Abstract
Auxin/indole acetic acids (Aux/IAAs) and auxin response factors (ARFs), major components of the Aux signaling network, are involved in many developmental processes in plants. Investigating their evolution will provide new sight on the relationship between the molecular evolution of these genes and the increasing morphotypes of plants. We constructed comparative analyses of the retention, structure, expansion, and expression patterns of Aux/IAAs and ARFs in Brassica rapa and their evolution in eight other plant species, including algae, bryophytes, lycophytes, and angiosperms. All 33 of the ARFs, including 1 ARF-like (AL) (a type of ARF-like protein) and 53 Aux/IAAs, were identified in the B. rapa genome. The genes mainly diverged approximately 13 Ma. After the split, no Aux/IAA was completely lost, and they were more preferentially retained than ARFs. In land plants, compared with ARFs, which increased in stability, Aux/IAAs expanded more rapidly and were under more relaxed selective pressure. Moreover, BraIAAs were expressed in a more tissue-specific fashion than BraARFs and demonstrated functional diversification during gene duplication under different treatments, which enhanced the cooperative interaction of homologs to help plants adapt to complex environments. In addition, ALs existed widely and had a closer relationship with ARFs, suggesting that ALs might be the initial structure of ARFs. Our results suggest that the rapid expansion and preferential retention of Aux/IAAs are likely paralleled by the increasingly complex morphotypes in Brassicas and even in land plants. Meanwhile, the data support the hypothesis that the PB1 domain plays a key role in the origin of both Aux/IAAs and ARFs.
Keywords: Brassica rapa, IAA, ARF, copy number variation, evolutionary pattern, expression divergence
Introduction
The plant hormone auxin (indole-3-acetic acid; Aux) is involved in many physiological and developmental processes in land plants, such as organogenesis, vascular tissue differentiation, cell elongation, apical dominance, gravitropism, and root patterning (Ludwig-Muller 2011; Gallavotti 2013; Shalom et al. 2014).
The auxin response factor (ARF) family and the Aux/indole acetic acid (IAA) family play key roles in regulating the expression of auxin response genes (Wright and Nemhauser 2015). During plant development, the short-lived nuclear-localized proteins Aux/IAA are able to interact with and repress ARF transcription factors under basal (i.e., low auxin) conditions (Ulmasov et al. 1997, 1999; Tiwari et al. 2004). Increased auxin reduces the levels of the Aux/IAA proteins by accelerating their degradation through SCFTIR1 complex, and therefore ARF activity is derepressed and numerous auxin-mediated transcriptional changes occur (supplementary fig. S1A, Supplementary Material online; Gray et al. 2001; Korasick et al. 2014). Most Aux/IAAs have three recognizable domains: An N-terminal EAR-motif (domain I), a short degron domain (domain II), and a C-terminal PB1 (Phox and Bem1) protein–protein interaction domain (previously referred to as domain III/IV or domains III and IV; Guilfoyle 2015). The PB1 domain also exists in most of the ARFs to facilitate the formation of ARF–ARF, ARF–Aux/IAA, and Aux/IAA–Aux/IAA homo- and hetero-oligomers; thus their interactions form a complex network in Arabidopsis thaliana (supplementary fig. S1B, Supplementary Material online). Additionally, most ARFs contain a middle region that functions as an activation domain, such as Arabidopsis ARF5-8 and 19 (Ulmasov et al. 1999; Tiwari et al. 2003), or repression domain. The N-terminal region on ARFs can also serve as a binding domain DNA binding domain (DBD) that is related to the B3 domain family and recognizes the TGTCTC sequence (auxin response elements) (Guilfoyle and Hagen 2007, 2012). The B3 domain family mainly includes ABI3, HSI, RAV, and ARF families that they are mainly involved in hormone signaling pathways (Romanel et al. 2009). However, the Ar. thaliana Aux/IAA protein family has diversified in degradation and Aux responsiveness. For example, AXR2/IAA7 has half-lives of 5–12 min; IAA8, IAA9, and IAA28 can degrade more slowly than previously characterized family members (Dreher et al. 2006).
During their evolution, plants have substantially altered their phenotypes to adapt to environmental changes by transforming the form and function of genes. Gene duplication, even a whole genome duplication (WGD), offers the chance for genes to change themselves (Rensing 2014). After duplication, one copy of the gene might either become nonfunctional (pseudogenized or silenced, also called gene death) or acquire a novel function (neofunctionalization). Alternatively, the two duplicates might divide the original function of the gene (subfunctionalization; Innan and Kondrashov 2010). During the long history of adjusting to the environment, it is unclear what happened to the Aux/IAA and ARF genes and how this affected the plant phenotypes. In previous reports of the moss (Physcomitrella patens) genome, the Aux signaling machinery originated before the divergence of moss and vascular plants (Rensing et al. 2008). In 2013, Finet et al. inferred that the ARF domain is a land plant innovation and found at least two ARF genes, corresponding to the precursors of the A and C clades, which already existed in the last common ancestor of the extant land plants. Further analyses of the charophyte expressed sequence tag library identified several Aux/IAAs and ARFs already present in the charophyte genomes (Lau et al. 2009; De Smet et al. 2011; Viaene et al. 2013). Although the Aux/IAAs and ARFs of several charophytes (Klebsormidium flaccidum, Nitella mirabilis) did not contain all of the typically conserved domains, they may nonetheless affect algal development (Wang et al. 2015).
The Brassica genus is composed of many diverse species, and each species contains rich morphotypes showing extreme traits. Brassica rapa (AA genome), a diploid species, shared a complex history with Ar. thaliana (γ, β, and α events) and experienced an additional whole genome triplication (WGT) event 13–17 Ma (Wang et al. 2011; Cheng et al. 2013). Since its divergence from Ar. thaliana from their most recent common ancestor (MRCA), B. rapa has undergone considerable fractionation. Compared with the 24 conserved ancestral genomic blocks (GBs, labeled A–X) in Ar. thaliana, 71 GBs were identified in B. rapa by syntenic ortholog analysis (Schranz et al. 2006; Wang et al. 2011). To identify the degree of fractionation (gene loss), the B. rapa genome was divided into three subgenomes: LF, MF1, and MF2 (Wang et al. 2011; Cheng et al. 2012). The specific and well-studied genome of B. rapa was then used to study the evolution of the Aux/IAA and ARF gene families and the relationship between these two gene families and morphotype diversity of Brassicas plants. In addition, the genome sequences for many plant species, such as algae, the ancestor of land plants, containing both protistan and higher taxa (Debashish and Linda 1998); bryophytes, the closest extant relatives of early land plants (Rensing et al. 2008); and lycophytes, early vascular plants with a dominant sporophyte generation (Banks et al. 2011), were recently made available. Moreover, the ARF gene family was identified and analyzed in B. rapa by Mun et al. (2012). All these data enable comparative genome analyses so that inferences can be made regarding the origin and evolution history of the Aux/IAA and ARF genes, especially in B. rapa.
In this study, we identified 53 Aux/IAAs, 32 ARFs, and 1 AL (which was defined by us) in B. rapa. Then, we constructed a comprehensive comparative analysis of Aux/IAAs and ARFs, including gene retentions, gene structures, gene expansions, and gene expression patterns, in different tissues to characterize the divergences in composition, expansion, and expression. In addition, the expression patterns of Aux/IAAs in response to stress treatments were explored. Furthermore, a comparative genomic analysis of the Aux/IAA and ARF genes in 8 other plant species, including 4 eudicots (Ar. thaliana, Carica papaya, Populus trichocarpa and Vitis vinifera), 1 basal angiosperm (Amborella trichopoda), 1 lycophyte (Selaginella moellendorffii), 1 moss (Ph. patens), and 1 chlorophyta (Chlamydomonas reinhardtii), demonstrated that AL genes widely exist, providing a new path to understand the origin of the Aux/IAA and ARF genes. The divergence of the expression patterns and their rapid expansion suggest that they play key roles during the creation of increasingly complex morphotypes in Brassicas.
