Abstract
Casposons are a superfamily of putative self-synthesizing transposable elements that are predicted to employ a homolog of Cas1 protein as a recombinase and could have contributed to the origin of the CRISPR-Cas adaptive immunity systems in archaea and bacteria. Casposons remain uncharacterized experimentally, except for the recent demonstration of the integrase activity of the Cas1 homolog, and given their relative rarity in archaea and bacteria, original comparative genomic analysis has not provided direct indications of their mobility. Here, we report evidence of casposon mobility obtained by comparison of the genomes of 62 strains of the archaeon Methanosarcina mazei. In these genomes, casposons are variably inserted in three distinct sites indicative of multiple, recent gains, and losses. Some casposons are inserted into other mobile genetic elements that might provide vehicles for horizontal transfer of the casposons. Additionally, many M. mazei genomes contain previously undetected solo terminal inverted repeats that apparently are derived from casposons and could resemble intermediates in CRISPR evolution. We further demonstrate the sequence specificity of casposon insertion and note clear parallels with the adaptation mechanism of CRISPR-Cas. Finally, besides identifying additional representatives in each of the three originally defined families, we describe a new, fourth, family of casposons.
Keywords: casposons, self-synthesizing transposons, CRISPR-Cas, mobile genetic elements, transposition
Introduction
The genomes of most bacteria, archaea, and eukaryotes contain multiple, integrated mobile genetic elements (MGEs), such as transposons, proviruses, and integrative plasmids. In many eukaryotes, in particular plants, the MGE-derived sequences comprise the majority of the genomic DNA. Most of these elements are “dead,” that is, inactive and disrupted to various extents (Kazazian 2004; Venner et al. 2009; Tollis and Boissinot 2012). The MGEs are less abundant in archaea and bacteria, conceivably due to the intense purifying selection that constrains the spread of selfish elements but nevertheless constitute up to 30% of some bacterial genomes (Casjens 2003; Carle et al. 2010). The MGE insertion can be deleterious when an element disrupts an essential host gene, nearly neutral when it inserts into an intergenic region or a nonessential gene, or beneficial to the host, through the gain of new phenotypes, such as antibiotic resistance or toxin production (Roberts and Mullany 2009; Cambray et al. 2010; Carle et al. 2010; Hua-Van et al. 2011).
Transposons are a major type of MGE that move from one location in the host genome to another. Transposons are naturally divided into two classes (Wicker et al. 2007; Kapitonov and Jurka 2008; Hua-Van et al. 2011; Piégu et al. 2015). Class I includes retrotransposons which transpose via an RNA intermediate that prior to integration is converted into the DNA form by the transposon-encoded reverse transcriptase. Class II consists of DNA transposons that are mobilized via the cut-and-paste mechanism, that is, excision of the transposon from its initial location and insertion into a new genomic locus. The excision and insertion are catalyzed by an element-encoded transposase (or by a transposase supplied by another element, in the case of nonautonomous transposons) and typically require terminal inverted repeats (TIR) that flank the transposon. Class II transposons show remarkable diversity with respect to the specific mechanisms of transposition, the identity of the transposase, the element size, and gene content (Jurka et al. 2007; Wicker et al. 2007; Hua-Van et al. 2011; Piégu et al. 2015). Most transposases belong to the DDE superfamily, named after the amino acid residues that form the catalytic triad (Rice and Baker 2001; Wicker et al. 2007; Hua-Van et al. 2011), but some transposons possess transposases homologous to the rolling-circle replication initiation endonucleases (Ilyina and Koonin 1992; Chandler et al. 2013; Krupovic 2013), phage integrase-like tyrosine recombinases (Goodwin et al. 2003; Goodwin and Poulter 2004), or serine integrases/invertases (Boocock and Rice 2013).
A distinct group of Class II elements that is widespread in diverse unicellular and multicellular eukaryotes includes large (15–20 kb), self-synthesizing DNA transposons, known as Mavericks or Polintons (Kapitonov and Jurka 2006; Feschotte and Pritham 2007; Pritham et al. 2007). These transposons encode their own protein-primed family B DNA polymerase that is implicated in the transposon replication (Kapitonov and Jurka 2006). In addition, Polintons encode several homologs of viral proteins that perform key functions in virion morphogenesis, namely the genome packaging ATPase and capsid maturation protease, and as recently shown, also major and minor capsid proteins (Krupovic et al. 2014a). Thus, these elements (that perhaps more appropriately could be denoted polintoviruses) combine features of viruses and transposons (Krupovic and Koonin 2015), although their virions and the conditions that trigger the predicted switch to the virus life style remain to be identified. Polintons have apparently played a key role in the emergence of several groups of eukaryotic DNA viruses and plasmids (Koonin et al. 2015; Krupovic and Koonin 2015).
Until recently, Polintons remained the only known (super)family of self-synthesizing transposons. However, in the course of an in depth investigation of bacterial and archaeal genomic neighborhoods that contain homologs of the cas1 gene (Makarova et al. 2014), a key component of the CRISPR-Cas adaptive immunity systems (Makarova et al. 2011, 2015), we identified a novel superfamily of predicted self-synthesizing transposons that we named casposons (Krupovic et al. 2014b). The homolog of the Cas1 protein encoded by these elements is predicted to function as a transposase. Indeed, the series of reactions that is catalyzed by the Cas1 protein during the adaptation (spacer acquisition) phase of the CRISPR-Cas response is closely analogous to transposition (Nuñez et al. 2014, 2015; Rollie et al. 2015). Recently, the integrase activity of the Cas1 homolog (also referred to as casposase) encoded by a casposon from the archaeon Aciduliprofundum boonei has been demonstrated experimentally (Hickman and Dyda 2015a). Notably, the integration was accompanied by generation of characteristic target site duplications (TSDs), fully consistent with our original prediction (Krupovic et al. 2014b).
The discovery of the casposons and phylogenetic analysis of Cas1 prompted an evolutionary scenario under which a casposon was the ancestor of the adaptation module of CRISPR-Cas systems, having contributed cas1 and possibly additional genes, whereas the TIR of that ancestral element gave rise to CRISPR repeats (Koonin and Krupovic 2015a). This scenario is closely parallel to the proposed scheme for the origin of the vertebrate adaptive immunity systems that involves a distinct family of Transib transposons (Kapitonov and Jurka 2005; Kapitonov and Koonin 2015), suggesting that in the evolutionary arms race between pathogens and hosts, transposons are regularly recruited as assault weapons for cellular defense (Koonin and Krupovic 2015b).
The casposons have been identified in a relatively small number of archaeal and only a handful of bacterial genomes. In the absence of direct experimental evidence, the original comparative genomic analysis provided no specific evidence that any of the casposons were active MGE. Here, we update the genomic census of the casposons and take advantage of the expanding collection of microbial genomes to analyze the distribution of these elements in 62 strains of the archaeon Methanosarcina mazei. The results of this analysis reveal recent mobility of the casposons and yield additional clues for the casposon involvement in the evolution of CRISPR-Cas.
