Abstract
In mycobacteria, various type VII secretion systems corresponding to different ESX (ESAT-6 secretory) types, are contributing to pathogenicity, iron acquisition, and/or conjugation. In addition to the known chromosomal ESX loci, the existence of plasmid-encoded ESX systems was recently reported. To investigate the potential role of ESX-encoding plasmids on mycobacterial evolution, we analyzed a large representative collection of mycobacterial genomes, including both chromosomal and plasmid-borne sequences. Data obtained for chromosomal ESX loci confirmed the previous five classical ESX types and identified a novel mycobacterial ESX-4-like type, termed ESX-4-bis. Moreover, analysis of the plasmid-encoded ESX loci showed extensive diversification, with at least seven new ESX profiles, identified. Three of them (ESX-P clusters 1–3) were found in multiple plasmids, while four corresponded to singletons. Our phylogenetic and gene-order-analyses revealed two main groups of ESX types: 1) ancestral types, including ESX-4 and ESX-4-like systems from mycobacterial and non-mycobacterial actinobacteria and 2) mycobacteria-specific ESX systems, including ESX-1-2-3-5 systems and the plasmid-encoded ESX types. Synteny analysis revealed that ESX-P systems are part of phylogenetic groups that derived from a common ancestor, which diversified and resulted in the different ESX types through extensive gene rearrangements. A converging body of evidence, derived from composition bias-, phylogenetic-, and synteny analyses points to a scenario in which ESX-encoding plasmids have been a major driving force for acquisition and diversification of type VII systems in mycobacteria, which likely played (and possibly still play) important roles in the adaptation to new environments and hosts during evolution of mycobacterial pathogenesis.
Keywords: mycobacteria, tuberculosis, virulence, horizontal gene transfer, phylogeny, ESX/type VII secretion systems, mycobacterial evolution
Introduction
Mycobacteria represent a prokaryotic genus with a vast diversity of lifestyles, ranging from major human pathogens, such as Mycobacterium tuberculosis, opportunistic pathogens, such as Mycobacterium abscessus, to environmental saprophytes that represent the great majority of mycobacterial species (Magee and Ward 2012; Boritsch et al. 2014). Mycobacteria are classified into rapidly growing mycobacteria (RGM), making visible colonies on solid media in <1 week of incubation, and slowly growing mycobacteria (SGM), which is a monophyletic cluster (Mignard and Flandrois 2008) that contains the major mycobacterial pathogens. Mycobacteria are characterized by an impermeable diderm cell envelope formed by a cytoplasmic membrane, a peptidoglycan and an arabinogalactan-layer, long-chain mycolic acids, and extractable lipids (Kaur et al. 2009; Le Chevalier et al. 2014), which contributes to the natural resistance of mycobacteria to many environmental stresses, biocides, and antibiotics.
Protein transport across this thick and complex mycobacterial cell envelope is carried out by different secretion systems, including the so-called ESX systems. These systems were named after the first identified substrate, the 6 kDa early secretory antigenic target (ESAT-6) (Brodin et al. 2004), and more recently were also termed type VII secretion systems (Abdallah et al. 2007; Majlessi et al. 2015). The typical ESX-secretion apparatus is constituted of a membrane-linked complex of at least four ESX-conserved-components (EccB, EccC, EccD, and MycP), ESX-type-specific associated proteins (EspA, EspB, EspC, EspG, etc.), and secreted/exported proteins, such as ESAT-6 and CFP-10, and/or PE and PPE proteins (Majlessi et al. 2015). The variation in the genetic organization of individual ESX systems defines five ESX subtypes in M. tuberculosis that are named ESX-1 to ESX-5 (Bitter et al. 2009).
The ESX-1 secretion system is a key component involved in M. tuberculosis pathogenicity (Majlessi et al. 2015), which is nonfunctional in the attenuated, closely related Mycobacterium bovis BCG vaccine due to the partial loss of the ESX-1-encoding genomic region, named region of difference 1 (RD1) (Mahairas et al. 1996; Behr et al. 1999; Pym et al. 2002). Protein secretion via this specialized ESX-1 system plays an important role for host–pathogen interaction of M. tuberculosis (Majlessi et al. 2015) and other pathogenic mycobacteria (Abdallah et al. 2007), enabling vacuolar rupture and cytosolic contact within host macrophages (Houben et al. 2012; Simeone et al. 2015). Moreover, other ESX systems are also involved in important biological functions of mycobacteria. Whereas ESX-3 plays a role in iron and zinc uptake (Serafini et al. 2009; Siegrist et al. 2014) the function of ESX-5 is linked to the export of PE and PPE proteins and pathogenicity (Abdallah et al. 2009; Bottai et al. 2012; Sayes et al. 2012).
Apart from protein secretion, ESX-1 systems are also involved in chromosomal DNA transfer through conjugation (Coros et al. 2008; Gray et al. 2013). In Mycobacterium smegmatis, the chromosomally encoded ESX-1 system enables unconventional genome-wide genetic exchanges between donor and recipient strains, named “distributive conjugal transfer” (Gray et al. 2013; Mortimer and Pepperell 2014). A recent study also describes conjugation events between strains of Mycobacterium marinum that involve a new class of plasmids-encoding elements of type VII and IV secretion systems and a relaxase (Ummels et al. 2014).
Thus, in mycobacteria, ESX systems govern diverse important biological functions for host–pathogen interaction and interstrain genetic transfer. Although some insights into the genetic organization and distribution of ESX systems among selected mycobacteria are available from previous studies (Cole et al. 1998; Tekaia et al. 1999; Gey Van Pittius et al. 2001), systematic large-scale screening data for typeVII/ESX components in mycobacterial genomes are not yet available. This led us to use an extensive, pan-genome-wide approach together with a large-scale Hidden Markov Model profile-based screen, to investigate the distribution of ESX systems in the large variety of mycobacterial chromosomes and plasmids. This study allowed us to identify a wide diversity of ESX systems in mycobacteria and to identify new, plasmid-encoded ESX-systems. Moreover, the generated deep phylogeny data and results from synteny analyses of the different ESX systems suggested that plasmid-encoded ESX clusters were substantially contributing to ESX diversification as well as to plasmid-chromosome genetic exchanges of ESX-associated genes and systems. Our results thus suggest an important contribution of ESX-encoding plasmids in long-term mycobacterial evolution, and more specifically in the evolution of ESX-mediated M. tuberculosis pathogenicity determinants, such as ESX-1 and/or ESX-5.
Materials and Methods
Mycobacterial Genome and Plasmids Database
The National Center for Biotechnology Information (NCBI) public database was used to build a representative set of mycobacterial sequences. When several sequences were available for a given species, the genome with the highest level of completeness was chosen based on the NCBI Genome Assembly and Annotation report (supplementary table S1, Supplementary Material online). All available mycobacterial plasmids that were fully assembled at the time of the database interrogation (July 2014) were introduced into the database (supplementary table S2, Supplementary Material online).
Strains from This Study
In addition to the sequences retrieved from the NCBI database, we added the sequences from two additional strains. One of them was taken from a clinical collection of M. abscessus isolates, and this strain was called M. abscessus subsp. bolletii strain 5625. The other strain was an environmental isolate from the Paris tap water network, belonging to the phylogenetic group of RGM M. aubagnense, and was called M. sp. 960. M. abscessus subsp. bolletii strain 5625 was introduced into the study because initial analysis of the assembled genome contig-sequences suggested that this strain contained a plasmid-encoding ESX elements. Strain M. sp. 960 was added, because no other genome information on the M. aubagnense phylogenetic group was yet published/available. NGS-derived sequence (SRA) reads from M. sp. 960 and M. abscessus subsp. bolletii strain 5625 were submitted to NCBI, and their accession numbers are, respectively, SRR1951096 and SRR1951800.
Genome Sequencing and Assembly
Paired-end Illumina libraries were constructed from 50 ng of genomic DNA according to the Epicenter Nextera protocol. A set of Nextera-compatible adaptor primers containing index sequences was used. Template amplification was performed using a cBot automated cluster generation system. Sequencing was performed on an Illumina HiSeq 2000 instrument (Illumina), using a read length of 50 or 100 bp. All library pools were treated as paired-end sequences. To ensure high data quality for various downstream analyses, such as sequence assembly, raw reads were subjected to a number of preprocessing “cleaning” steps: 1) reads bearing a number of bases with sufficiently high Phred quality score were selected (using Sanger quality > 20 and remaining read length > 30 nt as thresholds); 2) primer/adaptor sequence were excised from the remaining reads; 3) reads with lengths less than a given threshold were removed; 4) homopolymer-containing reads were trimmed; and 5) duplicated reads were identified and removed. Remaining reads were de novo assembled with CLC Genomics Workbench version 3 (CLC Bio, Cambridge, MA). Resulting sequence contigs were checked by comparison to the M. abscessus ATCC 19977 (GenBank ACC NC_010397) reference sequence, using the MUMmer package (Delcher et al. 2002).