Materials and Methods
Identification of the Aux/IAA and ARF Genes in Multiple Species
The Ar. thaliana Aux/IAA and ARF protein sequences were downloaded from the Arabidopsis Information Resource database (http://www.arabidopsis.org/; supplementary table S1, Supplementary Material online). All the files related to the B. rapa genome sequence data were obtained from the Brassica database (BRAD; http://brassicadb.org/brad/; Wang et al. 2011). The gene information for Am. trichopoda genes was retrieved from the Amborella Genome Database (http://www.amborella.org/; Albert et al. 2013). The gene information for V. vinifera, Ca. papaya, Po. trichocarpa, Ph. patens, S. moellendorffii, and Ch. reinhardtii were downloaded from Phytozome v9.1 (http://www.phytozome.net/; Goodstein et al. 2012).
To identify putative Aux/IAA and ARF family members, the Hidden Markov Model (HMM) profiles of Aux/IAA (PF02309), ARF (PF06507), and B3 (B3, PF02362) were retained from the Pfam database (http://pfam.xfam.org/) and were used to identify the putative Aux/IAA and ARF proteins with the best domain e-value cutoffs of < 1 × 10−4. The Arabidopsis Aux/IAA and ARF sequences were used as the query to perform a BLAST search in these species, with a cutoff e-value of < 10−10. To validate the HMM and BLAST search, these potential sequences were analyzed using the SMART tool (http://smart.embl-heidelberg.de/; Ludwig-Muller 2011) and the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov/). Then, the FGENESH program (http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind) was used to rectify incorrect start codon predictions, splicing errors, and missed or extra exons; manual reannotation was performed.
Synteny Analysis of the Aux/IAA and ARF Genes between Arabidopsis thaliana and Brassica rapa
The synteny between the Ar. thaliana and B. rapa genomes was constructed by McScanX (http://chibba.pgml.uga.edu/mcscan2/; MATCH_SCORE: 50, MATCH_SIZE: 5, GAP_SCORE: −3, E_VALUE: 1× 10−5; Wang, Deng, et al. 2012; Wang, Tang, et al. 2012; Duan et al. 2015). An all-against-all BLASTP comparison provided the pairwise gene information and the P value for a primary clustering. Paired segments were extended by identifying the clustered genes using dynamic programing. The potentially duplicated genes were also identified using MCScanX. The resulting BLAST hits were incorporated along with the chromosome coordinates of all of the protein-coding genes as an input for MCScanX and classified into segmental, tandem, proximal, and dispersed duplications under the default criteria.
Conservation of chromosomal synteny around the BraAL and ARF4 genes was derived from CoGe (http://www.genomevolution.org/CoGe/GEvo.pl). The positions of the B. rapa Aux/IAA and ARF genes in the blocks were verified by searching for homologous genes between Ar. thaliana and the three subgenomes of B. rapa (LF, MF1, and MF2) in BRAD (http://brassicadb.org/brad/searchSynteny.php; Cheng et al. 2012). Circos software was used to draw the syntenic diagram (Krzywinski et al. 2009).
Ks Analysis
The protein sequences of Aux/IAA and ARF from B. rapa were aligned with their syntenic genes in Ar. thaliana using MUSCLE (Edgar 2004). The protein alignments were translated into coding sequence alignments using an in-house Perl script. Ks (synonymous substitution rate) and Ka (nonsynonymous substitution rate) values were calculated based on the coding sequence alignments using the method of Nei and Gojobori as implemented in KaKs_calculator (Zhang et al. 2006). The Ks values of all the syntenic orthologs between B. rapa and Ar. thaliana were then plotted as the density and a boxplot using the R program (Ihaka and Gentleman 1996). The divergence time was calculated with the formula T = Ks/2r, with Ks being the synonymous substitutions per site and r being the rate of divergence for nuclear genes from plants. The r was taken to be 1.5 × 10−8 synonymous substitutions per site per year for dicotyledonous plants (Koch et al. 2000).
Phylogenetic and Molecular Evolution Analysis of the IAA and ARF Gene Families
The full-length protein sequences of Aux/IAAs and ARFs were aligned using the MUSCLE program with the default parameters (Edgar 2004). The phylogenetic relationship was constructed using the maximum-likelihood method in each analysis. Bootstrap values were calculated with 1,000 replications using MEGA5.2 (Tamura et al. 2011).
The variation in selective pressures between the classes of ARFs and Aux/IAAs were evaluated according to Yang et al. (2013). The branch models of CODEML in PAML were used to estimate ω = (dN/dS) under two assumptions: A one-ratio model that assumes the same ω ratio for the two classes and a two-ratio model in which the two classes were assigned to different ω ratios. To verify which of the models best fit the data, likelihood-ratio tests (LRTs) were performed by comparing twice the difference in log-likelihood values between pairs of the models using a χ2 distribution (Yang and Nielsen 2000).
Motif Identification and Exon–Intron Structural Analysis
To identify the conserved motifs of the Aux/IAAs and ARFs of B. rapa, the online Multiple Expectation-maximization for Motif Elicitation program version 4.9.0 (Bailey et al. 2009) was employed among the amino acid sequences with the default parameters, except for the following parameters: Maximum number of motifs, 12; optimum motif width ≥20 and ≤120.
The position information of the Aux/IAA, ARF, and B3 domains was obtained from the Pfam database, and the gene structure information of the IAAs and ARFs was parsed from the General Feature Format files of B. rapa using an in-house Perl script. Then, the domain and exon–intron structures positions were drawn using the online program GSDS (http://gsds.cbi.pku.edu.cn/; Hu et al. 2015).
Orthologous Aux/IAA and ARF Gene Analysis among Eight Plant Species
Among the eight plant species, the homologous Aux/IAA and ARF genes were identified by the program OrthoMCL (http://www.orthomcl.org/cgi-bin/OrthoMclWeb.cgi; Li et al. 2003) with the default settings, which initially required an all-against-all BLASTP, and then the relationships between the genes were deduced by the MCL clustering algorithm. The network of all the gene relationships was built using Cytoscape software (version 3.1; Shannon et al. 2003).
Expression Pattern Analysis for Aux/IAA and ARF Genes in Five Tissues
For expression profiling of the Aux/IAA and ARF genes in B. rapa, we utilized the Illumina RNA-seq data that were previously generated and analyzed by Tong et al. (2013). Five tissues from B. rapa accession Chiifu-401-42, including root, stem, leaf, flower, and silique, were analyzed. The transcript abundance is expressed as fragments per kilobase of the exon model per million mapped reads (FPKM) values. The Ar. thaliana development expression profiling was analyzed using the AtGenExpress Visualization Tool with mean normalized values (Schmid et al. 2005). The Venn diagrams were drawn using the R program (Ihaka and Gentleman 1996). Heat maps of the gene FPKM values in B. rapa and mean-normalized values in Ar. thaliana were visualized using Tree View (http://jtreeview.sourceforge.net/).
Plant Materials
One of the Chinese cabbage cultivar (B. rapa ssp. pekinensis cv. Chiifu) was used for the experiments. Seeds were surface sterilized in 12% sodium hypochlorite and were germinated on 0.5× Murashige and Skoog (MS) agar plates (0.7%) in a growth chamber at 22°C in the dark for 2 days. The germinated seeds were grown in pots containing a soil:vermiculite mixture (3:1) in the greenhouse of Nanjing Agricultural University, and the controlled environment growth chamber was programed for 75% humidity, light 16 h/25°C and dark 8 h/20°C. At 4 weeks after germination, the seedlings were placed onto new 1/2 MS liquid solution plates (pH 5.8, without agar and sugar) for 1 week of acclimatization and then were cultured in a control treatment or with treatments containing 100 μM auxin (indole-3-acetic acid), 100 μM 1-naphthylphthalamic acid (NPA), 100 μM abscisic acid (ABA), or 100 μM ethephon (Eth) for 0, 1, 4, and 12 h under the same growth conditions as described earlier. Another group of seedlings was treated at 4°C for 0, 1, 4, and 12 h. Each treatment consisted of three replicates. All the samples were frozen in liquid nitrogen and stored at −70°C for further analysis.
