomez M, Graham SW. 2015. The highly reduced plastome of mycoheterotrophic Sciaphila (Triuridaceae) is colinear with its green relatives and is under strong purifying selection. Genome Biol Evol. 7(8):2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leebens-Mack J, dePamphilis CW. 2002. Power analysis of tests for loss of selective constraint in cave crayfish and nonphotosynthetic plant lineages. Mol Biol Evol. 19(8):1292–1302. [DOI] [PubMed] [Google Scholar]
- Lemaire B, Huysmans S, Smets E, Merckx V. 2011. Rate accelerations in nuclear 18S rDNA of mycoheterotrophic and parasitic angiosperms. J Plant Res. 124:561–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, et al. 2013. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae). PLoS One 8(3):e58747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logacheva MD, Schelkunov MI, Nuraliev MS, Samigullin TH, Penin AA. 2014. The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements. Genome Biol Evol. 6(1):238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logacheva MD, Schelkunov MI, Penin AA. 2011. Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol Evol. 3:1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohan AJ, Wolfe KH. 1998. A subset of conserved tRNA genes in plastid DNA of nongreen plants. Genetics 150:425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274. [DOI] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass EE, Musselman LJ. 2001. Parasitic plants pummel pavement-Hydnora abyssinica (Hydnoraceae). Econ Bot. 55(1):7–8. [Google Scholar]
- Maréchal A, Brisson N. 2010. Recombination and the maintenance of plant organelle genome stability. New Phytol. 186:299–317. [DOI] [PubMed] [Google Scholar]
- Martin W, et al. 1998. Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393:162–165. [DOI] [PubMed] [Google Scholar]
- Martinez-Alberola F, et al. 2013. Balanced gene losses, duplications and intensive rearrangements led to an unusual regularly sized genome in Arbutus unedo chloroplasts. PLoS One 8(11):e79685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, dePamphilis CW. 2007. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal JR, Kuehl JV, Boore JL, Leebens-Mack JH, dePamphilis CW. 2009. Parallel loss of plastid introns and their maturase in the genus Cuscuta. PLoS One 4(6):e5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennes CB, et al. 2015. Ancient Gondwana break-up explains the distribution of the mycoheterotrophic family Corsiaceae (Liliales). J Biogeogr. 42(6):1123–1136. [Google Scholar]
- Molina J, et al. 2014. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Mol Biol Evol. 31:793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J, Cantor MH, Sheeler P, Kahn W. 1970. The ultrastructure of Polytomella agilis. J Protozool. 17:671–676. [Google Scholar]
- Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD. 2007. Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol. 7(1):135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman LJ, Visser JH. 1989. Taxonomy and natural history of Hydnora (Hydnoraceae). Aliso 12(2):317–326. [Google Scholar]
- Nakai M. 2015. YCF1: a green TIC: response to the de Vries et al. Commentary. Plant Cell 27:1834–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann J, et al. 2013. Single-copy nuclear genes place haustorial Hydnoraceae within Piperales and reveal a Cretaceous origin of multiple parasitic angiosperm lineages. PLoS One 8(11):e79204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickrent DL, et al. 2002. Molecular data place Hydnoraceae with Aristolochiaceae. Am J Bot. 89:1809–1817. [DOI] [PubMed] [Google Scholar]
- Nickrent DL, Yan OY, Duff RJ, dePamphilis CW. 1997. Do nonasterid holoparasitic flowering plants have plastid genomes? Plant Mol Biol. 34:717–729. [DOI] [PubMed] [Google Scholar]
- Ohyama K. 1996. Chloroplast and mitochondrial genomes from a liverwort, Marchantia polymorpha-gene organization and molecular evolution. J Mol Evol. 60:16–24. [DOI] [PubMed] [Google Scholar]
- Palmer JD. 1985. Comparative organization of chloroplast genomes. Annu Rev Genet. 19:325–354. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Herbon LA. 1988. Plant mitochondrial DNA evolved rapidly in structure, but slowly in sequence. J Mol Evol. 28:87–97. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Thompson WF. 1982. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29(2):537–550. [DOI] [PubMed] [Google Scholar]
- Park S, et al. 2014. Complete sequences of organelle genomes from the medicinal plant Rhazya stricta (Apocynaceae) and contrasting patterns of mitochondrial genome evolution across asterids. BMC Genomics 15:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry AS, Wolfe KH. 2002. Nucleotide substitution rates in legume chloroplast DNA depend on the presence of the inverted repeat. J Mol Evol. 55(5):501–508. [DOI] [PubMed] [Google Scholar]