ESX-Loci Annotation
Prokaryotic GeneMark.hmm (version 2.8) (Besemer and Borodovsky 2005) was used to predict ORFs on mycobacterial genomes and plasmids available in the GenBank database or sequenced in this study (supplementary tables S1 and S2, Supplementary Material online). Similarity searches were performed based on protein domains using HMMer package (Finn et al. 2011) (version 3.1b1), with ESX motifs previously identified in mycobacterial genomes: EccA (TIGR03922), EccB (TIGR03919), EccC (TIGR03924 and TIGR03925), EccD (TIGR03920), EccE (TIGR03923), MycP (TIGR03921), Esx (ESAT-6/CFP-10 proteins: TIGR03930), and PPE (PF00823). PE and EspG proteins were identified by similarity searches using BLASTP (Altschul et al. 1990) against mycobacterial genome sequences. An e-value threshold filter was introduced for each protein family to minimize the number of wrongly identified ESX-encoding genes. For each gene, the e-value threshold was determined based on the e-value distribution.
Phylogeny
Sequence alignments and curating was performed using MUSCLE and Gblocks, implemented in the Phylogeny.fr website (Dereeper et al. 2008). MEGA software (Tamura et al. 2013) was used for phylogenetic tree construction using a maximum-likelihood method with 250 bootstrap replicates and for generating best-fit models of evolution. Graphical representations of phylogenetic trees of individual genes were performed using iTOL (Letunic and Bork 2011). Phylogenetic trees at the gene level were obtained for EccB, EccC, MycP, followed by alignment of concatenated sequences of EccB, EccC, and MycP proteins, which was used to reconstruct the phylogeny of the ESX loci.
Comparative Genomics
Comparative genomics studies were performed on the Microbial Genome Annotation & Analysis Platform MaGe (Magnifying Genomes) (Vallenet et al. 2009), including synteny analysis, mobile elements identification, and genomic island detection. Composition bias detection was performed using Alien hunter software (Vernikos and Parkhill 2006). Plasmid and contig alignments at the nucleotide level were performed using Artemis Comparison Tool (Carver et al. 2005).
To estimate the level of gene conservation, pairwise dN/dS ratio ω (dN: nonsynonymous mutation substitution rate, dS: synonymous mutation substitution rate) were calculated using the program CODEML provided by the PAML (phylogenetic analyses by maximum likelihood) package version 4 (Yang 1997). Nucleotidique sequences have been aligned using TranslatorX (Abascal et al., 2010) guided by protein sequence alignments obtained using M-coffee (Wallace et al. 2006).
Recombination analysis was achieved using RDP4 version Beta 4.46 (Martin et al. 2015). Six methods including RDP, GENECONV, Bootscan, Maxchi, Chimaera, and SiScan implemented in RDP4 were used to detect recombination events, likely parental isolates and recombination break points under default settings.
Tree Topology Tests
As no a priori hypothesis for the phylogenetic placement of plasmid-borne ESX systems exists, we generated 85 bootstrapped trees of the concatenated alignment with PhyML (Guindon et al. 2010) under the LG+G+I+F amino acid substitution model (the best fit model according to ProtTest) (Abascal et al. 2005). Site-wise log-likelihood values of the 85 bootstrapped trees and the best tree obtained with MEGA were obtained with TreePuzzle (Schmidt et al. 2002) and fed into Consel (Shimodaira and Hasegawa 2001) to perform the approximately unbiased (AU) test of tree topologies. Monophyly of three plasmid-borne ESX clusters, the five ESX chromosomal types, and the sister relationships: ESX-P cluster 1—ESX-5; ESX-P cluster 2—(ESX-P cluster 3—ESX-2); ESX-P cluster 4—ESX-3 and ESX-4-bis—ESX-4 were assessed using Consel.
PFGE and Southern Blot Hybridization
Agarose plugs containing total genomic DNA from M. abscessus subsp. bolletii strain 5625 were prepared as previously described (Brosch et al. 2000). Non-digested DNA preparations were separated on a 1% (w/v) agarose gel by pulsed-field gel electrophoresis (PFGE) on a Biorad CHEF II apparatus with a pulse of 5 s ramping to 35 s for 23 h at 6 V/cm. Low-range PFG marker (NEB) was used as a size standard, and total genomic DNA from Mycobacterium canettii, known to lack plasmids (Supply et al. 2013), as negative control. Under such PFGE migration conditions, large plasmids migrate inside the gelmatrix, while intact chromosomal DNA is unable to migrate (Stinear et al. 2004). DNA was transferred onto a Hybond-C Extra nitrocellulose membrane (Amersham) as described in (Brosch et al. 2000) with some modifications. PCR-derived DNA probes were labeled with [α32P] dCTP using the Prime-It II kit (Stratagene), followed by hybridization at 68 °C in 6× SSC/0.5% SDS/0.01 M ethylenediaminetetraacetic acid/5 × Denhardt’s solution/100 μg/ml single-stranded salmon sperm DNA. The membrane was washed first for 5 min at room temperature in 2× SSC/0.5% SDS, then for 15 min in 2× SSC/0.1% SDS, and finally for 1 h in 1× SSC/0.5% SDS. The membrane was exposed on a phosphorimager screen and revealed using a STORM phosphorimager. The probes were amplified by PCR from M. bolletti pMBOL DNA with specific primers 5625-F (5′-AGGTACCAGCTCAAGGGAAC) and 5625-R (5′-GCATGGTGTTGGTGACGTTT), designed on M. bolletti pMBOL ESX-plasmid using Primer3 software (Koressaar and Remm 2007). PCR reaction conditions were as following: 100 ng of genomic DNA were added to the reaction mixture containing 2 μM of each primer, 0.5 units of Taq polymerase (ampliTaq, Applied Biosystems), PCR buffer (10% DMSO, 0.5 mM dNTP, 60 mM TrisHCl, pH 8.8, 2 mM MgCl2, 17 mM (NH4)2SO4, 10 mM β-mercaptoethanol), and sterile distilled water to 25 μl. PCR amplification conditions were as follows: initial denaturation of 94 °C for 5 min, 30 cycles of 94 °C for 30 s, 54 °C for 60 s, 72 °C for 45 s followed by a final extension of 72 °C for 10 min (Biorad). A PCR clean-up was performed (Macherey Nagel) and the purified fragments were double strand sequenced to confirm probes. To determine the contiguity and order of the two nonaligned contigs that contained ESX sequences in the M. abscessus subsp. bolletii 5625 strain, PCR primers were designed at the 5′ and 3′ ends of each contig covering all different contiguity possibilities. PCR products were then verified by sequencing, allowing to determine gene order and to confirm that each of these two nonaligned contigs contained a part of a single new ESX locus that was then probe-targeted for PFGE assay as described earlier.
Results
Identification of New ESX Loci in Mycobacterial Plasmids
To identify ESX-containing chromosomal loci and plasmids, we used the HMMER software (Finn et al. 2011) to launch motif searches corresponding to TIGR03922 (EccA), TIGR03919 (EccB), TIGR03924 (EccC), TIGR03920 (EccD), and TIGR3921 (MycP), in mycobacterial sequences from the NCBI database and in contig-sequences from our clinical collection of M. abscessus isolates. In total, we identified at least one or more full ESX locus/loci in 41 mycobacterial (22 RGM and 19 SGM strains) and in the 2 non-mycobacterial Actinobacteria (Nocardia farcinica and Gordonia bronchialis) genomes that were selected as out-group species (supplementary tables S1–S3, Supplementary Material online).