RNA Isolation and Quantitative Real-Time PCR Analysis
Total RNA was isolated from 100 mg of frozen tissue using an RNA kit (RNAsimply total RNA Kit; Tiangen, Beijing, China) according to the manufacturer’s instructions. The quality and quantity of every RNA sample was assessed by agarose gel electrophoresis. The cDNA was synthesized from the total RNA using a Prime Script RT reagent Kit (TaKaRa). The specific sequences of the primers used for real-time polymerase Chain Reaction (PCR) are listed in supplementary table S2, Supplementary Material online. To check the primer specificity, we used BLAST against the Brassica genome. The reactions were performed using a Step one plus Real-Time PCR System (Applied Biosystems, Carlsbad, CA). The PCR parameters were as follows: 94 °C for 30 s, 40 cycles at 94 °C for 10 s, and 60 °C for 30 s, and then a melting curve (61 cycles at 65 °C for 10 s) was generated to check the specificity of the amplification. Relative fold expression changes were calculated using the comparative Ct value method (Heid et al. 1996).
Pearson Correlation Analyses
Pearson correlation coefficients (PCCs) of stress-inducible Aux/IAA gene pairs were calculated using a house Perl script based on log2-transformed quantitative Real-Time (qRT)-PCR data (Tang et al. 2013). The entire gene pairs whose PCC was more than 0.8 and was significant at the 0.05 significance level (P value) were collected for a gene coregulatory network analysis. The coexpression networks were graphically visualized using Cytoscape based on the PCCs of these gene pairs (Shannon et al. 2003).
Results
Identification and Retention of Aux/IAA and ARF Genes in Brassica rapa
Totally, 53 BraIAAs and 33 BraARFs were obtained (supplementary table S3, Supplementary Material online) in B. rapa. Compared with Mun et al.’s (2012) reports, 2 ARF genes (Bra015374, Bra020243) were added.
To investigate the copy number variation in Aux/IAAs and ARFs during Brassica-specific WGT events, 87 and 69 B. rapa syntenic regions with Ar. thaliana Aux/IAAs and ARFs, respectively, were identified (supplementary fig. S2, Supplementary Material online). Among them, 50 (96%) Aux/IAA homologs and 27 (82%) ARF homologs were located in the syntenic regions and identified in the B. rapa subgenomes (fig. 1A and supplementary table S4, Supplementary Material online). The results demonstrated that 57% (50/87) of the Aux/IAA genes were retained in the syntenic regions, relative to 39% (27/69) of the ARF genes (fig. 1A). Additionally, all syntenic genes in Brassica oleracea and Brassica napa (Cn subgenome and An subgenome) were also identified, and the retentions of B. oleracea and B. napa were similar to B. rapa (supplementary table S4, Supplementary Material online).
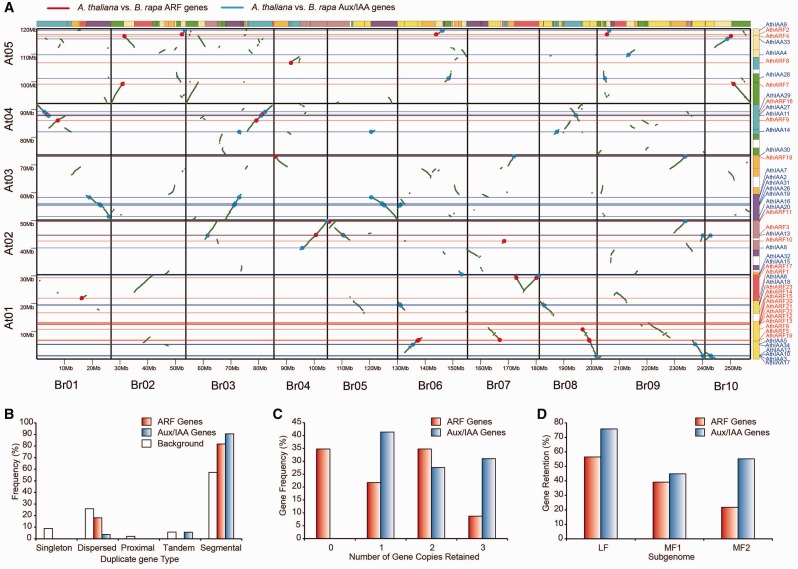
Fig. 1.
—Aux/IAA and ARF homologous genes in the segmental syntenic regions of the genomes of Brassica rapa and Arabidopsis thaliana and their different duplicated types and retentions. (A) Aux/IAA and ARF homologous genes in segmental syntenic regions of the genomes of B. rapa and Ar. thaliana. Conserved collinear blocks of genes (green irregular lines) are shown between the ten B. rapa chromosomes (horizontal axis) and the five Ar. thaliana chromosomes (vertical axis). The chromosomes are colored according to the inferred ancestral chromosomes following an established convention. Blue and red dots indicate the Aux/IAA and ARF homologs, respectively, in the two species. (B) The different duplicated types (singleton, dispersed, proximal, tandem, and segmental) of ARF and Aux/IAA were counted in B. rapa. Open boxes indicate the whole genome level. (C) Copy numbers of ARF and Aux/IAA genes after genome triplication and fractionation in B. rapa. (D) Retention of homoeologs of ARF and Aux/IAA genes in the three subgenomes (LF, MF1, and MF2) in B. rapa. LF: least fractionized subgenome; MF1: moderately fractionized subgenome; MF2: most fractionized subgenome. (B, C, and D) Red boxes indicate ARFs and blue boxes indicate Aux/IAAs.
Next, we specifically compared the retention of Aux/IAAs and ARFs by counting the gene numbers of different duplicated types, gene copies, and distributions of the three subgenomes. The results showed that both Aux/IAAs and ARFs contained more segmental duplication (91% and 82%) than other duplication types and above the B. rapa whole genome level (57%), suggesting that segmental duplication may be the major factor responsible for the expansion of these genes in B. rapa from the MRCA. ARFs contained more dispersed duplication than Aux/IAAs (fig. 1B). Importantly, all the Aux/IAAs were retained, and most (59%) were retained in two or three copies, which is significantly greater than the retention of the ARFs (43%). On the other hand, 35% of the ARFs were completely lost (fig. 1C). In addition, the proportion of Aux/IAA and ARF homologs retained was higher in the LF subgenome than in the MF1 and MF2 subgenomes, consistent with a previous report (Wang et al. 2011). Among all the subgenomes, more Aux/IAA gene homologs were retained than ARF gene homologs (fig. 1D). Overall, the results confirmed that Aux/IAAs were more preferentially retained than ARFs during diploidization following WGT in B. rapa.
Ks Analysis of the Aux/IAA and ARF Genes in Arabidopsis thaliana and Brassica rapa
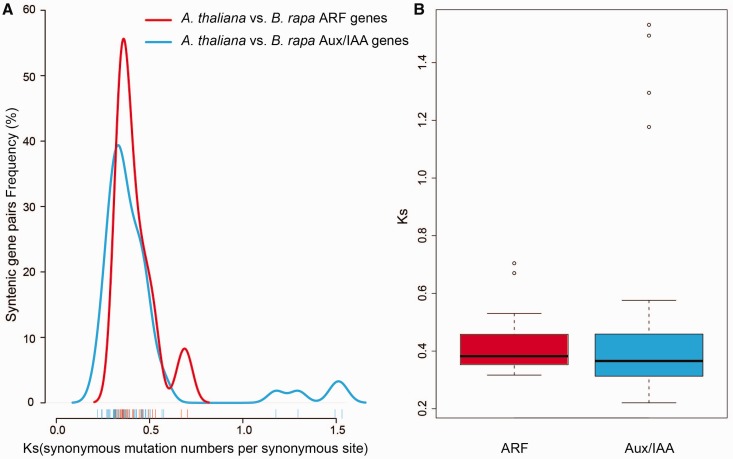
The Aux/IAA and ARF syntenic orthologs between B. rapa and Ar. thaliana were used to calculate the Ks and Ka values. In total, 50 Aux/IAA and 27 ARF syntenic gene pairs were analyzed (supplementary table S5, Supplementary Material online). The results indicated that the Ka/Ks ratios of all the Aux/IAA and ARF syntenic orthologs were less than 1, representing purifying selection on the Aux/IAA and ARF genes. To estimate the divergence time, the Ks values of the B. rapa Aux/IAA genes ranged from 0.2 to 0.6 and focused on approximately 3.9 (∼13 Myr), while the ARF genes ranged from 0.3 to 0.55 and also focused on approximately 3.8 (∼12.7 My; fig. 2A and B). Based on the Ks values of all the syntenic orthologs in B. rapa relative to Ar. thaliana (∼0.42–0.45, ∼14.5 Myr; Cheng et al. 2013), we concluded that the Aux/IAA and ARF genes diverged concurrently with the Brassica-specific WGT event (13–17 Ma; Wang et al. 2011). Specifically, the Ks values of the BraARFs had one more peak than the BraIAAs at approximately 0.7 (∼23.4 Myr; fig. 2A).