- Petersen G, Cuenca A, Seberg O. 2015. Plastome evolution in hemiparasitic mistletoes. Genome Biol Evol. 7(9):2520–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race HL, Hermann RG, Martin WF. 1999. Why have organelles retained genomes? Trends Genet. 15(9):364–370. [DOI] [PubMed] [Google Scholar]
- Randle CP, Wolfe AD. 2005. The evolution and expression of rbcL in holoparasitic sister-genera Harveya and Hyobanche (Orobanchaceae). Am J Bot. 92(9):1575–1585. [DOI] [PubMed] [Google Scholar]
- Rice DW, et al. 2013. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 342:1468–1473. [DOI] [PubMed] [Google Scholar]
- Ruhlman T, Jansen RK. 2014. The plastid genomes of flowering plants In: Maliga P, editor. Chloroplast biotechnology: methods and protocols, methods in molecular biology. New York: Humana Press; p. 3–38. [DOI] [PubMed] [Google Scholar]
- Ruiz-Nieto JE, et al. 2015. Photosynthesis and chloroplast genes are involved in water-use efficiency in common bean. Plant Physiol Biochem. 86:166–173. [DOI] [PubMed] [Google Scholar]
- Schelkunov MI, et al. 2015. Exploring the limits for reduction of plastid genomes: a case study of the, mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol Evol. 7(4):1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour RS, Maass E, Bolin JF. 2009. Floral thermogenesis of three species of Hydnora (Hydnoraceae) in Africa. Ann Bot. 104:823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, et al. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. Embo J. 5:2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Lee RW. 2014. A plastid without a genome: evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiol. 164(4):1812–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoehr HA, editor. 1919. The carbohydrate economy of cacti. Washington: Carnegie Institution of Washington, The Lord Baltimore Press. [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Stöver BC, Müller KF. 2010. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub SCK, Fishbein M, et al. 2011. Building a model: developing genomic resources for common milkweed (Asclepias syriaca) with low coverage genome sequencing. BMC Genomics 12:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub SCK, Parks M, et al. 2011. Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Am J Bot. 99:349–364. [DOI] [PubMed] [Google Scholar]
- Uribe-Convers S, Duke JR, Moore MJ, Tank DC. 2014. A long PCR–based approach for DNA enrichment prior to next-generation sequencing for systematic studies. Appl Plant Sci. 2(1):1300063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser JH, Musselman LJ. 1986. The strangest plant in the world! Veld Flora 71:109–111. [Google Scholar]
- Wagner ST, et al. 2014. Major trends in stem anatomy and growth forms in the perianth-bearing Piperales, with special focus on Aristolochia. Ann Bot. 113(7):1139–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, et al. 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25:3711–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, Müller KF, dePamphilis CW, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76:273–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, et al. 2008. Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis. Mol Biol Evol. 25:393–401. [DOI] [PubMed] [Google Scholar]
- Williams AV, Boykin LM, Howell KA, Nevill PG, Small I. 2015. The complete sequence of the Acacia ligulata chloroplast genome reveals a nighly divergent clpP1 gene. PLoS One 10(5):e0125768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf PG, et al. 2015. An exploration into fern genome space. Genome Biol Evol. 7(9):2533–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM. 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 84:9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD. 1992. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci U S A. 89:10648–10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CS, Wang YN, Hsu CY, Lin CP, Chaw SM. 2011. Loss of different inverted repeat copies from the chloroplast genomes of Pinaceae and cupressophytes and onfluence of heterotachy on the evaluation of Gymnosperm phylogeny. Genome Biol Evol. 3:1284–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21:1859–1875. [DOI] [PubMed] [Google Scholar]
- Wu Z, Cuthbert JM, Taylor DR, Sloan DB. 2015. The massive mitochondrial genome of the angiosperm Silene noctiflora is evolving by gain or loss of entire chromosomes. Proc Natl Acad Sci U S A. 112(33):10185–10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Xu B, Yang Z. 2013. PAMLX: a graphical user interface for PAML. Mol Biol Evol. 30(12):2723–2724. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zoschke R, et al. 2010. An organellar maturase associates with multiple group II introns. Proc Natl Acad Sci U S A. 107:3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.