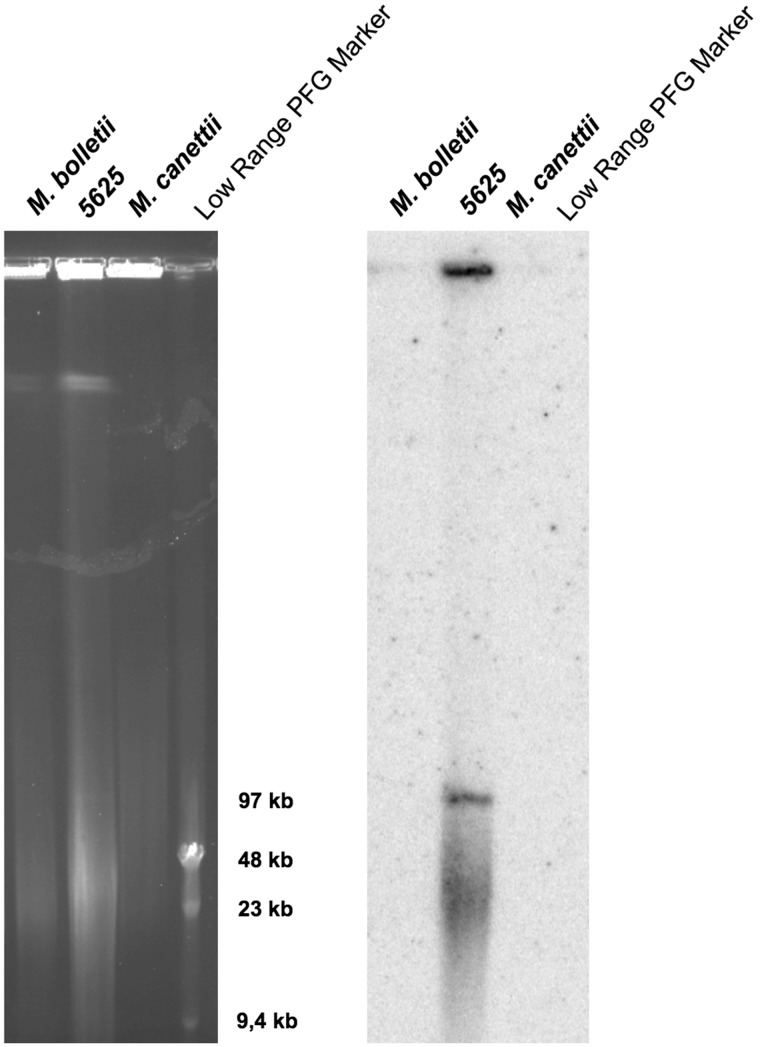
Interestingly, this approach identified ESX loci not only in numerous chromosomal segments (supplementary table S1, Supplementary Material online) but also revealed ESX motifs in 13 plasmids listed in the NCBI database (table 1), ranging from 97 kb to 615 kb in size. One of these plasmids corresponded to the recently described M. marinum pRAW-plasmid harboring an ESX-P1 system (Ummels et al. 2014). In addition, one isolate from our clinical strain collection, named M. abscessus subsp. bolletii strain 5625 was found to contain three distinct ESX loci. When we aligned the contig-sequences from this strain to the M. abscessus reference genome (Ripoll et al. 2009) and sorted the contigs based on to alignment or lack of such, a clear distinction was noticed. Within the group of the contig-sequences aligning to the reference, the sequences were identical to the ESX-3 and ESX-4 systems of the M. abscessus reference genome. In contrast, within the nonaligned contig-sequences, we noticed a new ESX locus that differed substantially from the known ESX-3 and ESX-4 systems of M. abscessus. To evaluate whether this latter locus was of plasmid origin, we prepared highly concentrated genomic DNA from strain 5625 and subjected it to PFGE analysis and Southern hybridization using a specific probe from the nonaligning ESX sequences. As shown in figure 1, PFGE analysis of non-digested genomic DNA from this strain revealed a band of approximately 100 kb that hybridized with the specific ESX probe. In agreement with previous observations reported for Mycobacterium ulcerans strains (Stinear et al. 2004), linearized forms of large, circular plasmids do migrate in PFGE gels, opposed to high molecular weight chromosomal DNA that remains immobilized in the wells. This example of strain 5625 shown in figure 1 thus serves as a proof of concept that ESX clusters identified by motif search in genome sequence databases may indeed be localized on large plasmids of mycobacterial species or strains. The plasmid in M. abscessus subsp. bolletii strain 5625 was named pMBOL. Overall, this information is also important for confirmation of the aforementioned results from NCBI database motif search, which identified 13 new, apparently plasmid-encoded ESX systems (table 1, fig. 2A, and supplementary table S3, Supplementary Material online) that are the main subject of our here presented study.
Table 1.
General characteristics of the identified mycobacterial ESX-plasmids
| Strain | Plasmid Name | Size (kb) | Reference | GI Number |
|---|---|---|---|---|
| M. abscessus subsp. bolletii strain 5625 | pMBOL | 97 | This study | |
| M. abscessus subsp. bolletii strain 50594 | plasmid 2 | 97 | (Kim et al. 2013a) | 506965416 |
| M. chubuense NBB4 | pMYCCH.01 | 615 | Lucas et al. unpublished dataa | 392405727 |
| M. chubuense NBB4 | pMYCCH.02 | 144 | Lucas et al. unpublished dataa | 392406268 |
| M. gilvum PYR-GCK | pMFLV01 | 321 | Copeland et al. unpublished dataa | 145225871 |
| M. kansasii ATCC 12478 | pMK12478 | 145 | (Wang et al. 2015) | 556559712 |
| M. marinum E11 | pRAW | 114 | (Ummels et al. 2014) | 641308534 |
| M. smegmatis | pMYCSM01 | 394 | Lucas et al. unpublished dataa | 433644115 |
| M. smegmatis | pMYCSM02 | 199 | Lucas et al. unpublished dataa | 433644438 |
| M. smegmatis | pMYCSM03 | 164 | Lucas et al. unpublished dataa | 433644684 |
| M. sp. KMS | pMKMS01 | 302 | Copeland et al. unpublished dataa | 119854889 |
| M. sp. KMS | pMKMS02 | 217 | Copeland et al. unpublished dataa | 119855174 |
| M. sp. MCS | plasmid 1 | 215 | Copeland et al. unpublished dataa | 108772792 |
| M. yongonense 05-1390 | pMyong1 | 123 | (Kim et al. 2013b) | 451770451 |
Note.—GI: identifier from NCBI database.
Publicly released genome sequence without associated publication.
Fig. 1.—
Ethidium bromide stained PFGE gel (left panel) and corresponding Southern hybridization blot (right panel) obtained with a PCR-derived, 32P-labeled probe from ESX-region of Mycobacterium abscessus subsp. bolletii strain 5625. Lane 1: M. abscessus subsp. bolletii type strain; lane 2: M. bolletii 5625. Lane 3: Mycobacterium canettii control strain STB-D (described in (Supply et al. 2013)), lane 4: low-range PFG Marker (NEB). PFGE conditions: 5 s ramping to 35 s; 6 V/cm; 23 h.