Fig. 2.
—Pairwise comparison of the Ks values for Aux/IAA and ARF homologous genes in Brassica rapa and Arabidopsis thaliana. (A) The distribution of the Ks values for Aux/IAA and ARF genes between Ar. thaliana and B. rapa. The lines representing the Aux/IAA and ARF genes are blue and red, respectively. (B) Box plot of the Ks values for the Aux/IAA and ARF genes between Ar. thaliana and B. rapa. Blue boxes indicate Aux/IAAs, and red boxes indicate ARFs.
Expansion and Structural Characteristics of the Aux/IAA and ARF Genes in Brassica rapa
All the Aux/IAA and ARF genes were distributed nonrandomly on the GBs of ten B. rapa chromosomes and were named BraIAA01 to BraIAA53 and BraARF01 to BraARF32 according to the position of their corresponding genes on chromosomes 1–10, from top to bottom, as well as the order of their scaffolds (supplementary fig. S2 and table S3, Supplementary Material online). BraAL was identified as an ARF gene by a BLAST search and was the orthologous gene of ARF4 in B. rapa, but lacked an ARF and even a B3 domain (supplementary table S3, Supplementary Material online).
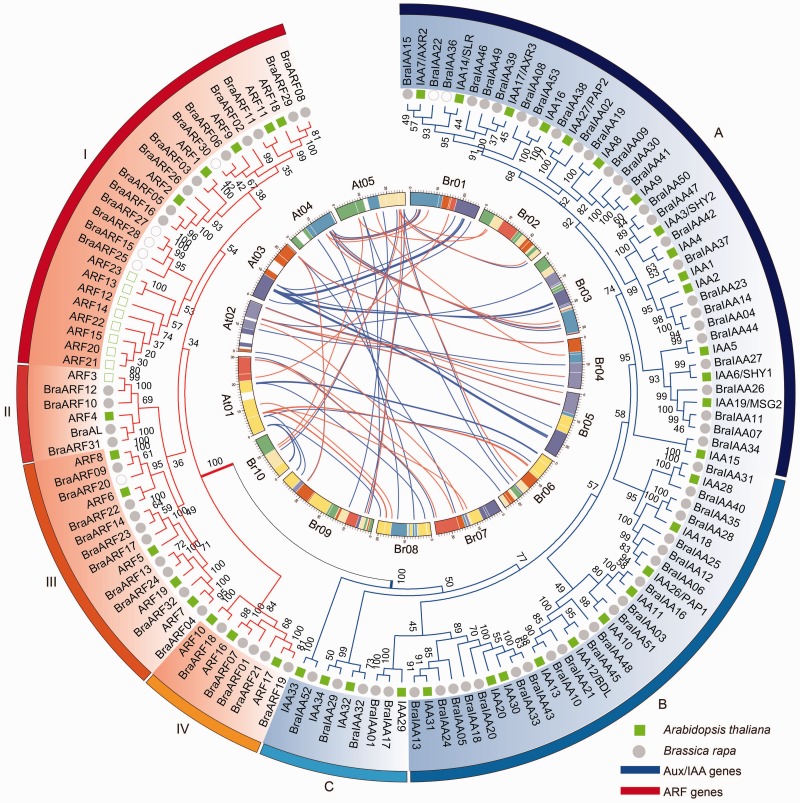
To investigate the extent of the lineage-specific expansion of the Aux/IAA and ARF genes in B. rapa and Ar. thaliana, we performed a joint phylogenetic analysis of all the Aux/IAAs and ARFs, and the homologous genes were marked on the tree. Then, the relationships of the Aux/IAA and ARF syntenic orthologs between B. rapa and Ar. thaliana were displayed (fig. 3). All the Aux/IAAs were clustered into three groups (A, B, and C), similar to previous reports in Ar. thaliana (Remington et al. 2004), while the ARFs were divided into four groups (I, II, III, and IV), consistent with the reports in B. rapa (Mun et al. 2012). In Finet et al’s (2013) report, the ARFs were clustered into three groups, in which groups I and II from our analysis were classified in the same group. Brassica rapa and Ar. thaliana had 53 and 29 Aux/IAAs, respectively. There were at least 28 ancestral IAAs in the MRCA of B. rapa and Ar. thaliana. Both IAA14 and IAA1 were homologs of BraIAA37, but instead of falling into the same clade as IAA1 and BraIAA37 in the phylogenetic tree, IAA14 paired with BraIAA36, which was gained after the split. Overall, B. rapa has gained two Aux/IAAs and lost almost no genes. However, there were eight ARFs from Ar. thaliana that were lost completely, and six ARFs from B. rapa were gained. All 14 genes except BraARF20 were clustered into group I (fig. 3 and supplementary table S3, Supplementary Material online). The results demonstrate that both the ARF and Aux/IAA genes expanded by segmental duplication mainly in B. rapa, although some ARF group I genes were expanded by dispersal. In addition, Aux/IAAs were more retained than ARFs (fig. 3).
Fig. 3.
—Phylogenetic tree and syntenic relationship among Aux/IAA and ARF genes in Brassica rapa and Arabidopsis thaliana. Collinear correlations of Aux/IAAs and ARFs in the Ar. thaliana and B. rapa genomes are displayed by Circos. The B. rapa and Ar. thaliana chromosomes are colored according to the inferred ancestral chromosomes following an established convention. In the phylogenetic tree, Aux/IAAs were clustered into three groups (A, B, and C) and ARFs were divided into four groups (I, II, III, and IV). Arabidopsis thaliana and B. rapa are indicated by green squares and grey circles, respectively. Open squares and circles mean that the genes are nonsyntenic orthologs.
Furthermore, the structural characteristics of BraIAAs and BraARFs were analyzed, including their motif locations and gene structures (fig. 4). Overall, the length of the BraARFs was longer than the BraIAAs and the structures of these genes were varied, but most of the genes were conserved in the same class. In detail, 12 motifs included in Aux/IAA, B3, ARF, and other domains were detected, demonstrating that motif 1 and motif 3 (PB1 domain) were widely present in BraIAAs and BraARFs. However, BraAL was short and contained only motif 1. To refine the relationship between BraAL and the ARFs, three conserved collinear blocks flanking the ARF4 orthologous gene were identified among three B. rapa subgenomes and then investigated (Short for comparative genomics; supplementary fig. S3, Supplementary Material online). BraAL was clearly the orthologous gene of ARF4, but might have lost domains or passed them on to sister genes of ARFs during the evolutionary history.
Fig. 4.