Materials and Methods
All genome sequences were downloaded from the NCBI sequence database. For comprehensive identification of cas1 genes, the TBLASTN program with the E-value cutoff of 0.01 and low complexity filtering turned off (Altschul et al. 1997) was used to search the NCBI WGS (whole-genome short gun contigs) database using the Cas1 profile (Makarova and Koonin 2015) as the query. Protein sequences were searched against the nonredundant sequence database at the NCBI using PSI-BLAST (Altschul et al. 1997; Marchler-Bauer et al. 2013) and HHpred (Soding et al. 2006). Inverted and direct repeats flanking the casposons were analyzed using Unipro UGENE (Okonechnikov et al. 2012). Casposons were compared to each other and visualized using EasyFig (Sullivan et al. 2011). Multiple alignments of protein sequences were constructed using MUSCLE (Edgar 2004). Phylogenetic analysis was performed using the FastTree program with the WAG evolutionary model and the discrete gamma model with 20 rate categories (Price et al. 2010).
Results
Identification of New Casposons in Genomic Databases
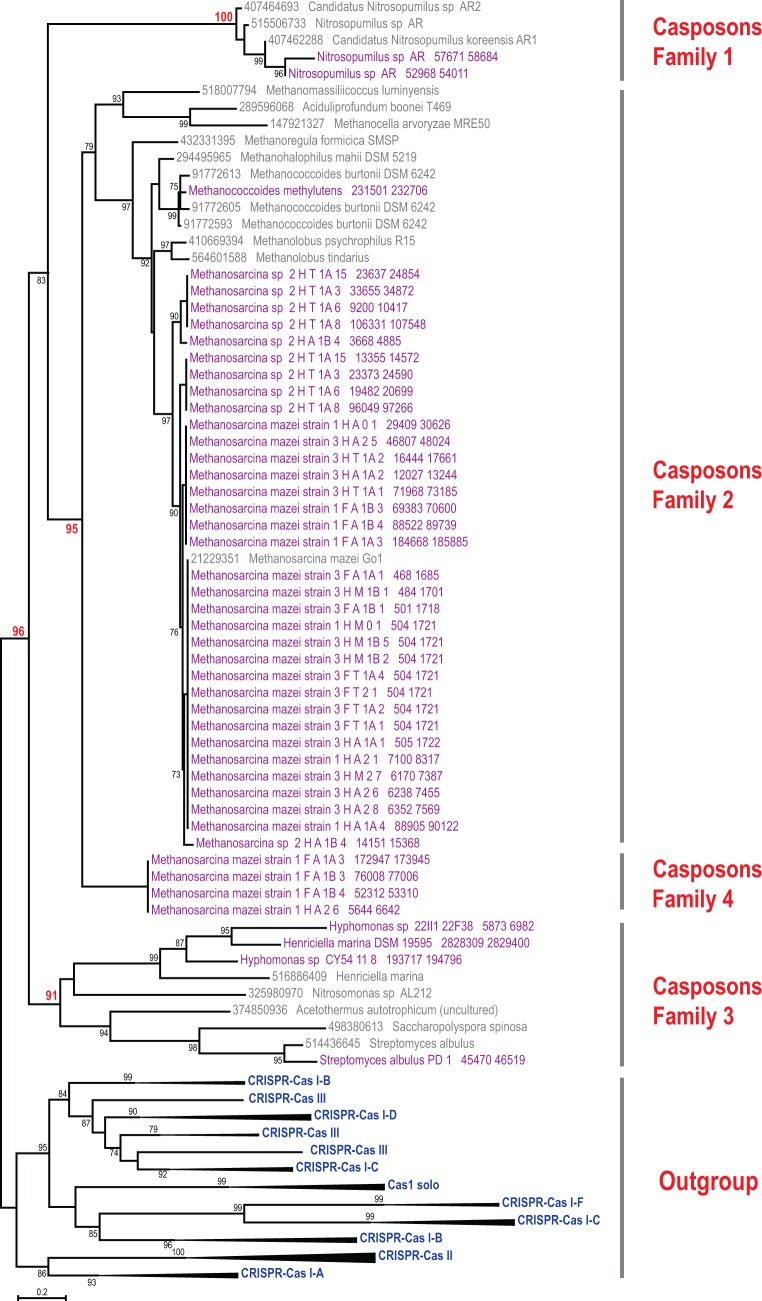
In order to further explore the properties and taxonomic distribution of casposons, we analyzed available genomic databases for the presence of genes coding for casposon-specific variant of Cas1 endonuclease. As a result, 52 new cas1 genes most similar to those present in previously identified casposons were identified (supplementary file S1, Supplementary Material online). Half of these genes were present within short genomic fragments or at the termini of genomic contigs, precluding delineation of the complete or near-complete casposons. The remaining 26 Cas1 proteins were encoded within genomic regions that exhibited all features of casposons, including TIR and, in all but two cases, TSD (supplementary file S2, Supplementary Material online). Phylogenetic analysis of the Cas1 proteins indicated that the newly identified elements represented all three previously defined casposon families and, in addition, revealed a new Cas1 clade which forms a sister group to Cas1 from family 2 casposons (fig. 1).
Fig. 1.—
Phylogenetic tree of Cas1. The maximum likelihood tree was constructed using FastTree from a multiple alignment of 116 Cas1 protein sequences, including 45 Cas1 sequences associated with newly identified casposons (highlighted in purple); 150 phylogenetically informative positions of this alignment was used for tree reconstruction. For the outgroup, 52 selected representatives of Cas1 associated with all known CRISPR-Cas systems (Makarova et al. 2015) were used; branches corresponding to each monophyletic group are collapsed and labeled. Casposon-derived Cas1 sequences are labeled with protein identification numbers and species names for the subset reported previously (Krupovic et al. 2014b) and by strain name and cas1 gene coordinates in the respective contig for the new ones (see also supplementary file S1, Supplementary Material online). The bootstrap support values are given as percentage points and are shown only for branches with >70% support; several key bootstrap values are highlighted in red. The complete tree in the Newick format and the underlying alignment are available as the supplementary file S5, Supplementary Material online.