Fig. 2.—
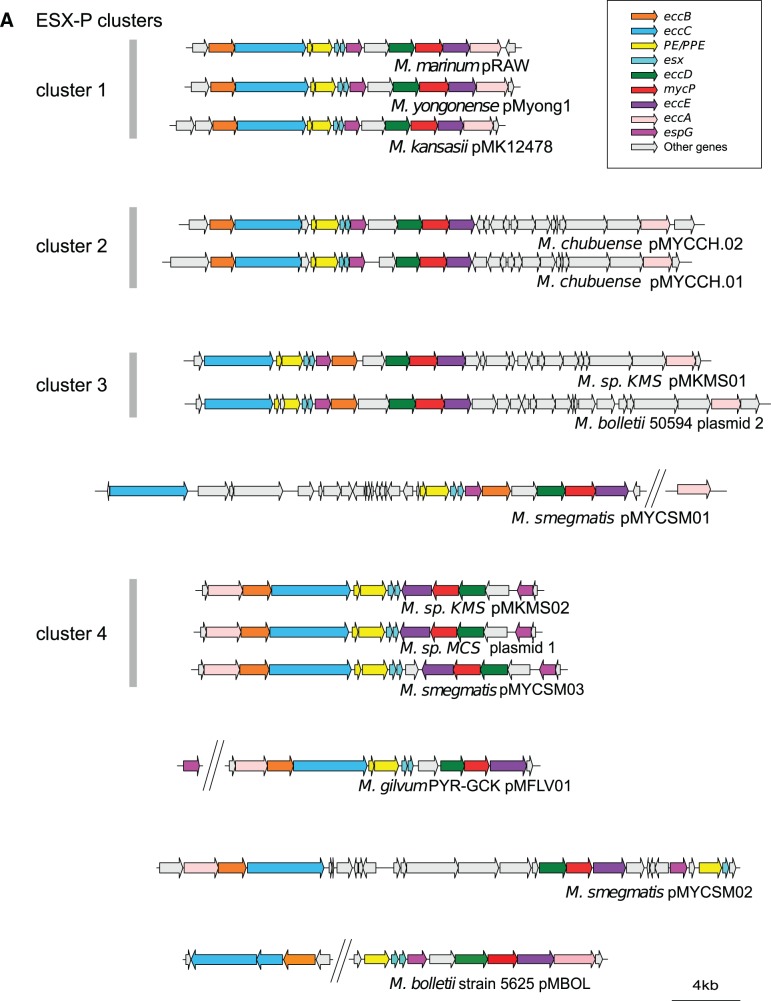
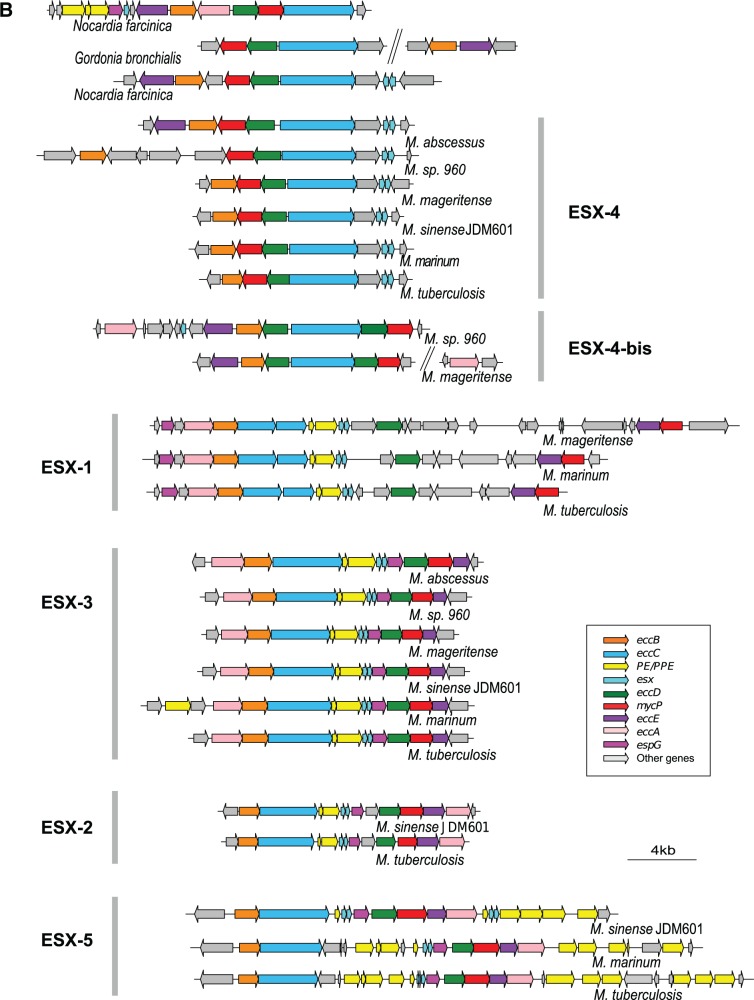
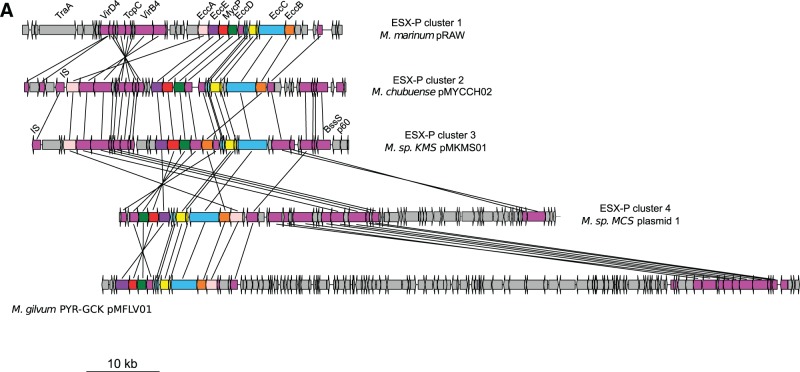
Genetic organization of ESX loci. (A) Genetic organization of ESX-P loci in mycobacterial plasmids. ESX-P clusters 2–4 represent new ESX types found in more than one plasmid. (B) Genetic organization of chromosomal ESX loci from a representative mycobacterial data set together with Nocardia farcinica and Gordonia bronchialis strains. Note that within the ESX-1 locus, the downstream gene of esxA was drawn according to the highest coding probability scores, which for Mycobacterium tuberculosis and Mycobacterium marinum were different from the original annotation.
Mycobacterial ESX Diversity
In parallel to our motif search-based analysis, manual gene annotation was applied to chromosomal ESX-loci of selected, representative species (M. abscessus, M. sp. 960, Mycobacterium mageritense, Mycobacterium sinense JDM601, M. marinum, and M. tuberculosis, respectively, belonging to three different RGM groups, to the intermediate Mycobacterium terrae complex group, and to two different SGM groups), and two closely related actinobacterial outgroups (N. farcinica and G. bronchialis) (fig. 2B). This screening showed that most ESX systems identified in the different mycobacteria and actinobacterial outgroups as being of chromosomal origin, displayed similarities in gene order and gene content with the previously described ESX types of M. tuberculosis (Bitter et al. 2009). However, this analysis also identified an additional, novel ESX variant in the chromosomal sequences of M. sp. 960, and M. mageritense, which showed some resemblance to ESX-4 systems and was termed ESX-4-bis (fig. 2B). The distinction between ESX-4-bis and classical ESX-4 was made on criteria concerning gene order and gene orientation. Whereas mycobacterial ESX-4 systems show a typical eccB/mycP/eccD/eccC/esx gene order, ESX-4-bis profiles contain two variants of EccD-encoding genes, situated up and downstream of eccC, displaying a yet unknown eccE/eccB/eccD/eccC/eccD/mycP gene order. Strikingly, ESX-4-bis loci, similar to ESX-4 systems, lacked PE/PPE-encoding genes and espG genes, whose gene products were reported to interact and play important roles in the biology of mycobacteria (Bottai et al. 2011; Ekiert and Cox 2014; Korotkova et al. 2014).
From all identified ESX types, the ESX-4 systems were the most widely distributed ESX system in mycobacterial species (supplementary table S1, Supplementary Material online), which is in agreement with previous reports (Tekaia et al. 1999; Gey Van Pittius et al. 2001). Other systems, such as ESX-1 and ESX-3 systems were also widely present in diverse species, whereas ESX-2 and ESX-5 systems were restricted to the SGM and M. terrae complex subgroups, in accordance with data in the literature (Gey van Pittius et al. 2006; Bitter et al. 2009; Bottai et al. 2014).
When the aforementioned plasmid-encoded ESX clusters were subjected to manual annotation, several novel ESX gene-organization profiles were found (fig. 2A, supplementary table S2, Supplementary Material online), which could be regrouped in ESX-P clusters or represented singletons (fig. 2A). Interestingly, members of ESX-P cluster 1 were present in plasmids from SGM species M. marinum, Mycobacterium kansasii, and Mycobacterium yongonense, whereas members of ESX-P clusters 2–4 or singletons were found in plasmids from various RGM species (fig. 2A).
Classification and Phylogeny of Ancestral and Mycobacteria-Specific ESX Types
To define the similarity/distance of these new ESX systems relative to the five classical chromosomal ESX-1-2-3-4-5 types, we reconstructed the individual phylogenies of selected ESX proteins, namely the ESX-conserved-components EccB and EccC (Bitter et al. 2009), as well as the MycP protease across the different mycobacterial species. Comparison of their amino acid sequences revealed that EccB, EccC, and MycP proteins from different species formed distinct clusters, in which orthologous proteins of each of the five chromosomal ESX types were grouped together, in agreement with the current ESX classification scheme (supplementary fig. S1, Supplementary Material online) (Gey Van Pittius et al. 2001; Bitter et al. 2009). Moreover, within each ESX type, the ESX protein-based phylogeny was found to be congruent with the mycobacterial species-based phylogeny, separating RGM and SGM within ESX clusters ESX-4-3-1 into different sub-clades (supplementary fig. S1, Supplementary Material online). The results further showed that ESX-4 and ESX-3 systems are ubiquitously distributed within the genus Mycobacterium, present in almost all RGM and SGM species analyzed. Moreover, ESX-1 systems were also frequently found both in SGM and a subgroup of RGM species. In contrast, the remaining two chromosomal ESX systems (ESX-2 and ESX-5) showed a more restricted distribution. ESX-5 systems were present exclusively in SGM and M. terrae complex species, whereas ESX-2 systems were restricted to one particular sub-group of the SGM and M. terrae complex. The characteristic gene order in each ESX-cluster (fig. 2B), the similar phylogenetic clustering of each of the three tested proteins (supplementary fig. S1, Supplementary Material online), together with the results from the AU test of phylogenetic tree selection (supplementary fig. S2, Supplementary Material online), suggest that the ESX loci are encoded by stably associated blocks of homologous genes. These findings encouraged us to concatenate the EccB, EccC, and MycP sequences with the aim to calculate a global phylogeny of ESX loci and to investigate the long-term evolutionary relationships among the different ESX types.