—An analytical view of the Aux/IAA and ARF genes in Brassica rapa. The following parts are shown from left to right. (A) Protein maximum-likelihood tree: The tree was constructed using a maximum-likelihood method, and bootstrap values were calculated with 1,000 replications using MEGA 5.2. (B) Protein structure: The search for common motifs shared among the Aux/IAA–ARF proteins of each group was performed with Multiple Expectation-maximization for Motif Elicitation. Motifs that were included in the Aux/IAA, ARF, and B3 domains are classified on the right. (C) Gene structure: The B3, ARF, and Aux/IAA domains are highlighted by green, red, and blue boxes, respectively. Introns are shown as lines.
Evolutionary History of the Aux/IAA and ARF Families in Plants
To further understand the evolutionary history of Aux/IAAs and ARFs in plants, seven other plant species, including three eudicots (Ca. papaya, Po. trichocarpa, and V. vinifera), one basal angiosperm (Am. trichopoda), one lycophyte (S. moellendorffii), on physcomitrella (Ph. patens), and one chlorophyta (Ch. reinhardtii), were used. Except in Ch. reinhardtii, we obtained 96 (20, 35, 23, 12, 4, and 2) Aux/IAAs and 101 (11, 36, 19, 13, 8, and 14) ARFs in the six other species. Furthermore, 25 (5, 2, 2, 8, 5, and 3) AL genes were also found (supplementary table S3, Supplementary Material online). The results supported the previous report that Aux/IAAs and ARFs may have begun to function in land plants (Finet et al. 2013) and confirmed the widespread existence of ALs in plants.
Phylogenetic trees for each species, including B. rapa, Ar. thaliana, and six other species, were constructed, and the motif locations were identified (supplementary fig. S4, Supplementary Material online). In addition, the phylogenetic relationships among all the genes of these plant species were analyzed (fig. 5A). Based on the phylogenetic trees, Aux/IAAs and ARFs separated into two significant classes, while ALs were more similar to ARFs than to Aux/IAAs. To further determine the relationship among the three groups, the genetic distance was analyzed (fig. 5B). The box plot showed that the genetic distance of ARFs versus ALs was smaller than Aux/IAAs versus ALs and Aux/IAAs versus ARFs, indicating that ALs had a closer relationship with ARFs. In addition, the genetic distances of Aux/IAAs, ARFs, and ALs were similar, but the scope of ARFs was the smallest, suggesting that the degree of sequence divergence of ARFs was lower than Aux/IAAs (fig. 5B). Simultaneously, the gene length, exon number, and exon length of all the genes were analyzed (supplementary fig. S5, Supplementary Material online). ARFs had the most exons (focused on approximately 13) and the longest gene length (focused on approximately 5,000 bp) among the three groups.
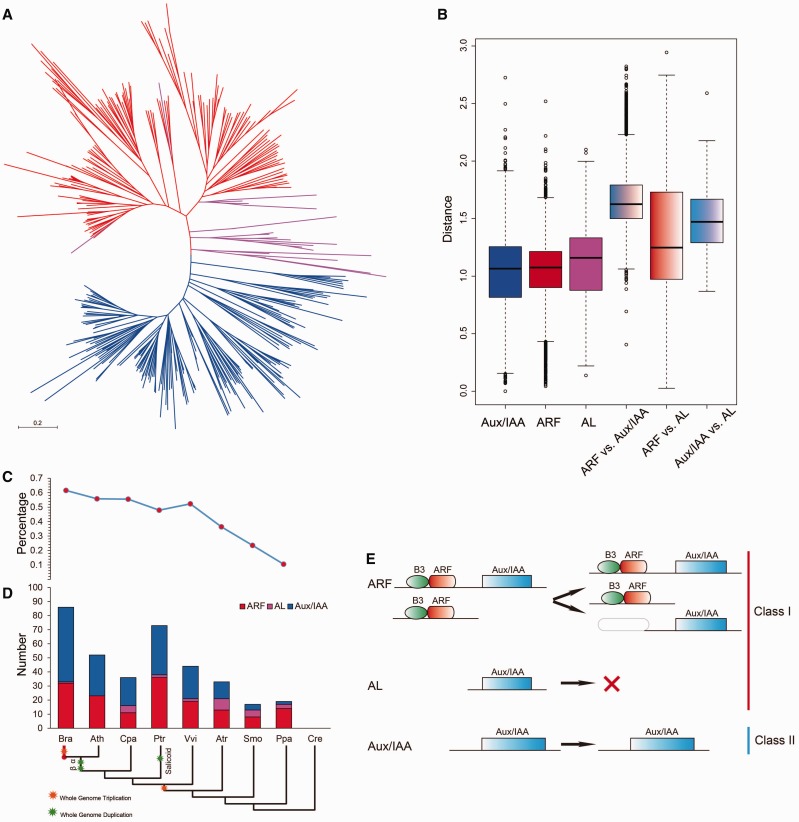
Fig. 5.
—Phylogenetic relationships among 360 Aux/IAA and ARF genes (A); genetic distance among the different clades of Aux/IAAs and ARFs (B); comparison of the percentage of Aux/IAAs and copy numbers of Aux/IAAs, ALs, and ARFs in representative species (C, D); and the evolutionary pattern of Aux/IAA–ARFs (E). Aux/IAAs, ALs, and ARFs are shaded blue, purple, and red, respectively. In (B), the box plot shows the median (black line), interquartile range (box), and maximum and minimum scores (whiskers) of each data set. In (D), the short species names are Chlamydomonas reinhardtii (Cre), Physcomitrella patens (Ppa), Selaginella moellendorffii (Smo), Amborella trichopoda (Atr), Vitis vinifera (Vvi), Populus trichocarpa (Ptr), Carica papaya (Cpa), B. rapa (Bra), and Arabidopsis thaliana (Ath). The α, β, γ, salicoid duplications, and the Brassica-specific triplication are indicated on the branches of the trees according to the Plant Genome Duplication Database. In (E), the green ellipse indicates the B3 domain, the rounded red box indicates the ARF domain, the blue box indicates the Aux/IAA domain, and the noncolored rounded box indicates the lost domain.
The gene family sizes and the percentages of Aux/IAAs in eight plants, including bryophytes, lycophytes, and angiosperms (fig. 5C and D), suggested that Aux/IAAs expanded rapidly during evolution and further expanded in the Brassicaceae. WGD is known to have significant impacts on the expansion and evolution of gene families in plant genomes. Compared with Aux/IAAs, the expansion of ARFs was more stable. However, along with the gradual increase in the Aux/IAA percentage, the number of ALs grew smaller. After the divergence of B. rapa and Ar. thaliana from MRCA, only 1 BraAL existed.
The orthologous groups of Aux/IAAs, ARFs, and ALs among all eight species were analyzed (Li et al. 2003). All 1,078 orthologous gene pairs and 825 coorthologous gene pairs were identified (supplementary table S7, Supplementary Material online). Then, the networks of all the orthologous gene pairs were constructed (supplementary fig. S6, Supplementary Material online). The five AL genes were clustered with ARF genes, and none paired with Aux/IAA genes, confirming the closer relationship between ARF and AL. In addition, there were less orthologous gene pairs in the Aux/IAA genes than in the ARF genes, suggesting that the IAA genes may have diversified and obtained more functions following the rapid expansion. According to our findings, the evolutionary history of Aux/IAAs and ARFs in the plant kingdom was constructed (fig. 5E).
Expansion History of the Aux/IAA and ARF Genes in Eudicots
Based on the two phylogenetic trees of all of the Aux/IAAs and ARFs, we attempted to reconstruct the expansion history of the two gene families (supplementary fig. S7, Supplementary Material online). In all eight species, ARFs and Aux/IAAs were mainly divided into four and three groups, respectively, which we named groups I to IV and groups A to C according to the classification of Ar. thaliana (Remington et al. 2004). In supplementary figure S7A, Supplementary Material online, we found that the S. moellendorffii and Ph. patens Aux/IAAs (yellow squares and green triangles) were independent and clustered primarily into group A (supplementary fig. S7C and D, Supplementary Material online), while all three groups existed in Am. trichopoda (red balls). All the major subgroups of Aux/IAAs existed in angiosperms and then expanded rapidly, accompanied by WGD. Meanwhile, unlike the Am. trichopoda ARFs (red balls), which were distributed into all four groups, the ARFs of S. moellendorffii and Ph. patens (yellow squares and green triangles) were mainly clustered into groups III and group IV, indicating that groups III and IV may have appeared prior to groups I and II and that all four groups originated from duplication events prior to the γ event (supplementary fig. S7A and B, Supplementary Material online). After the γ WGD, rounds of duplication, loss, and rearrangement took place in the ARFs.