The newly detected family 1 casposons include two elements, NitAR-C2 and NitAR-C3, from thaumarchaeon Nitrosopumilus sp. AR. These two elements are closely related to the previously described casposons of Nitrosopumilus, in particular NitAR1-C1 of Candidatus Nitrosopumilus koreensis AR1, but differ considerably from NitAR-C1 which we have previously identified in the genome of Nitrosopumilus sp. AR (Krupovic et al. 2014b). Thus, Nitrosopumilus sp. AR appears to contain three distinct family 1 casposons. The only other known organism which contains three (family 2) casposons is Methanococcoides burtonii DSM 6242 (Krupovic et al. 2014b). However, in Methanoc. burtonii, the casposons lack TIRs and TSDs and some of the core genes are fragmented, suggesting that these elements are inactive. In contrast, all three casposons of Nitrosopumilus sp. AR are flanked by TIRs and contain apparently intact genes, suggesting that these are functional elements.
The majority of the new casposons (20) belong to family 2, the most abundant of the three originally defined families that is dominated by casposons from methanogenic archaea (Krupovic et al. 2014b). All new family 2 elements are from different members of the euryarchaeal order Methanosarcinales (supplementary file S2, Supplementary Material online). Family 3 includes bacterial casposons; four new representatives of this family were identified in the genomes of Henriciella marina DSM 19595, Hyphomonas sp. CY54-11-8, Streptomyces albulus PD-1, and Citromicrobium bathyomarinum JL354. The latter bacterium belongs to the order Sphingomonadales (class Alphaproteobacteria), which until now has not been known to carry casposons.
Besides the Cas1 endonuclease, all new casposons contain other conserved genes characteristic of their respective families. Family 1 casposons encode protein-primed family B DNA polymerases (PolB), whereas those of families 2 and 3 contain genes for RNA-primed PolBs (Krupovic et al. 2014b). In addition, all family 2 casposons encompass a conserved set of genes encoding two helix-turn-helix proteins and an HNH endonuclease. Other notable functions encoded by nonconserved casposon genes include bacterial retrotransposon/retron-like reverse transcriptase (cd03487; Met2HT1A3-C1); ATP-dependent 26S proteasome regulatory subunit (CitBat-C1); transglutaminase (Met2HT1A3-C2); SGNH hydrolase, adenylyltransferase and glycosyltransferase (HenMar-C2); ParB/RepB/Spo0J family protein involved in plasmid partitioning and a serine resolvase (HypCY54-C1); AbiF-like abortive infection system protein (Met2HA1B4-C2); RES (Met2HT1A3-C1) and HEPN (Met2HA1B4-C1) domain proteins; tetratricopeptide (MetMaz1FA1A3-C1); and pentapeptide (MetMazS6-C1) repeat proteins. These functions further expand the already rich pangenome of the casposons. However, the roles of these genes in the propagation of casposons remain enigmatic.
Integration Target Sites
According to the proposed mechanism, Cas1-mediated casposon integration (referred to as casposition) results in staggered cut within the target site. Subsequently, the single-stranded overhangs are fill-in repaired resulting in duplication of the target site. Identification of the TSDs thus helps to pinpoint the location within the genome that was recognized and cut by the Cas1 endonuclease. Analysis of the TSD locations showed that, consistent with previous results, casposon integration targets can be broadly categorized into three groups: 1) intergenic regions, 2) tRNA genes, and 3) a protein-coding gene.
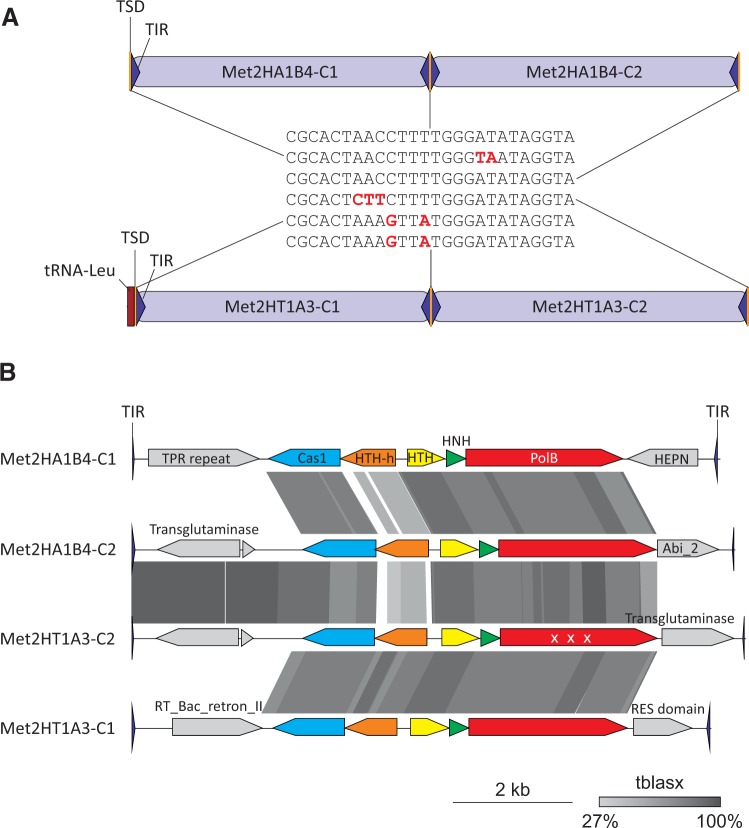
All bacterial casposons (family 3) are inserted within intergenic regions. Thaumarchaeal casposon NitAR-C2 (family 1) has targeted the 3′-distal region of the gene encoding translation elongation factor aEF-2, as previously observed for other thaumarchaeal casposons (Krupovic et al. 2014b). In contrast, family 2 casposons are integrated either into intergenic regions or into the 3’-distal region of tRNA genes. Two nonorthologous tRNA-Leu genes are targeted in different species of Methanosarcina, one in various strains of M. mazei and the other in Methanosarcina sp. 2.H.T.1A.15, 2.H.T.1A.3, 2.H.T.1A.6 and 2.H.T.1A.8 (the four strains contain identical casposons; supplementary file S2, Supplementary Material online). Notably, Methanosarcina sp. 2.H.T.1A.15, 2.H.T.1A.3, 2.H.T.1A.6, and 2.H.T.1A.8 contain two closely related casposon copies that are located adjacent to each other and are separated only by a shared TSD sequence (fig. 2). This arrangement suggests that the same target site was utilized twice, resulting in an array of tandemly integrated elements. In all cases, one of the two adjacent copies was apparently inactivated based on the fact that the gene encoding for DNA polymerase contained multiple internal stop codons. Such repeated integration into the same target site (out of all possible positions in the genome) suggests that casposition occurs in a sequence-specific manner, as opposed to random target selection. This apparent specificity is reminiscent of the transposition of bacterial transposon Tn7 where multiple elements, some of which are inactivated, accumulate within the same locus, leading to the formation of genomic islands (Parks and Peters 2009).