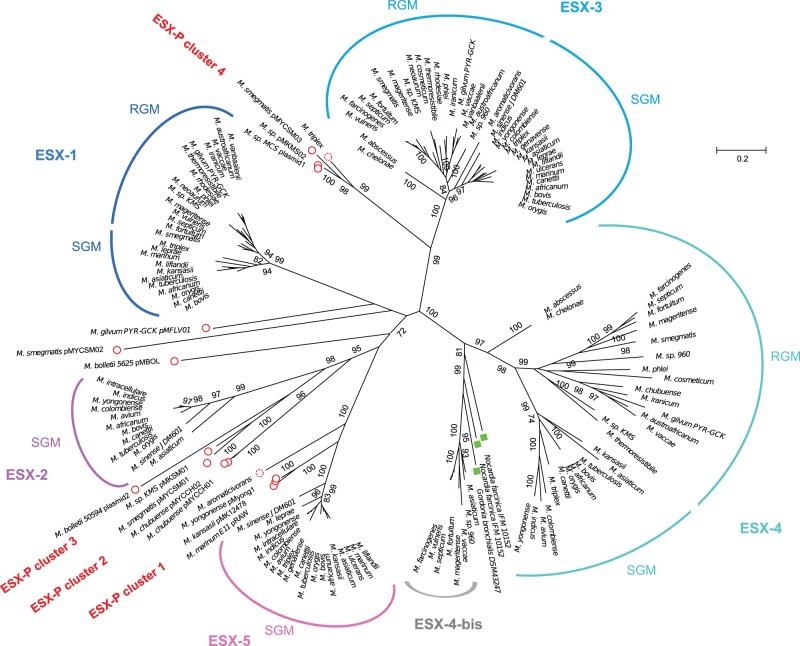
In the phylogenetic tree obtained (fig. 3), the chromosomal ESX-1-ESX-5 loci form the major branches supported by bootstrap values ranging from 99% to 100%. As seen for the analysis of the single EccB/C or MycP proteins, the branches of ESX-4, ESX-3, and ESX-1 are sub-divided into systems from RGM and SGM species. The concatenated sequence-based tree (fig. 3), together with the AU test (supplementary fig. S2, Supplementary Material online), also supports the monophyletic relationship of the mycobacterial ESX-4 and ESX-4-bis systems, with the non-mycobacterial actinobacterial ESX-4-like systems from N. farcinica, and G. bronchialis. Indeed, the ESX-4 associated types were clearly separated from the other, mycobacteria-specific chromosomal and plasmid-borne ESX types (bootstrap value 100%), emphasizing the ancestral character of ESX-4 and ESX-4-bis systems. Interestingly, several mycobacterial strains harbored both a classical ESX-4 system and an ESX-4-bis system (fig. 3). Moreover, our phylogenetic analysis revealed that ESX-1 and ESX-3 systems each formed clearly separated clusters with little intracluster diversity, whereas the ESX-5 and ESX-2 clusters share a common root and thus form a subgroup within the proposed phylogeny. However, the most interesting novel insights from the study come from inspection of the plasmid-borne ESX-systems, which branch at deep rooting positions next to the ESX-1, ESX-3, and ESX-2-5 systems.
Fig. 3.—
Phylogenetic tree of mycobacterial ESX loci from concatenated sequences of EccB, EccC, and MycP. Red circles: plasmid-borne ESX types. Dotted red circles: putative ESX-containing plasmids. Green boxes: Nocardia farcinica and Gordonia bronchialis. Best evolution model identified by MEGA software (WAG+G+I). Tree constructed with maximum-likelihood method with 250 bootstrap replications. Values > 70% are indicated. Note that for some species only selected chromosomal ESX systems are shown, as defined in supplementary table S1, Supplementary Material online. RGM, rapidly growing mycobacteria; SGM, slowly growing mycobacteria.
Classification of Plasmid-Borne ESX Families (ESX-P)
Analysis of the data presented in the phylogenetic tree of ESX concatenated sequences showed that the different ESX-P types are grouping together with certain chromosomal ESX families (fig. 3 and supplementary fig. S2, Supplementary Material online). Members of the ESX-P cluster 4, for example, were found to group together with the ESX-3 family (bootstrap value 99%, confirmed by AU test). Two consecutively branching groups constituted by members of ESX-P clusters 2 and 3 were found at the root of the chromosomal ESX-2 types (bootstrap values 95% and 98%, respectively). Moreover, the members of the ESX-P cluster 1 branched at the root of the ESX-5 systems (bootstrap value 100%) (fig. 3 and supplementary fig. S2, Supplementary Material online). Each ESX-P cluster was characterized by a specific gene organization. However, all plasmid-borne ESX types shared a minimal common gene order defined by eccC/PE/PPE/esx/esx and eccD/mycP/eccE. Interestingly, the gene order in ESX-P profiles differed markedly from ancestral ESX types and was closer to the ESX-2, ESX-3, and ESX-5 organization, which also showed an eccD/mycP/eccE organization.
Finally, the distribution of the different ESX systems on the phylogenetic tree also allowed us to classify two yet nonclassified ESX clusters that were found on nonaligned contigs from Mycobacterium aromaticivorans and Mycobacterium triplex. These sequences grouped with ESX-P clusters 1 and 4, respectively, (fig. 3, dotted red circles and supplementary fig. S2, Supplementary Material online) and also shared similar genetic organization containing both elements of type IV and type VII secretion systems (supplementary figs. S3 and S4, Supplementary Material online), suggesting that they might represent yet unknown plasmid-borne ESX systems.
To exclude that branches supporting the different plasmid-borne ESX families, which are located between chromosomal ESX groups were the result of genetic mosaicism and recombination between chromosomal ESX systems, we analyzed our data set using the Recombination Detection Program RDP4 (Martin et al. 2015). No recombination events between the chromosomal ESX and plasmid-borne ESX-P systems were detected, limiting the possibility of an artifact due to mosaic ESX genes to a minimum. Moreover, to see if plasmid-borne ESX-P clusters were under purifying selection, we calculated the ratios of synonymous and nonsynonymous substitutions (supplementary fig. S5, Supplementary Material online). This analysis showed that EccB-, EccC-, and MycP-encoding genes from ESX-P clusters 1, 2, and 3 were under purifying selection (dn/ds < 1). This result advocates for a diversification process leading to the observed ESX-P diversity (rather than lack of selection pressure leading to various degenerated ESX systems). Taken together, all these data suggest that ESX-P systems represent genuine functional and diversified, plasmid-specific ESX families.
Finally, the gene order within the various ESX-P systems was also consistent in most cases with the phylogeny obtained from concatenated sequences (figs. 2A and 3). Similarly to ESX-P cluster 4, in ESX-1 and ESX-3 the position of eccA was found to be located at the upstream part of the ESX locus, followed by eccB/eccC. On the contrary, in chromosomal ESX-2 and ESX-5 systems EccA is encoded in the most downstream part of the ESX locus, after eccD/mycP/eccE, a constellation, which is also observed in the plasmid-borne ESX-P clusters 1-2-3. These similarities are consistent with the ESX-P positions within the distance tree based on concatenated sequences (fig. 3) and thus suggest a plausible evolutionary link between ESX-P cluster 4 and ESX-3/ESX-1 on the one side, as well as ESX-P clusters 1-2-3 and ESX-2/ESX-5 on the other side.
Phylogeny of ESX-P Families and Mycobacteria-Specific Genomic ESX
To investigate the evolutionary history of plasmid-borne and chromosomal mycobacteria-specific ESX systems, we performed synteny analysis of the ESX loci using the Microbial Genome Annotation & Analysis Platform MaGe (Vallenet et al. 2009). This tool was used to determine synteny blocks in the vicinity of ESX-P loci. In all investigated ESX-encoding plasmids, ESX loci were embedded within larger synteny blocks including plasmid-specific type IV secretion system genetic elements, such as genes encoding VirD4, TrpC, and VirB4 (fig. 4A and supplementary figs. S3 and S4, Supplementary Material online). Within the different plasmids, ESX-P and type-IV loci were contiguous and displayed various relative positions probably due to plasmid rearrangements. The observation of these large synteny blocks involving both ESX and type IV systems strongly support that ESX-P systems are phylogenetically related. The ESX-encoding plasmids thus form a group that seems to derive from a single common origin. Variation of gene order most probably diversified through plasmid rearrangements. Moreover, the observation of the eccA position within the various clusters suggests that this gene co-migrates with type IV genetic rearrangements. Thus, the differences observed in eccA positions among the various ESX-P clusters might be explained by these local rearrangements. Interestingly, as mentioned earlier, the position of eccA is a feature that differentiates ESX-1/ESX-3 from ESX-2/ESX-5 systems. Thus, plasmid rearrangements may have made a phylogenetic link between these genomic ESX systems, suggesting that diversification of ESX-coding plasmids might be at the origin of—at least some—mycobacteria-specific chromosomal ESX types. This hypothesis is consistent with observed phylogeny, showing ESX-P systems branching at the root of mycobacteria-specific chromosomal ESX systems (fig. 3). Such scenario would thus necessitate a chromosomal integration of ESX-P systems through horizontal gene transfer (HGT) into mycobacteria.