Divergence of the Selective Pressure between the Class I and Class II Genes
In their evolutionary footprint, compared with class I (ARFs and ALs), class II (Aux/IAAs) showed rapid expansion in plant species, including B. rapa, Ar. thaliana, and 6 other species. Especially in B. rapa, the number of Aux/IAAs was the largest. To infer the influence of selection on the expansion of Aux/IAA genes, we evaluated the different selective pressures between the two classes (Yang 2007; supplementary table S8, Supplementary Material online). For the B. rapa genes, the log-likelihood values under the one-ratio and two-ratio models were logeL = −90597.557345 and −90558.660972, respectively. The LRT showed that the two-ratio model was not equal to the null model (one-ratio model), suggesting that the selective pressure differed significantly between the two classes (P < 0.0001). Under the two-ratio model, the ω values for B. rapa classes I and II were 0.2263 and 0.3355, respectively, indicating that the B. rapa class II (Aux/IAA) genes were under more relaxed selection constraints than the class I genes (ARFs and ALs). Except for Am. trichopoda, six other species were similar to B. rapa, where class II (Aux/IAA) genes had higher ω values than those of their class I genes. The relaxed pressure may lead to the Aux/IAA genes expanding rapidly and diversifying to adopt more functions.
Expression Divergence of the Aux/IAA and ARF Genes in Different Tissues of Brassica rapa and Arabidopsis thaliana
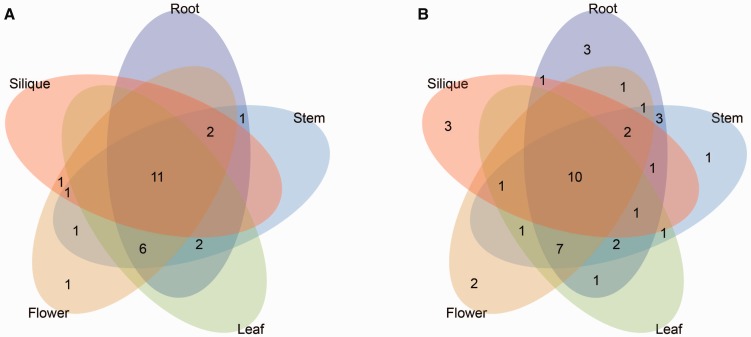
To investigate the divergence of homologs, the expression patterns of Aux/IAAs and ARFs in five tissues, including the roots, stems, leaves, flowers, and siliques of Ar. thaliana and B. rapa, were analyzed in this study (supplementary fig. S8 and tables S9 and S10, Supplementary Material online). Among the 56 ARFs (including 23 AthARFs and 33 BraARFs), 9 (ARF6, ARF8, ARF9, ARF21, ARF22, BraARF25, BraARF27, BraARF28, and BraAL) were not expressed and 2 (BraARF15 and BraARF30) were barely expressed in any tissue. Notably, eight of the ARFs belonged to group I, and five BraARFs were nonsyntenic orthologs. The rest of the AthARFs and BraARFs were expressed in at least one tissue. In addition, a total of 26 (81%) BraARFs had high expression levels (FPKM value > 5) in at least one tissue, 11 BraARFs were expressed highly in all 5 tissues, and only 1 (BraARF1) gene was selectively expressed highly in a specific tissue (flower; fig. 6 and supplementary table S10, Supplementary Material online). However, among all 82 Aux/IAAs, only 4 (IAA15, BraIAA5, BraIAA29, and BraIAA38) were not expressed and 1 gene (BraIAA18) was barely expressed in any tissues. In contrast, 42 (79%) BraIAAs were highly expressed. Among them, ten were expressed in all five tissues and nine showed high expression in specific tissues (fig. 6). Above all, the expression of BraIAAs was more tissue-specific than the BraARFs, suggesting that Aux/IAAs might play more specific and important roles in tissue development.
Fig. 6.
—Venn diagram depicting the distribution of shared expression of the ARF (A) and Aux/IAA (B) genes among five Brassica rapa tissues, that is, root, steam, leaf, flower, and silique. The FPKM values are more than five counted in the different tissues.
Expression Divergence and Coregulatory Networks of IAAs Under Multiple Treatments in Brassica rapa
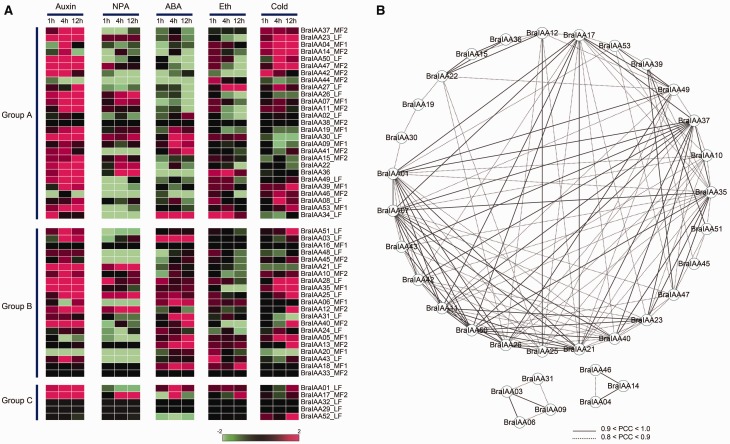
Gene expression is not only usually tissue specific but is also hormone or stress induced. Additionally, parts of the growth and developmental processes were regulated by crosstalk between auxin and other hormones (ethylene and ABA) according to previous reports (Muday et al. 2012; Zhao et al. 2015). Moreover, auxin was confirmed as a regulator of cold stress response (Rahman 2013). To recover all the divergence information of BraIAA homolog genes, the expression patterns following different hormone, including auxin (indole-3-acetic acid), NPA, ABA, and Eth, and cold treatments, were analyzed (fig. 7A). We used exogenous auxin and NPA (an auxin transport inhibitor) to control the auxin level of B. rapa leaves. Among the 53 isolated Aux/IAAs, most of them were upregulated by auxin and downregulated by NPA. In contrast to NPA treatment, most genes were upregulated following the cold treatment. Meanwhile, the counts of upregulated and downregulated genes were similar between the ABA and Eth treatments. During all four hormones and cold treatments, except BraIAA16, 29, 32, 33, and 38, all the BraIAAs were induced to some extent. We found that BraIAA35, 30, 23, 16, and 20 were significantly induced in response to auxin, NPA, ABA, Eth, and cold treatments, respectively. Then, we investigated the expression trends among 14 duplicate gene groups which identified based phylogenetic tree and syntenic relationship (supplementary fig. S9, Supplementary Material online). Among these duplicate genes, there were four types of expression patterns. First, all duplicate genes were expressed in the same trend under all treatments, for example, BraIAA26/07/11, suggesting that the duplicate genes might have similar functions. Second, all duplicate genes were expressed under the same treatment but in different trends (e.g., BraIAA01/17). Third, the duplicate genes were induced under different treatments (e.g., BraIAA21/10). These two types of expression patterns suggested that the functions of these duplicate genes might diverge. Finally, one gene was expressed in response to one or all specific treatments, while the other was not detected under any treatments, for example, BraIAA02/38, suggesting that one duplicate gene may have become a pseudogene or evolved into a new function which was not identified in this study. These data were consistent with the hypothesis that Aux/IAA genes gained functional diversification and were retained to adapt to different environments.
Fig. 7.