Fig. 2.—
Comparison of tandemly integrated casposons in different Methanosarcina species. (A) Schematic representation of the genomic loci within Methanosarcina sp. 2.H.A.1B.4 (top) and Methanosarcina sp. 2.H.T.1A.3 (bottom) containing casposons integrated into the intergenic region and tRNA-Leu gene, respectively. TIR, TSD, and tRNA gene are depicted with blue triangles, yellow bars, and a red rectangle, respectively. The sequences of the corresponding TSDs are shown in the middle and substitutions with respect to the top sequence are highlighted in red. (B) Comparison of the genome maps of casposon depicted in panel A. Pairwise tBLASTx hits between casposons are indicated by different shades of gray (the identity scale is included). “X X X” indicates that the gene is fragmented in Met2HT1A3-C2. TPR, tetratricopeptide; HTH-h, helix-turn-helix protein with a C-terminal HEAT repeat domain; RT_Bac_retron_II, retrotransposon/retron-like reverse transcriptase; PolB, family B DNA polymerase; HNH, HNH endonuclease.
Methanosarcina sp. 2.H.A.1B.4 also contains two tandemly integrated casposons (Met2HA1B4-C1 and Met2HA1B4-C2). The two elements, both of which appear to be intact, are related to each other as well as to the casposons found in Methanosarcina sp. 2.H.T.1A.3, 2.H.T.1A.15, 2.H.T.1A.6, and 2.H.T.1A.8. Both groups of elements are flanked by nearly identical TSDs (fig. 2). However, in the 2.H.A.1B.4 strain, the casposons are integrated not into the tRNA-Leu gene but into an intergenic region within a different genomic locus, strongly suggesting that in Methanosarcina casposons have been mobilized relatively recently. In the Cas1 phylogeny, casposons of 2.H.A.1B.4 that are integrated within the intergenic region are basal with respect to those within tRNA-Leu genes (fig. 1). Further evidence of casposon mobility was obtained in the course of detailed comparative genome analysis of multiple M. mazei strains as described below.
Casposons of M. mazei
We have previously identified one casposon, MetMaz-C1, in M. mazei Go1, which was integrated within an intergenic region (Krupovic et al. 2014b). Recently, a large collection of M. mazei strains isolated from a Columbia River sediment has been sequenced (56 isolates, averaging <1% nucleotide divergence) (Youngblut et al. 2015), providing a particularly useful resource for investigating potential (recent) mobility of integrated elements. This collection was complemented with the six previously sequenced M. mazei genomes (SarPi, Lyc, Go1, S-6, WWM610, C16), resulting in a final genomic dataset from 62 closely related strains. Our analysis shows that M. mazei strains other than Go1 also contain full-length casposons some of which are closely related or identical to MetMaz-C1 (supplementary file S2, Supplementary Material online). Interestingly, however, these elements are inserted not only into the intergenic region corresponding to the one occupied by MetMaz-C1 but also into other genomic loci, and some strains contain more than one casposon. However, none of the latter strains displayed tandem insertions as in the case of Methanosarcina spp. described above (fig. 2).
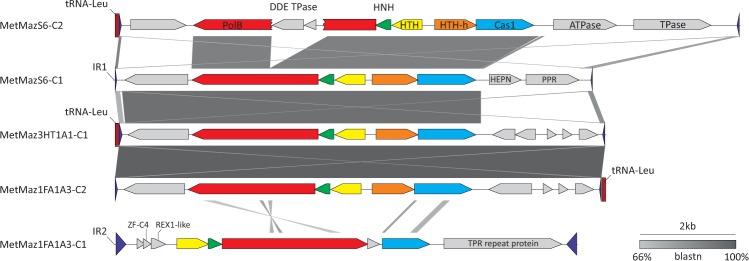
Three distinct casposon-containing loci were identified. In two of these, casposons were inserted into intergenic regions, whereas in the third locus, the integration target was within a tRNA-Leu gene (see supplementary file S2, Supplementary Material online, for precise coordinates). The intergenic region containing MetMaz-C1 in M. mazei Go1 is denoted as IR1, whereas the other one is referred to as IR2. The casposons inserted into IR1 and tRNA-Leu genes are closely related (see below). In contrast, the casposons within IR2 differed considerably in both sequence and the arrangement of the core casposon genes (figs. 1 and 3). Notably, in phylogenetic analysis of Cas1, M. mazei casposons integrated within IR2 form a sister group to all other known family 2 casposons (fig. 1). Furthermore, these elements, exemplified by MetMaz1FA1A3-C1 in figure 3, do not contain one of the family 2 core genes encoding a helix-turn-helix protein with a C-terminal HEAT repeat domain. Instead, they encode a Zn-finger protein, a DNA repair REX1-like protein (PF14966; HHpred, P = 96) and a large (827 aa), tetratricopeptide repeat-containing protein. Another feature that sets apart casposons residing within IR2 from those in IR1 and tRNA-Leu gene is their TIRs, which are considerably longer (225 bp) than those of other M. mazei casposons (31–59 bp) (supplementary file S4, Supplementary Material online). On the basis of the phylogenetic analysis of Cas1 proteins as well as distinct genomic features described above, we classify MetMaz1FA1A3-C1-like casposons into a new family 4 (fig. 1).
Fig. 3.—
Comparison of casposons integrated into the three different genomic loci of M. mazei. TIR and tRNA genes are depicted with blue triangles and red rectangle, respectively. Pairwise BLASTn hits between the casposons are indicated by different shades of gray (the identity scale is included). TPR, tetratricopeptide; PolB, family B DNA polymerase; HNH, HNH endonuclease; DDE TPase, transposase of the DDE superfamily (named after two aspartate and one glutamate residues that form the catalytic triad of these enzymes); HTH-h, helix-turn-helix protein with a C-terminal HEAT repeat domain; PPR, pentapeptide repeat.