Fig. 4.—
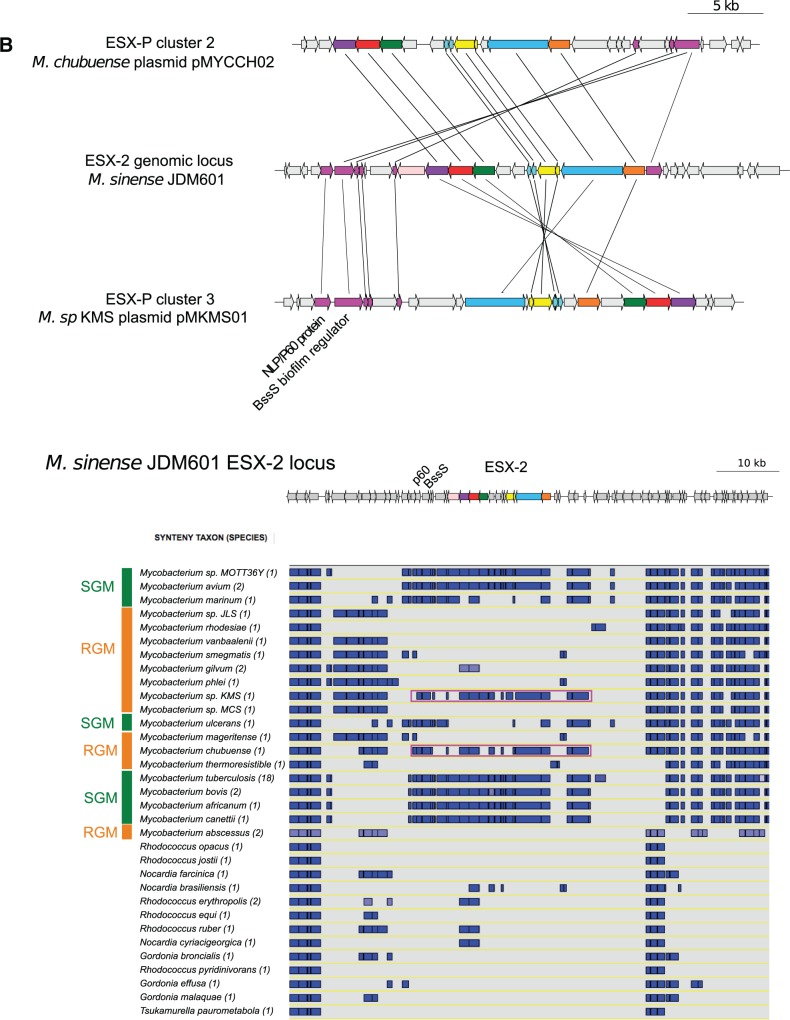
ESX loci within synteny blocks. Purple arrows: genes involved in synteny block within contiguous region of ESX loci. (A) Synteny blocks involving ESX loci of mycobacterial plasmids from each ESX-P plasmid cluster and Mycobacterium gilvum pMFLV01. (B) Synteny blocks within Mycobacterium sinense JDM601 ESX-2 genomic region. Upper panel: genetic organization of ESX plasmids pMYCCH02 and pMKMS02 in synteny with M. sinense JDM601 ESX-2 locus. Lower panel: a representative subset of SGM and RGM, respectively, and other closely related actinobacteria were investigated. Blue boxes: synteny blocks. Surrounding purple box: plasmid.
Clues of HGT between ESX-P and Genomic ESX
To investigate further the hypothesis of a plasmid-borne origin of mycobacteria-specific chromosomal ESX systems, we focused on ESX-2 and ESX-5. These ESX systems are absent from RGM and present in SGM and M. sinense JDM601 (a species that belongs to the intermediate group of the M. terrae complex). Interestingly, genomic comparison and synteny analysis of the ESX-2 locus in M. sinense JDM601 showed that this gene cluster is embedded into a larger synteny block which is present in SGM chromosomes and ESX-encoding plasmids pMKMS01 and pMYCCH02 of the RGM species M. sp KMS and M. chubuense, respectively, whereas it is absent from RGM chromosomes (fig. 4B). Interestingly, this synteny block harbors a gene bearing an NLP/p60 domain, which is found in most ESX-P systems (supplementary table S4, Supplementary Material online), and which is involved in B. subtilis conjugation functions (DeWitt and Grossman 2014). The synteny block also contained a gene encoding BssS, a biofilm regulator that has homologs only in ESX-2 loci and in mycobacterial plasmids (supplementary table S5, Supplementary Material online). Taken together, these findings may serve as one example how a chromosomal ESX system might have emerged by genetic exchange that apparently occurred between ESX-encoding plasmids and mycobacterial chromosomes.
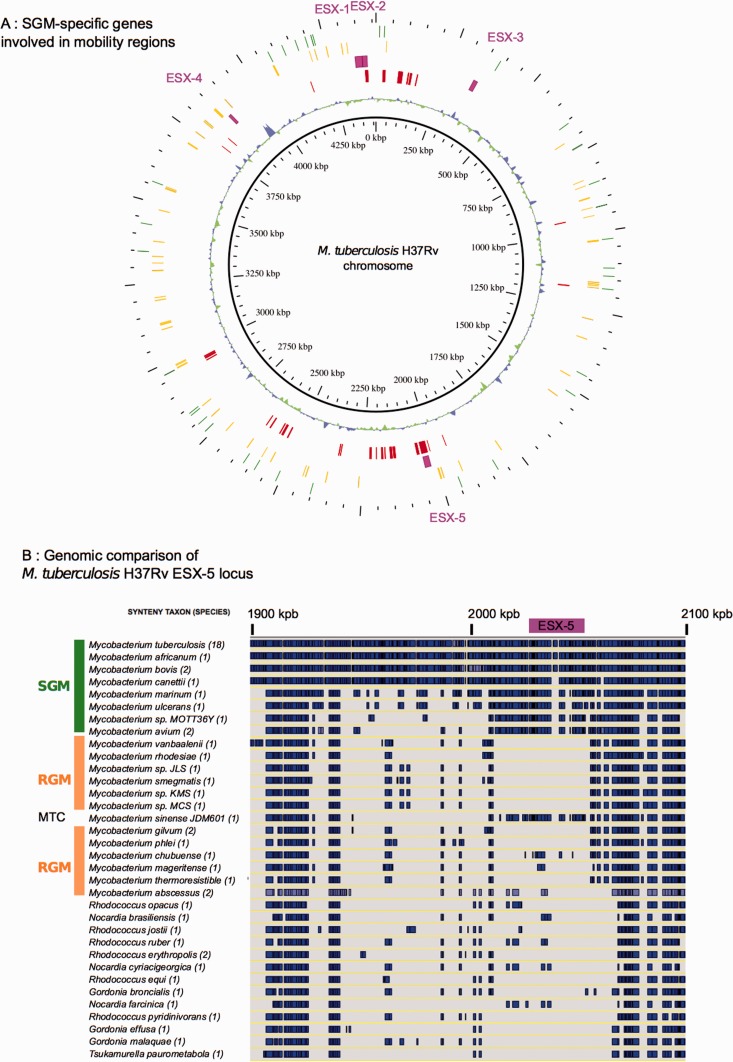
To identify putative regions of exogenous DNA integration, specific for SGM and members of the M. terrae complex (that specifically contained ESX-2 and ESX-5), we used the MaGe platform (Vallenet et al. 2009) to find regions of genomic plasticity. Selection criteria for members of these regions were the following: M. tuberculosis H37Rv genes should share homologs in both Mycobacterium avium and M. sinense JDM601 but without any homolog in M. smegmatis, Mycobacterium gilvum PYR-GCK, and M. abscessus (best bidirectional hit with 30% identity threshold). Furthermore, the selected genes should also be present within a synteny group showing compositional bias (Vernikos and Parkhill 2006), as identified by the Alien Hunter software (fig. 5A). Interestingly, ESX-2 and ESX-5 were both found in genomic regions that were detected as regions of genomic plasticity (fig. 5A). Moreover, the putative genomic island region flanking ESX-5 also contained mobile genetic elements, such as insertion sequences as well as tRNA genes known to serve as potential integration sites (fig. 5A). Moreover, this ESX system was found to be embedded within a larger 50 kb synteny block present in SGM and M. terrae complex species but absent from all tested RGM species (fig. 5B). Taken together, these findings support the hypothesis that ESX-2 and ESX-5 might have been acquired by SGM and M. terrae complex species via independent HGT episodes during mycobacterial evolution, most probably from ESX-encoding plasmids.