—Expression analysis of the Aux/IAA genes under five treatments in Brassica rapa, that is, indole-3-acetic acid, NPA, ABA, Eth and cold and coregulatory networks of the stress-inducible BraIAAs. (A) Heat map representation of BraIAAs under these five treatments displays the log2-transformed relative expression values. These expression profile data were obtained using qRT-PCR. (B) The coregulatory networks of stress-inducible BraIAAs were established based on the PCCs of stress-inducible BraIAA gene pairs using log2-transformed qPCR data, which involved 34 nodes and 144 regulatory edges. The different edge line styles indicate the different significance levels of the coregulated gene pairs.
Coregulatory networks were established based on the PCCs of stress-inducible BraIAA gene pairs (supplementary table S11, Supplementary Material online). All the BraIAAs appeared to have different degrees of positive correlations. Next, 34 BraIAAs with PCC values that were significant at the 0.05 significance level and were greater than 0.8 were collected and visualized to construct hormones and cold stress coregulatory networks (fig. 7B). Most correlations occurred among members belonging to the same group, suggesting that the gene duplication not only led to functional divergence but also enhanced the cooperative interaction of homologs to help plants adapt to their complex environments.
Discussion
During their long evolution history, all extant angiosperms are believed to have undergone at least one and often multiple WGD events, which provided opportunities for duplicated genes to gain functional diversification, resulting in more complex organisms (Ohno 1970; Edger and Pires 2009; Jiao et al. 2011). Typically, WGD is followed by substantial gene loss (fractionation). However, based on the gene balance hypothesis, genes whose products participate in macromolecular complexes or in transcriptional or signaling networks are more likely to be retained, thus avoiding network instability caused by the loss of one member (Birchler and Veitia 2007; Lou et al. 2012). The Brassica genus is composed of many diverse species, and each species contains rich morphotypes showing extreme traits. Brassica rapa experienced a complex WGD history, including γ, β, and α events and an additional WGT event, providing an excellent chance to study the relationship between gene family fractionation and changes in plant morphotypes (Wang et al. 2011; Cheng et al. 2013). As we know, two related gene families of interest in high plants, which are the main components of the auxin signaling pathway that plays a key role during plant development, code for the ARF and Aux/IAA proteins. The evolutionary history of ARF in land plants has been investigated in detail (Finet et al. 2013). In previous reports, ARF genes were divided into three groups in Ar. thaliana and even land plants or four groups in B. rapa (Mun et al. 2012; Wang, Deng, et al. 2012; Wang, Tang et al. 2012; Finet et al. 2013). In this study, 53 Aux/IAAs and 33 ARFs were identified in B. rapa. The Aux/IAA genes were divided into three groups, but ARFs were clustered into four groups, consistent with Mun et al.’s (2012) reports.
By comparing the number of different duplicated types, gene copies, and distribution of the three subgenomes, we found that all the AthIAA orthologs were retained in B. rapa; in contrast, eight AthARF orthologs were completely lost. Aux/IAAs were more preferentially retained than ARFs since the split between B. rapa and Ar. thaliana from the MRCA. What is the relationship between them? Were the Aux/IAAs more retained because they were more important than the ARFs in the network, or were there other reasons? To answer these questions, we analyzed all the Aux/IAAs and ARFs in eight species. Each of the duplicated genes, including the Aux/IAA and ARF genes, could subsequently follow one of three broad fates: Subfunctionalization, neofunctionalization, and nonfunctionalization (deletion or pseudogenization) (Innan and Kondrashov 2010). In previous reports, even the most recently diverged paralogs differed in their catalytic efficiency, expression, and/or substrate spectrum, suggesting that duplicates have a relatively high rate of rapid divergence in function (Lan et al. 2009). Furthermore, the genes that acquired new functions were more preferentially retained. Compared with ARFs, Aux/IAAs expanded rapidly from moss (Ph. patens) to high plants and even expanded more extensively in B. rapa than in the other eight species. Moreover, the Aux/IAAs were under more relaxed selection constraints than the ARFs in most of the eight species, which allowed new duplicate genes to diverge into new tissue-specific expression patterns or functions (Yang et al. 2013). In addition, comparing the tissue expression pattern among the Aux/IAA and ARF genes in B. rapa and Ar. thaliana, most of the genes maintained similar expression patterns, but Aux/IAAs demonstrated higher tissue specialization, suggesting that Aux/IAAs obtained more subfunctionalization and neofunctionalization than ARFs, which suggest why plants need to retain more Aux/IAAs. However, gene duplication has also played a major role in the functional diversification of the ARF family. Alternative splicing, a mechanism previously found to operate on the ortholog of ARF4 in Am. trichopoda (Finet et al. 2010) was found to operate more widely in the ARF family (Finet et al. 2013). Therefore, ARFs could achieve more functions using fewer genes than Aux/IAAs to adapt to the increasingly complex network. This may explain why more Aux/IAAs were retained than ARFs.
Both Aux/IAAs and ARFs have a common domain, the PB1 protein–protein interaction domain (previously referred to domain III and domain IV as or domain III/IV), in their C termini (supplementary fig. S4, Supplementary Material online). In addition, most ARF proteins also have a B3 domain and a middle domain (Guilfoyle 2015). Based on previous reports, both Aux/IAAs and ARFs were found in the Ph. patens genome, indicating that aspects of Aux/IAA function have also been conserved in land plants and that the ARF domain is a land plant innovation (Imaizumi et al. 2002; Finet et al. 2013). In 2015, Wang et al. found the homologs or orthologs of Aux/IAAs and ARFs in several charophytes (N. mirabilis, P. margaritaceum, and K. flaccidum). In addition, the Aux/IAA ortholog of K. flaccidum contained all of the typically conserved domains, I, II, and PB1, of the Aux/IAA proteins, but in N. mirabilis, the orthologs only contained the domain PB1. The ARF ortholog was not identified in the K. flaccidum genome, but was found in the genome of N. mirabilis. However, ARF genes found in chlorophyta did not completely encode ARF-like proteins; while they had a B3 domain and PB1 domain, they lacked the Auxin_resp domain (De Smet et al. 2011; Finet et al. 2013). In our study, we identified these two gene families in B. rapa using both the HMM software package and the BLAST program. In addition to typical Aux/IAAs and ARFs, a type of ARF-like protein that lacks the ARF domain and even the B3 domain was also obtained and named AL. We identified AL in seven other species when we identified the Aux/IAAs and ARFs. Most of them were identified as Aux/IAAs by the HMM software package, but our results suggest that AL had a closer relationship with the ARFs through the following analyses: 1) phylogenetic relationships, 2) ortholog groups, 3) synteny analysis, and 4) genetic distance. Thus, the orthologs of Aux/IAAs and ARFs in charophytes could be considered to be ALs. Meanwhile, we found that in contrast to Aux/IAAs and ARFs, the number and percent of ALs were decreasing over the evolutionary history. In B. rapa, BraAL was the orthologous gene of ARF4, but it was not expressed in any tissue or under any treatment evaluated in this experiment, suggesting that it might be pseudogenized. Overall, we estimated that perhaps, at the beginning, both Aux/IAAs and ARFs originated from the PB1 domain, and then Aux/IAAs become stable (fig. 5E). At the same time, ALs, as incompletely evolved ARFs, were involved in the development of the ARF genes and were increasingly lost, until no AL gene existed in Ar. thaliana and only one gene existed in BraAL. However, the relationship between the decrease in ALs and the increase in Aux/IAAs must be further explored.