Casposon Mobility in M. mazei
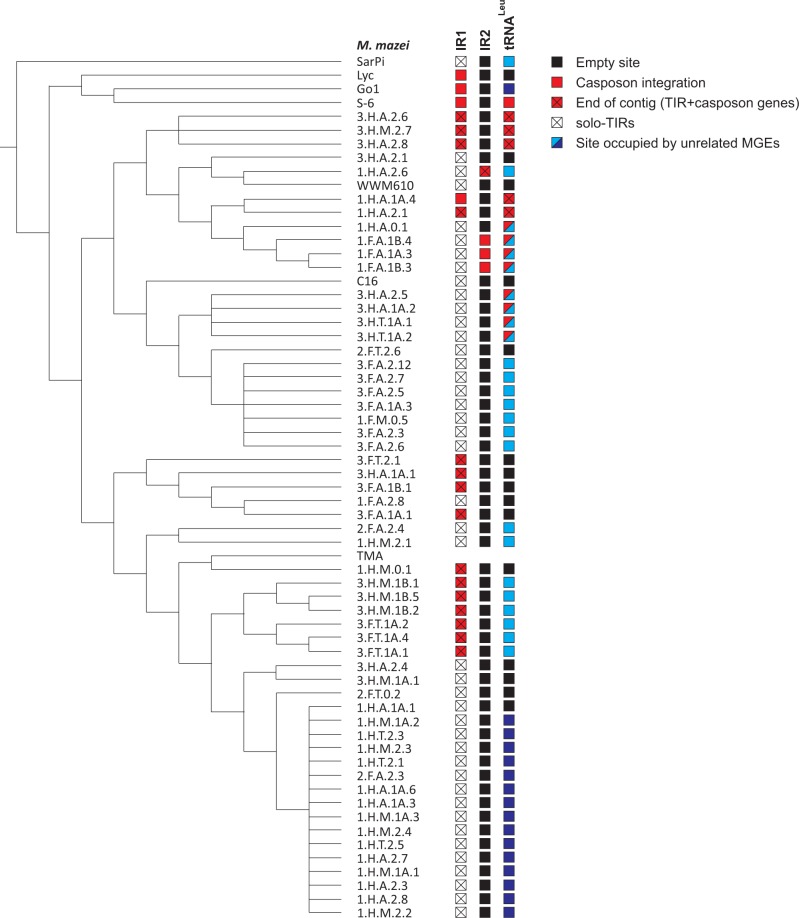
The results of the systematic analysis of the three loci across all 62 strains were projected on the previously reported M. mazei core gene phylogeny (Youngblut et al. 2015), revealing a complex evolutionary history of casposons in M. mazei species (fig. 3). Strikingly, all analyzed M. mazei strains display a genomic record of casposon encounter, even if not all of these strains currently contain full-length casposons. Nevertheless, of the 62 strains, nearly half (28) harbor casposons in at least one of the three insertion loci. Typically, these elements are found at the extremities of the genomic contigs and are split between two contigs, suggesting that casposons are difficult to assemble, as is often the case with integrated, repetitive elements (Tang 2007). In 43 strains that did not contain casposons within the IR1 region, we could identify remnants of the casposons in this region, that is, sequences matching one of the TIRs, hereinafter referred to as solo-TIRs (fig. 4; supplementary file S3, Supplementary Material online). Notably, M. mazei SarPi, which occupies the basal position in the M. mazei phylogenetic tree (fig. 4), contains three solo-TIRs within different genomic loci, including IR1, but not a single full-length casposon. Similar remnants of transposable elements (e.g., solo-LTRs [long terminal repeats] of retroelements) are frequently found in various genomes, where they are at least as abundant but typically outnumber the complete elements from which they are derived (Shirasu et al. 2000; Devos et al. 2002; Yin et al. 2014). There was no segregation of complete casposons and solo-TIRs within the M. mazei tree (fig. 4), suggesting that the solo-TIRs have emerged by degradation of full-length casposons on multiple occasions during the evolution of M. mazei strains.
Fig. 4.—
Distribution of casposons and integrated MGEs within 62 M. mazei strains. The cladogram is based on the M. mazei core gene phylogeny reported by Youngblut et al. (2015). The genome sequence of the strain TMA was not publicly available; thus, the information on casposon in this strain is lacking. IR1 (intergenic region 1), IR2, and tRNA-Leu genes correspond to the three sites targeted by casposons. Empty sites are indicated with black boxes; sites containing casposons are denoted with red boxes, whereas red boxes that are crossed indicate that TIR sequence and several casposon genes were identified but casposon is incomplete due to termination of the genomic contig (notably, the rest of casposon genes and the second TIR could be typically found on other contigs); open crossed boxes show that only solo-TIRs, without any casposons genes, were identified within the corresponding target site. The two groups of MGEs integrating using tyrosine recombinases are indicated by light and dark blue boxes, respectively.
In contrast to the ubiquity of casposon integrations within IR1, family 4 elements residing within IR2 are represented in only four M. mazei strains, whereas the rest of the strains contain intact, empty target sites. This observation, along with the finding that the latter elements are only distantly related to the other M. mazei casposons (figs. 1 and 3), suggests that they have been introduced into M. mazei relatively recently, following the divergence of the major M. mazei clades (fig. 4).
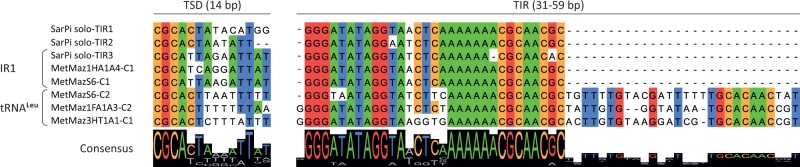
Casposons within tRNA-Leu genes were identified in 14 M. mazei strains. Six of these, in addition, contained casposons within the IR1 locus (fig. 4). The elements inserted into the tRNA and IR1 sites are closely related (fig. 3), suggestive of casposon amplification and mobility in these strains. To verify the latter possibility, we compared all nonredundant TIR and TSD sequences of casposons from the two loci (fig. 5). We found that casposons present in IR1 and tRNA-Leu genes display conservation not only within their TIR but also in the TSD sequences. In both cases, the TSDs are 14-bp long and are perfectly conserved within the four 5’-terminal positions, whereas the five 3’-terminal positions are always AT-rich. The six central nucleotides are more variable, suggesting that target selection is predominantly determined by the terminal nucleotides. Notably, all three solo-TIRs in the SarPi strain are also adjacent to sequences containing the conserved CGCA motif, in accord with the conservation patterns of the target site (fig. 5).
Fig. 5.—
Comparison of the TSD and TIR sequences from casposons integrated into different genomic loci.
Considering that all M. mazei strains, including the basal ones, contain casposons or remnants thereof within the IR1 locus, it appears highly likely that this was the ancestral site of casposon insertion, whereas the considerably less abundant elements inserted into the tRNA-Leu genes are derived from those inserted into IR1. Furthermore, phylogenetic relationship between the casposon-containing strains is best consistent with several independent casposition events (fig. 4). This possibility is further supported by the observation that casposons integrated into the tRNA-Leu genes in different strains are present in two alternative orientations with respect to the target site. Specifically, MetMaz1FA1A3-C2 is inverted when compared to MetMaz3HT1A1-C1 (note that in fig. 3 the tRNA genes are located on the opposite sides of the two elements). Such variation could result from either independent insertion of casposons in the two strains or inversion of the casposon in one of the strains by intramolecular recombination between the TIRs. The fact that the two strains belong to different M. mazei clades (fig. 4) better supports the independent insertion scenario.