Fig. 5.—
SGM-specific genes of Mycobacterium tuberculosis H37Rv and ESX-5 genomic organization. (A) SGM-specific genes involved in mobility regions. First inner circle: GC%. Second inner circle: red boxes represent M. tuberculosis genes with homologs in Mycobacterium avium and Mycobacterium sinense JDM501 but without any homolog in Mycobacterium smegmatis, Mycobacterium gilvum PYR-GCK, or Mycobacterium abscessus (BBH 30% identity) and identified within synteny groups containing composition bias according to Alien Hunter software. Third inner circle: purple boxes represent ESX loci. First outer circle: tRNAs. Secound outer circle: insertion sequences. (B) Synteny blocks within ESX-5 region of M. tuberculosis H37Rv. A representative subset of SGM and RGM, respectively, and other closely related actinobacteria were investigated. MTC, Mycobacterium terrae complex. Blue boxes: synteny blocks. Purple box: ESX-5 locus.
Discussion
Gene flow is an important factor of bacterial niche adaptation and speciation through the acquisition of foreign genetic material by HGT. The mechanisms that mediate this process comprise phage transduction, natural transformation, and plasmid conjugation, and these events are especially important for the transfer of antibiotic resistance and acquisition of virulence factors (Blair et al. 2015). However, for mycobacteria the impact of HGT in the pathogenomic evolution of its members remains largely unknown. Some insights have been gained from the analysis of genomic islands in the genome of M. tuberculosis (Rosas-Magallanes et al. 2006; Becq et al. 2007) but the question remains how in the earlier evolution of the pathogenic SGM species HGT might have been organized and by which mechanisms gene flow was enabled to occur. In many bacterial species, transfer of plasmids is one of the key driving forces of HGT. For mycobacteria, it is known for long time that plasmids are present in some species (Le Dantec et al. 2001; Stinear et al. 2004, 2008; Ripoll et al. 2009; Leao et al. 2013; Uchiya et al. 2015), although the classical OriT/type IV conjugative systems do not seem to play a role in this genus. However, recent experiments showed that conjugal transfer of plasmids can be observed in certain M. marinum strains, involving a novel type of conjugative plasmid that possesses an ESX system and elements of a classical type IV system, located on the same plasmid, named pRAW (Ummels et al. 2014). The identification of this plasmid-mediated conjugation mechanism is supported by a previous report that described a putative plasmid transfer between the SGM species M. avium and M. kansasii in a mixed infection in a patient (Rabello et al. 2012). Apart from these rare reports on HGT mediated by plasmids, it is also known that mycobacterial HGT may be organized via chromosomally encoded conjugation systems. This is the case for a process driven by the ESX-1 system of M. smegmatis, resulting in genome-wide recombination clusters and mosaicism that was named distributive conjugal transfer (Gray et al. 2013). Together, the examples presently described in the literature suggest that conjugative processes might have been—or still are—responsible for certain episodes of HGT among mycobacterial strains, thereby driving mycobacterial evolution. However, it should also be mentioned that overall, the insights into the mechanisms of HGT and gene flow in mycobacteria are scarce, which was one of the main motivations for us to undertake this study and use recently available mycobacterial pan-genome data to elucidate the question of potential mobile conjugation systems and HGT in mycobacteria.
This analysis allowed us to explore the diversity and the putative origin of ESX systems in mycobacterial chromosomes and plasmids. Our approach confirmed that the five previously designated chromosomal ESX types (ESX-1-5) (Gey Van Pittius et al. 2001; Bitter et al. 2009) constitute meaningful groups from a phylogenetic point of view, because they represent well-defined phylogenetic clusters, each of them being in accordance with mycobacterial phylogeny. We also confirmed the ancestral nature of the ESX-4 type and identified novel ESX-4 like type, named ESX-4-bis, which were present in various mycobacterial strains in addition to the classical ESX-4 systems.
Importantly, our analysis resulted in the identification of new, divergent ESX systems encoded on plasmids, thereby largely expanding the current knowledge on ESX-type diversity. The description of new genetic ESX organization-schemes in mycobacterial mega-plasmids (>100 kb) suggests that, besides the well-known type VII secretion system functions, that is pathogenicity, metal ion uptake, and conjugation, other yet unknown functions may potentially be provided by these yet unexplored ESX systems. In addition, their location on mega-plasmids with large coding capacities suggests that the presence of ESX-carrying plasmids in selected strains might modulate the phenotype of the concerned mycobacteria, and thus might be an important factor for promoting niche adaptation in new environments. In a more practical perspective, the identification of a 100 kb sized plasmid pMBOL in M. abscessus subsp. bolletii strain now provides the possibility for experimentally addressing such questions on evolution and transfer of ESX-containing plasmids and the involved mechanisms.
During data analysis and the generation of the phylogenetic tree (fig. 3), the question arose whether our analysis could have been biased by genetic saturation and “long branch attraction” phenomena (Philippe and Forterre 1999; Raymann et al. 2015), as for the comparison of distantly-related ESX types such effects cannot be excluded a priori. However, we are confident that these phenomena did not have a measurable impact on the results of our analysis because several other characteristics such as AU test results, gene order, synteny, and lacking gene sequence mosaicisms in different ESX clusters also support the obtained phylogeny. Based on this converging body of evidence an evolutionary scenario can be proposed in which ancestral ESX-4-like systems present in mycobacteria and/or other actinobacterial species had been transferred onto plasmids, and underwent extensive rearrangement processes, leading to diverse forms of new ESX types. Some of these rearranged ESX types were subsequently transferred to the chromosomes of certain mycobacteria, resulting in the mycobacteria-specific ESX types. Because ESX-5 and ESX-2 are found exclusively in SGM and M. terrae complex species (M. sinense JDM601), their putative chromosomal acquisition by HGT seems to have occurred in episodes prior or during SGM differentiation. It is tempting to hypothesize that some HGT episodes like, for example, the acquisition of ESX-5 might have contributed to acquisition of new functions to SGM species that are now exploited by pathogenic SGM species during host–pathogen interaction (Abdallah et al. 2011; Bottai et al. 2012; Sayes et al. 2012; Ates et al. 2015).
In conclusion, our study provides new insights on the diversity and conservation of ESX systems in a broad range of mycobacteria and proposes a unique model in which ESX-carrying plasmids play a key role in the distribution and refinement of type VII secretion-related processes during the long-term evolution of the mycobacterial genus. As observed from the different ESX-related gene distribution profiles, the involved plasmids might not only have acted as mycobacterial gene-exchange vectors but might also have served as accelerators of adaptation and biodiversity with probable impact on the emergence of mycobacterial pathogenicity.
Supplementary Material
Acknowledgments
The authors are grateful to Guillaume Achaz, Sophie Brouillet, Joël Pothier (Atelier de Bioinformatique de Jussieu), Philippe Lopez (UMR 7138—Université Pierre et Marie Curie), and to Jean-Louis Herrmann (INSERM U1173, UFR Simone Veil, Versailles-Saint-Quentin University) for help and advice. Support by grants from the European Community (260872 and H2020-PHC-643381), the Agence National de Recherche (ANR-14-JAMR-001-02) and the Fondation pour la Recherche Médicale (DEQ20130326471) are gratefully acknowledged. E.C.B. was supported by a stipend from the Pasteur–Paris University (PPU) International PhD program and the Institut Carnot Pasteur Maladies Infectieuses.