In the evolution of land plants, the differential distribution of auxin results in the diverse morphology of plants (Ludwig-Muller 2011; Wang et al. 2015). After complex gene duplications, in most high plant genomes, multiple paralogs exist for each part of the auxin nuclear signaling pathway. This potential to combine diverse portions of signaling pathways likely contributes to the myriad of context-specific responses to auxin and increases the pathway complexity (Rensing 2014; Wright and Nemhauser 2015). Brassicas plants have rich diversity with respect to both speciation and the abundant morphotypes in each Brassicas species (Cheng et al. 2014). The species share many traits, but developed independently and in parallel. For example, B. rapa, B. oleracea, and B. napa all have the distinct feature of a leafy head; turnip develops enlarged roots as storage organs; caixin and purple caitai have long and tender stems; and some morphotypes of B. rapa and B. oleracea have developed beautiful leaf patterns and colors and are therefore used as ornamental plants (Cheng et al. 2014). In our study, nearly all the Aux/IAAs and most of the ARF genes were retained in B. rapa. In addition, the Aux/IAAs of B. oleracea and B. napa were also retained completely. Compared with Ar. thaliana, the expression patterns of the BraIAAs exhibited more tissue-specific expression and functional diversification under different stress treatments. Their coregulatory networks demonstrated that the gene duplication enhanced the cooperative interaction of homologs to adapt to the environment. Following WGT, the BraARFs obtained new genes and new functions by alternative splicing, which should be further explored. The large number of Aux/IAAs and the diverse transcriptional mechanism of ARFs created a large complex auxin signaling network, which may lead to more complex organs and more morphotypes. Overall, we firmly believe that the changes in Aux/IAAs and ARFs during the WGT event played a crucial role in the expansion of rich morphotypes in the genus Brassicas.
Supplementary Material
Supplementary figures S1–S9 and tables S1–S11 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the National Program on Key Basic Research Projects (The 973 Program: 2012CB113900), National Natural Science Foundation of China (Key Program, No. 31330067), and National High Technology Research and Development Program of China (863 Program, No. 2012AA100101).
Literature Cited
- Albert VA, et al. 2013. The Amborella genome and the evolution of flowering plants. Science 342:1241089. [DOI] [PubMed] [Google Scholar]
- Bailey TL, et al. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks JA, et al. 2011. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332:960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA. 2007. The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19:395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Wu J, Fang L, Wang X. 2012. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front Plant Sci. 3:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Wu J, Wang X. 2014. Genome triplication drove the diversification of Brassica plants. Hortic Res. 1:14024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, et al. 2013. Deciphering the diploid ancestral genome of the Mesohexaploid Brassica rapa. Plant Cell 25:1541–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, et al. 2011. Unraveling the evolution of auxin signaling. Plant Physiol. 155:209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debashish B, Linda M. 1998. Algal phylogeny and the origin of land plants. Plant Physiol. 116:9–15. [Google Scholar]
- Dreher KA, Brown J, Saw RE, Callis J. 2006. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18:699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W, et al. 2015. Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Mol Genet Genomics. 290:239–255. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Pires JC. 2009. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 17:699–717. [DOI] [PubMed] [Google Scholar]
- Finet C, Berne-Dedieu A, Scutt CP, Marlétaz F. 2013. Evolution of the ARF gene family in land plants: old domains, new tricks. Mol Biol Evol. 30:45–56. [DOI] [PubMed] [Google Scholar]
- Finet C, et al. 2010. Parallel structural evolution of auxin response factors in the angiosperms. Plant J. 63:952–959. [DOI] [PubMed] [Google Scholar]
- Gallavotti A. 2013. The role of auxin in shaping shoot architecture. J Exp Bot. 64:2593–2608. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. 2001. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414:271–276. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ. 2015. The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. 2007. Auxin response factors. Curr Opin Plant Biol. 10:453–460. [DOI] [PubMed] [Google Scholar]
- Guilfoyle TJ, Hagen G. 2012. Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions. Plant Sci. 190:82–88. [DOI] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. 1996. Real time quantitative PCR. Genome Res. 6:986–994. [DOI] [PubMed] [Google Scholar]
- Hu B, et al. 2015. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. 1996. R: a language for data analysis and graphics. J Comput Graph Stat. 5:299–314. [Google Scholar]
- Imaizumi T, Kadota A, Hasebe M, Wada M. 2002. Cryptochrome light signals control development to suppress auxin sensitivity in the moss Physcomitrella patens. Plant Cell 14:373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 11:97–108. [DOI] [PubMed] [Google Scholar]
- Jiao Y, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–100. [DOI] [PubMed] [Google Scholar]
- Koch MA, Haubold B, Mitchell-Olds T. 2000. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol Biol Evol. 17:1483–1498. [DOI] [PubMed] [Google Scholar]
- Korasick DA, et al. 2014. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc Natl Acad Sci U S A. 111:5427–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T, et al. 2009. Extensive functional diversification of the Populus glutathione S-transferase supergene family. Plant Cell 21:3749–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S, Shao N, Bock R, Jurgens G, De Smet I. 2009. Auxin signaling in algal lineages: fact or myth? Trends Plant Sci.. 14:182–188. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou P, et al. 2012. Preferential retention of circadian clock genes during diploidization following whole genome triplication in Brassica rapa. Plant Cell 24:2415–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Muller J. 2011. Auxin conjugates: their role for plant development and in the evolution of land plants. J Exp Bot. 62:1757–1773. [DOI] [PubMed] [Google Scholar]
- Muday GK, Rahman A, Binder BM. 2012. Auxin and ethylene: collaborators or competitors?. Trends Plant Sci. 17:181–195. [DOI] [PubMed] [Google Scholar]
- Mun JH, et al. 2012. Auxin response factor gene family in Brassica rapa: genomic organization, divergence, expression, and evolution. Mol Genet Genomics. 287:765–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Berlin and New York: Springer-Verlag. [Google Scholar]
- Rahman A.. 2013. Auxin: a regulator of cold stress response. Physiol Plant. 147:28–35. [DOI] [PubMed] [Google Scholar]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW. 2004. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 135:1738–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA. 2014. Gene duplication as a driver of plant morphogenetic evolution. Curr Opin Plant Biol. 17:43–48. [DOI] [PubMed] [Google Scholar]
- Rensing SA, et al. 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319:64–69. [DOI] [PubMed] [Google Scholar]
- Romanel EA, Schrago CG, Couñago RM, Russo CA, Alves-Ferreira M. 2009. Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS One 4:e5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, et al. 2005. A gene expression map of Arabidopsis thaliana development. Nat Genet. 37:501–506. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Lysak MA, Mitchell-Olds T. 2006. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11:535–542. [DOI] [PubMed] [Google Scholar]
- Shalom L, et al. 2014. Fruit load induces changes in global gene expression and in abscisic acid (ABA) and indole acetic acid (IAA) homeostasis in citrus buds. J Exp Bot. 65:3029–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, et al. 2013. Characterization and co-expression analysis of WRKY orthologs involved in responses to multiple abiotic stresses in Pak-choi (Brassica campestris ssp. chinensis). BMC Plant Biol. 13:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Gretchen H, Tom G. 2003. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15:533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. 2004. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16:533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong C, et al. 2013. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genomics 14:689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ. 1999. Dimerization and DNA binding of auxin response factors. Plant J. 19:309–319. [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9:1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaene T, Delwiche CF, Rensing SA, Friml J. 2013. Origin and evolution of PIN auxin transporters in the green lineage. Trends Plant Sci. 18:5–10. [DOI] [PubMed] [Google Scholar]
- Wang C, Liu Y, Li SS, Han GZ. 2015. Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 167:872–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. 2011. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 43:1035–1039. [DOI] [PubMed] [Google Scholar]
- Wang Y, Deng D, et al. 2012. Diversification, phylogeny and evolution of auxin response factor (ARF) family: insights gained from analyzing maize ARF genes. Mol Biol Rep. 39:2401–2415. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang H, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RC, Nemhauser JL. 2015. New tangles in the auxin signaling web. F1000Prime Rep. 7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 17:32–43. [DOI] [PubMed] [Google Scholar]
- Yang ZL, Liu HJ, Wang XR, Zeng QY. 2013. Molecular evolution and expression divergence of the Populus polygalacturonase supergene family shed light on the evolution of increasingly complex organs in plants. New Phytol. 197:1353–1365. [DOI] [PubMed] [Google Scholar]
- Zhang Z, et al. 2006. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4:259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao FY, et al. 2015. ABA plays essential roles in regulating root growth by interacting with auxin and MAPK signaling pathways and cell-cycle machinery in rice seedlings. Plant Growth Reg. 75(2):535–547 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.