Cohabitation of Casposons and Unrelated Mobile Elements within the Same Target Sites
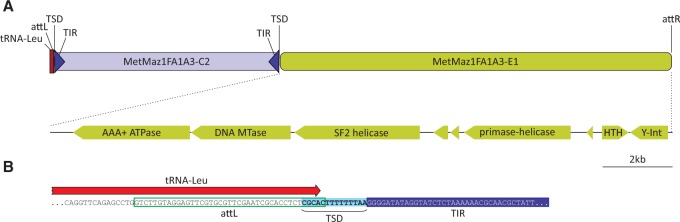
Genomic context analysis showed that casposons are not the only mobile elements that integrate into the tRNA-Leu gene of M. mazei. The same tRNA gene was targeted by two other groups of MGEs, MGE1 and MGE2, in different M. mazei strains. Elements from both these groups are not closely related to previously described archaeal viruses or plasmids but encode typical integrases of the tyrosine recombinase superfamily (Grindley et al. 2006) and appear to have recombined site-specifically with the 3’-distal region of the tRNA-Leu gene. The latter process involves recombination between homologous regions, known as attachment sites (att), that are present on the circular dsDNA molecule of the mobile element and the cellular genome. As a result, the mobile element is inserted site specifically into the host chromosome and is flanked by direct repeats corresponding to the att sites (attL and attR) (Grindley et al. 2006; Krupovic and Forterre 2015). Interestingly, elements from one of the two groups (MGE1) co-occur with and are adjacent to casposons in some of the M. mazei strains (fig. 3). The corresponding locus in M. mazei strain 1.F.A.1A.3 contains the casposon MetMaz1FA1A3-C2 and the integrated element MetMaz1FA1A3-E1 (fig. 6). Gene content analysis of MetMaz1FA1A3-E1 shows that its gene complement is typical of mobile elements. Specifically, besides the tyrosine recombinase, MetMaz1FA1A3-E1 encodes several proteins that are likely to be involved in its replication, including an AAA+ ATPase, superfamily 2 helicase and a primase-helicase fusion protein (fig. 6A). The latter contains the N-terminal primpol domain homologous to the small subunit of archaeo-eukaryotic primases and the C-terminal superfamily 3 helicase domain. The two domains, either separately or in combination, are frequently found in various plasmids and viruses from all three cellular domains (Iyer et al. 2005; Lipps 2011; Krupovic et al. 2013; Gill et al. 2014).
Fig. 6.—
Co-integration of a casposon and an unrelated MGE into the same tRNA-Leu gene. (A) Schematic representation of the genomic locus of Methanosarcina mazei strain 1.F.A.1B.3, containing casposon MetMaz1FA1A3-C2 and integrating element MetMaz1FA1A3-E1. The genome map of the latter is also shown with open reading frames being shown as arrows, indicating the direction of transcription. attL and attR, left and right attachment sites, respectively; MTase, methyltransferase; SF2, superfamily 2; Y-Int, integrase of the tyrosine recombinase superfamily. (B) Detailed view of the tRNA-Leu gene (red arrow) region encompassing the attL (green outline) and TSD (light blue shading) sequences as well as a fragment of TIR (dark blue shading).
Detailed analysis of the recombination signals (att, TIR, TSD) of the two adjacent elements from the 1.F.A.1A.3 strain (fig. 6B) showed that the site targeted by the casposon partially overlaps the attachment site employed by MetMaz1FA1A3-E1. Importantly, the positions of the two elements with respect to each other and the target tRNA gene indicates that casposon insertion in this strain occurred subsequent to the site-specific integration of MetMaz1FA1A3-E1. Consistent with this assessment is the finding that some of the M. mazei strains, in particular the basal SarPi, contain the integrated MGE1 but not the casposon (fig. 4).
Discussion
Casposons represent one of the most recent additions to the vastly diverse repertoire of transposons and other integrative MGEs. Here, we describe new members of this transposon superfamily from a range of bacterial and archaeal genomes and introduce a new family of casposons. These elements not only expand the taxonomic distribution of casposons, in particular to the bacterial order Sphingomonadales, but more importantly, for the first time, provide evidence of casposons mobility. Furthermore, the identification of a new casposon family implies that the genetic diversity of these elements remains largely unexplored, and many additional groups of casposons are likely to be discovered in the future.
We took advantage of the large collection of M. mazei strains for which genomic data have become available recently (Youngblut et al. 2015). Systematic analysis of the 62 sequenced strains led to the identification of three distinct genomic loci targeted by casposons, with some of the strains containing more than one casposon. Sequence conservation between casposons integrated into distinct loci of the same genome and homology between the utilized target sites strongly suggest that the elements have been active in the recent history of M. mazei species and that casposition is, at least to some extent, sequence specific in vivo. The latter feature is rather unusual among typical transposons but is characteristic of MGEs that utilize tyrosine or serine recombinases to promote homologous recombination between the element (most commonly plasmid or viral DNA) and the host chromosome (Grindley et al. 2006). Nevertheless, mechanistically, casposition is likely to mirror the integration mediated by various transposases (Hickman and Dyda 2015b). In this mechanism, linear ends of the casposon would be joined into the target site through two consecutive transesterification reactions, followed by fill-in repair of the single-stranded overhangs resulting from a staggered cut within the target site which produces the characteristic TSDs flanking the casposon. Notably, support for this succession of events comes from recent biochemical studies on spacer acquisition by CRISPR-associated Cas1 endonucleases which show that Cas1 can catalyze both the integration and the reverse, disintegration reactions similar to those of retroviral integrases and other DDE transposases (Nuñez et al. 2014, 2015; Rollie et al. 2015). Consistent with the analysis of casposon integration described here, spacer acquisition by Cas1 has been found to be sequence-specific (Nuñez et al. 2015; Rollie et al. 2015). In contrast, in vitro integration of oligonucleotides or mini-casposons into the target DNA catalyzed by the purified casposase from Aciduliprofundum boonei appears to occur at random sites (Hickman and Dyda 2015a). This discrepancy awaits further investigation. It seems a distinct possibility that the in vivo specificity of the casposase is conferred by an additional casposon-encoded protein.
At the first glance, it might appear surprising how much the mechanism of spacer integration mediated by CRISPR-associated Cas1 resembles the transposition reaction catalyzed by DDE transposases. However, considering that the original function of Cas1, in all likelihood, is mobilization of genetic elements, specifically casposons, all the parallels between Cas1 and transposases become only natural. The key feature of Cas1 proteins, from both casposons and CRISPR-Cas systems, is the sequence dependence of their nuclease activity. Conceivably, it is this sequence specificity that rendered these enzymes suitable for controlled genome remodeling, eventually leading to the emergence of prokaryotic adaptive CRISPR-Cas immunity (Koonin and Krupovic 2015a, 2015b). There is, however, a striking duality in Cas1 specificity toward DNA substrates. On the one hand, acquisition of new spacers from the invading DNA merely depends on the presence of a di- to pentanucleotide, known as the protospacer adjacent motif (PAM), without any discernible sequence requirements within the protospacer itself (Mojica et al. 2009; Shah et al. 2013; van der Oost et al. 2014). On the other hand, spacer insertion into the CRISPR array occurs at a strictly predefined site at the Leader-end of the CRISPR locus (van der Oost et al. 2014). How this dualism in specificity has emerged is one of the least clear steps in the evolutionary scenario of the origin of CRISPR-Cas systems from casposons, due mainly to the current lack of understanding of the exact mechanistic details underlying casposon integration and excision. However, the observations presented here might hold a clue to this riddle.