Supplementary Material
Supplementary tables S1–S5 and figures S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Literature Cited
- Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. [DOI] [PubMed] [Google Scholar]
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:W7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah AM, et al. 2011. Mycobacterial secretion systems ESX-1 and ESX-5 play distinct roles in host cell death and inflammasome activation. J Immunol. 187:4744–4753. [DOI] [PubMed] [Google Scholar]
- Abdallah AM, et al. 2007. Type VII secretion system of mycobacteria show the way. Nat Rev Microbiol. 5:883–891. [DOI] [PubMed] [Google Scholar]
- Abdallah AM, et al. 2009. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol. 73:329–340. [DOI] [PubMed] [Google Scholar]
- Altschul S, Gish W, Miller W, Myers E, Lipman D. 1990. A basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Ates LS, et al. 2015. Essential role of the ESX-5 secretion system in outer membrane permeability of pathogenic mycobacteria. PLoS Genet. 11:e1005190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becq J, et al. 2007. Contribution of horizontally acquired genomic islands to the evolution of the tubercle bacilli. Mol Biol Evol. 24:1861–1871. [DOI] [PubMed] [Google Scholar]
- Behr MA, et al. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520–1523. [DOI] [PubMed] [Google Scholar]
- Besemer J, Borodovsky M. 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33:W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter W, et al. 2009. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 5:e1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 13:42–51. [DOI] [PubMed] [Google Scholar]
- Boritsch EC, et al. 2014. A glimpse into the past and predictions for the future: the molecular evolution of the tuberculosis agent. Mol Microbiol. 93:835–852. [DOI] [PubMed] [Google Scholar]
- Bottai D, et al. 2012. Disruption of the ESX-5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Mol Microbiol. 83:1195–1209. [DOI] [PubMed] [Google Scholar]
- Bottai D, et al. 2011. ESAT-6 secretion-independent impact of ESX-1 genes espF and espG1 on virulence of Mycobacterium tuberculosis. J Infect Dis. 203:1155–1164. [DOI] [PubMed] [Google Scholar]
- Bottai D, Stinear TP, Supply P, Brosch R. 2014. Mycobacterial Pathogenomics and Evolution. Microbiol Spectrum. 2: MGM2-0025-2013. [DOI] [PubMed] [Google Scholar]
- Brodin P, Rosenkrands I, Andersen P, Cole ST, Brosch R. 2004. ESAT-6 proteins: protective antigens and virulence factors? Trends Microbiol. 12:500–508. [DOI] [PubMed] [Google Scholar]
- Brosch R, et al. 2000. Comparative genomics uncovers large tandem chromosomal duplications in Mycobacterium bovis BCG Pasteur. Yeast 17:111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TJ, et al. 2005. ACT: the artemis comparison tool. Bioinformatics 21:3422–3423. [DOI] [PubMed] [Google Scholar]
- Cole ST, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. [DOI] [PubMed] [Google Scholar]
- Coros A, Callahan B, Battaglioli E, Derbyshire KM. 2008. The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis. Mol Microbiol. 69:794–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Phillippy A, Carlton J, Salzberg SL. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30:2478–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt T, Grossman AD. 2014. The bifunctional cell wall hydrolase CwlT is needed for conjugation of the integrative and conjugative element ICEBs1 in Bacillus subtilis and B. anthracis. J Bacteriol. 196:1588–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Cox JS. 2014. Structure of a PE-PPE-EspG complex from Mycobacterium tuberculosis reveals molecular specificity of ESX protein secretion. Proc Natl Acad Sci U S A. 111:14758–14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gey Van Pittius NC, et al. 2001. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2:RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gey van Pittius NC, et al. 2006. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol Biol. 6:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray TA, Krywy JA, Harold J, Palumbo MJ, Derbyshire KM. 2013. Distributive conjugal transfer in mycobacteria generates progeny with meiotic-like genome-wide mosaicism, allowing mapping of a mating identity locus. PLoS Biol. 11:e1001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Houben D, et al. 2012. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol. 14:1287–1298. [DOI] [PubMed] [Google Scholar]
- Kaur D, Guerin ME, Skovierova H, Brennan PJ, Jackson M. 2009. Chapter 2: biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv Appl Microbiol. 69:23–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BJ, et al. 2013a. Complete genome sequence of Mycobacterium massiliense clinical strain Asan 50594, belonging to the type II genotype. Genome Announc. 1(4):e00429–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BJ, et al. 2013b. Whole-genome sequence of a novel species, Mycobacterium yongonense DSM 45126T. Genome Announc. 1(4):e00604–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koressaar T, Remm M. 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291. [DOI] [PubMed] [Google Scholar]
- Korotkova N, et al. 2014. Structure of the Mycobacterium tuberculosis type VII secretion system chaperone EspG5 in complex with PE25-PPE41 dimer. Mol Microbiol. 94:367–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Chevalier F, Cascioferro A, Majlessi L, Herrmann JL, Brosch R. 2014. Mycobacterium tuberculosis evolutionary pathogenesis and its putative impact on drug development. Future Microbiol. 9:969–985. [DOI] [PubMed] [Google Scholar]
- Le Dantec C, Winter N, Gicquel B, Vincent V, Picardeau M. 2001. Genomic sequence and transcriptional analysis of a 23-kilobase mycobacterial linear plasmid: evidence for horizontal transfer and identification of plasmid maintenance systems. J Bacteriol. 183:2157–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leao SC, et al. 2013. The detection and sequencing of a broad-host-range conjugative IncP-1beta plasmid in an epidemic strain of Mycobacterium abscessus subsp. bolletii . PLoS One 8:e60746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39:W475–W478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee JG, Ward AC. 2012. Genus I. Mycobacterium In: Goodfellow M, Kämpfer P, Busse HJ, Trujillo ME, Suzuki KI, Ludwig W, Whitman WB, editors. Bergey’s manual of systematic bacteriology, the actinobacteria. New York: Springer; p. 312–375. [Google Scholar]
- Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, Stover CK. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol. 178:1274–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majlessi L, Prados-Rosales R, Casadevall A, Brosch R. 2015. Release of mycobacterial antigens. Immunol Rev. 264:25–45. [DOI] [PubMed] [Google Scholar]
- Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 1:vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignard S, Flandrois JP. 2008. A seven-gene, multilocus, genus-wide approach to the phylogeny of mycobacteria using supertrees. Int J Syst Evol Microbiol. 58:1432–1441. [DOI] [PubMed] [Google Scholar]
- Mortimer TD, Pepperell CS. 2014. Genomic signatures of distributive conjugal transfer among mycobacteria. Genome Biol Evol. 6:2489–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Forterre P. 1999. The rooting of the universal tree of life is not reliable. J Mol Evol. 49:509–523. [DOI] [PubMed] [Google Scholar]
- Pym AS, Brodin P, Brosch R, Huerre M, Cole ST. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol Microbiol. 46:709–717. [DOI] [PubMed] [Google Scholar]
- Rabello MC, et al. 2012. First description of natural and experimental conjugation between mycobacteria mediated by a linear plasmid. PLoS One 7:e29884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymann K, Brochier-Armanet C, Gribaldo S. 2015. The two-domain tree of life is linked to a new root for the Archaea. Proc Natl Acad Sci U S A. 112:6670–6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll F, et al. 2009. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Magallanes V, et al. 2006. Horizontal transfer of a virulence operon to the ancestor of Mycobacterium tuberculosis. Mol Biol Evol. 23:1129–1135. [DOI] [PubMed] [Google Scholar]
- Sayes F, et al. 2012. Strong immunogenicity and cross-reactivity of Mycobacterium tuberculosis ESX-5 Type VII Secretion- encoded PE-PPE proteins predicts vaccine potential. Cell Host Microbe. 11:352–363. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502–504. [DOI] [PubMed] [Google Scholar]
- Serafini A, Boldrin F, Palu G, Manganelli R. 2009. Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. J Bacteriol. 191:6340–6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246–1247. [DOI] [PubMed] [Google Scholar]
- Siegrist MS, et al. 2014. Mycobacterial Esx-3 requires multiple components for iron acquisition. MBio. 5:e01073–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone R, et al. 2015. Cytosolic access of Mycobacterium tuberculosis: critical impact of phagosomal acidification control and demonstration of occurrence in vivo. PLoS Pathog. 11:e1004650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear TP, et al. 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci U S A. 101:1345–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinear TP, et al. 2008. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 18:729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P, et al. 2013. Genomic analysis of smooth tubercle bacilli provides insights into ancestry and pathoadaptation of Mycobacterium tuberculosis. Nat Genet. 45:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekaia F, et al. 1999. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber Lung Dis. 79:329–342. [DOI] [PubMed] [Google Scholar]
- Uchiya K, et al. 2015. Characterization of a novel plasmid, pMAH135, from Mycobacterium avium Subsp. hominissuis. PLoS One 10:e0117797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ummels R, et al. 2014. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. MBio. 5:e01744–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D, et al. 2009. MicroScope: a platform for microbial genome annotation and comparative genomics. Database 2009:bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos GS, Parkhill J. 2006. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22:2196–2203. [DOI] [PubMed] [Google Scholar]
- Wallace IM, O’Sullivan O, Higgins DG, Notredame C. 2006. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 34:1692–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. 2015. Insights on the emergence of Mycobacterium tuberculosis from the analysis of Mycobacterium kansasii. Genome Biol Evol. 7:856–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 13:555–556 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.