On the basis of the sequence and secondary structure similarity between the TIRs of certain casposons and CRISPR, we have recently proposed that CRISPR arrays have evolved from solo-TIRs, in parallel to the evolution of recombination signal sequences utilized in the eukaryotic V(D)J recombination from the TIRs of Transib transposons (Kapitonov and Jurka 2005; Koonin and Krupovic 2015a). However, until now, casposon-derived solo-TIRs have not been observed in any bacterial or archaeal genomes. Here, we show that all M. mazei strains that do not contain full-length casposons, carry solo-TIRs in one of the genomic loci (fig. 4), indicating that such sequences can and do emerge repeatedly in the course of evolution. Furthermore, we found that closely related casposons from different loci target homologous sites in M. mazei genome. However, strict sequence conservation within the target sequences is limited to the tetranucleotide CGCA. We propose that this conserved target sequence is the counterpart of the PAM sequences recognized by the CRISPR-Cas systems, whereas the rest of the target site sequence is equivalent to the protospacer. Then, the parallel between the activities of Cas1 in CRISPR-Cas and casposons becomes increasingly transparent. In the CRISPR-Cas systems, the protospacer is selected based on a short PAM motif which, by definition, is adjacent to the protospacer sequence. Similarly, in the case of casposons, the target sequence appears to be defined by the presence of a tetranucleotide motif which is found at the extremity of the processed sequence. In contrast, integration of the protospacer into the CRISPR locus is highly specific, being determined by the nucleotides flanking the integration site at the Leader-repeat 1 boundary (Rollie et al. 2015). In the case of casposons, TIRs can be predicted to contain the Cas1-binding sites which would ensure the specificity of integration into the target sites, as is the case in other transposon systems (Hickman and Dyda 2015b). Importantly, there is apparent coevolution between Cas1 endonucleases and the sequences of the CRISPR and PAMs in CRISPR-Cas systems (Shah et al. 2013). Similarly, the sequences of TIRs and TSDs are casposon specific and seem to vary for distinct casposons, even those present in the same host.
The lack of strict sequence conservation within the M. mazei TSDs (beyond the invariant tetranucleotide) is likely to allow for a degree of flexibility in casposon target selection. It has been suggested that the length of the casposon target site is dictated by the distance between and the orientation of the catalytic sites of the putative multimeric Cas1 caspososome (Hickman and Dyda 2014). Although the length of the TSDs is indeed conserved among closely related casposons (fig. 4), it can vary considerably in distantly related ones. For example, the length of the TSDs of related casposons integrated into IR1 and the tRNA-Leu gene in M. mazei is 14 nucleotides, whereas the TSDs flanking the distantly related casposon inserted into the IR2 site of the same M. mazei strain are 35-nucleotide long (supplementary file S2, Supplementary Material online). Thus, the length of the TSD appears to be casposon specific, and the mechanism of its determination remains to be characterized. Similarly, the length of TIRs varies substantially between different casposons (Krupovic et al. 2014b), and the same is true for CRISPR repeats and spacers: the size of the repeat can vary between 24 and 47 bp, whereas that of the spacer between 26 and 72 bp (Sorek et al. 2008).
We have previously observed a patchy taxonomic distribution of casposons in archaeal and bacterial genomes (Krupovic et al. 2014b). Here, we show that the same holds true at short evolutionary distances, namely at the level of strains of the same archaeal species. This observation is clearly indicative of recent casposon mobility. Phylogenetic analysis of the casposon PolB proteins has suggested that casposons emerged in archaea and were horizontally transferred to bacteria subsequent to the divergence of the casposon families 1 and 2 (Krupovic et al. 2014b). However, given that none of the currently identified casposons encodes viral capsid proteins or other proteins that could facilitate intercellular transfer (e.g., conjugative apparatus), the mechanism of horizontal transfer of casposons between different organisms especially archaea and bacteria, remains unclear (apart from passive transfer following host lysis). In this context, the finding that casposons occasionally integrate into the genomes of other MGEs, as is the case in some M. mazei strains, might be indicative of a transfer route. Notably, in the case depicted in figure 6A, homologous recombination mediated by the element-encoded tyrosine recombinase between the attL and attR sites would lead to excision of a circular chimeric DNA molecule encompassing both the original element and the casposon. Such excision would potentially liberate the strain from the resident casposon. Indeed, several M. mazei strains are devoid of either element in the tRNA-Leu gene (fig. 4). Conversely, transient integration of casposons into other MGEs might provide the means for their intercellular transfer, possibly over large phylogenetic distances.
Conclusions
Casposons remain to be experimentally characterized although the recent demonstration of the casposase activity in vitro provides an important validation of the predictions made by sequence analysis. In anticipation of further experiments, comparative genomics, and especially comparative analysis of multiple, closely related genomes of the same microbial species, can provide substantial clues into the biology of these elements. Here, we demonstrate striking variation of the casposon insertion sites among 62 strains of M. mazei and insertion of closely related casposons into different sites. These findings leave no doubt in the recent mobilization of casposons. The presence of solo casposon-derived TIRs in many M. mazei strains shows that processes similar to those that likely gave rise to the CRISPR-Cas systems are rather common during evolution. We also demonstrate the sequence specificity of casposon insertion sites and Cas1 recognition sites in the TIRs that parallel the specificities observed at different steps of adaptation during the CRISPR-Cas response. Collectively, the observations presented here add credence to the key role of casposons in the emergence of CRISPR-Cas.
Supplementary Material
Supplementary files S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
P.F. was supported by the European Union's Seventh Framework Program (FP/2007-2013)/Project EVOMOBIL - ERC Grant Agreement no. 340440. E.V.K. and K.S.M. are supported by intramural funds of the US Department of Health and Human Services (to the National Library of Medicine).
Literature Cited
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boocock MR, Rice PA. 2013. A proposed mechanism for IS607-family serine transposases. Mob DNA. 4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambray G, Guerout AM, Mazel D. 2010. Integrons. Annu Rev Genet. 44:141–166. [DOI] [PubMed] [Google Scholar]
- Carle P, et al. 2010. Partial chromosome sequence of Spiroplasma citri reveals extensive viral invasion and important gene decay. Appl Environ Microbiol. 76:3420–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 49:277–300. [DOI] [PubMed] [Google Scholar]
- Chandler M, et al. 2013. Breaking and joining single-stranded DNA: the HUH endonuclease superfamily. Nat Rev Microbiol. 11:525–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Brown JK, Bennetzen JL. 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12:1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. 2007. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 41:331–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S, et al. 2014. A highly divergent archaeo-eukaryotic primase from the Thermococcus nautilus plasmid, pTN2. Nucleic Acids Res. 42:3707–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin TJ, Butler MI, Poulter RT. 2003. Cryptons: a group of tyrosine-recombinase-encoding DNA transposons from pathogenic fungi. Microbiology 149:3099–3109. [DOI] [PubMed] [Google Scholar]
- Goodwin TJ, Poulter RT. 2004. A new group of tyrosine recombinase-encoding retrotransposons. Mol Biol Evol. 21:746–759. [DOI] [PubMed] [Google Scholar]
- Grindley ND, Whiteson KL, Rice PA. 2006. Mechanisms of site-specific recombination. Annu Rev Biochem. 75:567–605. [DOI] [PubMed] [Google Scholar]
- Hickman AB, Dyda F. 2014. CRISPR-Cas immunity and mobile DNA: a new superfamily of DNA transposons encoding a Cas1 endonuclease. Mob DNA. 5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman AB, Dyda F. 2015a. The casposon-encoded Cas1 protein from Aciduliprofundum boonei is a DNA integrase that generates target site duplications. Nucleic Acids Res. 43:10576–10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman AB, Dyda F. 2015b. Mechanisms of DNA transposition. Microbiol Spectr. 3:MDNA3-0034-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua-Van A, Le Rouzic A, Boutin TS, Filee J, Capy P. 2011. The struggle for life of the genome's selfish architects. Biol Direct. 6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyina TV, Koonin EV. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20:3279–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Leipe DD, Aravind L. 2005. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 33:3875–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Kohany O, Jurka MV. 2007. Repetitive sequences in complex genomes: structure and evolution. Annu Rev Genomics Hum Genet. 8:241–259. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. 2005. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 3:e181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. 2006. Self-synthesizing DNA transposons in eukaryotes. Proc Natl Acad Sci U S A. 103:4540–4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. 2008. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 9:411–412. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Koonin EV. 2015. Evolution of the RAG1-RAG2 locus: both proteins came from the same transposon. Biol Direct. 10:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr. 2004. Mobile elements: drivers of genome evolution. Science 303:1626–1632. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Dolja VV, Krupovic M. 2015. Origins and evolution of viruses of eukaryotes: the ultimate modularity. Virology 479480:2–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Krupovic M. 2015a. Evolution of adaptive immunity from transposable elements combined with innate immune systems. Nat Rev Genet. 16:184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Krupovic M. 2015b. A movable defense. Scientist 29:46–53. [Google Scholar]
- Krupovic M. 2013. Networks of evolutionary interactions underlying the polyphyletic origin of ssDNA viruses. Curr Opin Virol. 3:578–586. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Bamford DH, Koonin EV. 2014a. Conservation of major and minor jelly-roll capsid proteins in Polinton (Maverick) transposons suggests that they are bona fide viruses. Biol Direct. 9:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Forterre P. 2015. Single-stranded DNA viruses employ a variety of mechanisms for integration into host genomes. Ann N Y Acad Sci. 1341:41–53. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Gonnet M, Hania WB, Forterre P, Erauso G. 2013. Insights into dynamics of mobile genetic elements in hyperthermophilic environments from five new Thermococcus plasmids. PloS One 8:e49044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Koonin EV. 2015. Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution. Nat Rev Microbiol. 13:105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Makarova KS, Forterre P, Prangishvili D, Koonin EV. 2014b. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 12:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps G. 2011. Structure and function of the primase domain of the replication protein from the archaeal plasmid pRN1. Biochem Soc Trans. 39:104–106. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Koonin EV. 2015. Annotation and classification of CRISPR-Cas systems. Methods Mol Biol. 1311:47–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, et al. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 9:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, et al. 2014. Dark matter in archaeal genomes: a rich source of novel mobile elements, defense systems and secretory complexes. Extremophiles 18:877–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, et al. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. 2013. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41:D348–D352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155:733–740. [DOI] [PubMed] [Google Scholar]
- Nuñez JK, et al. 2014. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol. 21:528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez JK, Lee AS, Engelman A, Doudna JA. 2015. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 519:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K, Golosova O, Fursov M. 2012. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28:1166–1167. [DOI] [PubMed] [Google Scholar]
- Parks AR, Peters JE. 2009. Tn7 elements: engendering diversity from chromosomes to episomes. Plasmid 61:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piégu B, Bire S, Arensburger P, Bigot Y. 2015. A survey of transposable element classification systems—a call for a fundamental update to meet the challenge of their diversity and complexity. Mol Phylogenet Evol. 86:90–109. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritham EJ, Putliwala T, Feschotte C. 2007. Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene 390:3–17. [DOI] [PubMed] [Google Scholar]
- Rice PA, Baker TA. 2001. Comparative architecture of transposase and integrase complexes. Nat Struct Biol. 8:302–307. [DOI] [PubMed] [Google Scholar]
- Roberts AP, Mullany P. 2009. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 17:251–258. [DOI] [PubMed] [Google Scholar]
- Rollie C, Schneider S, Brinkmann AS, Bolt EL, White MF. 2015. Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. Elife 4:e08716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SA, Erdmann S, Mojica FJ, Garrett RA. 2013. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 10:891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P. 2000. A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 10:908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J, Remmert M, Biegert A, Lupas AN. 2006. HHsenser: exhaustive transitive profile search using HMM-HMM comparison. Nucleic Acids Res. 34:W374–W378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 6:181–186. [DOI] [PubMed] [Google Scholar]
- Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. 2007. Genome assembly, rearrangement, and repeats. Chem Rev. 107:3391–3406. [DOI] [PubMed] [Google Scholar]
- Tollis M, Boissinot S. 2012. The evolutionary dynamics of transposable elements in eukaryote genomes. Genome Dyn. 7:68–91. [DOI] [PubMed] [Google Scholar]
- van der Oost J, Westra ER, Jackson RN, Wiedenheft B. 2014. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 12:479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venner S, Feschotte C, Biemont C. 2009. Dynamics of transposable elements: towards a community ecology of the genome. Trends Genet. 25:317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 8:973–982. [DOI] [PubMed] [Google Scholar]
- Yin H, et al. 2014. Comparative genomic analysis reveals multiple long terminal repeats, lineage-specific amplification, and frequent interelement recombination for Cassandra retrotransposon in pear (Pyrus bretschneideri Rehd.). Genome Biol Evol. 6:1423–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblut ND, et al. 2015. Genomic and phenotypic differentiation among Methanosarcina mazei populations from Columbia River sediment. ISME J. 9:2191–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.