Abstract
Transposable elements (TEs) are mobile genetic elements with the ability to replicate themselves throughout the host genome. In some taxa TEs reach copy numbers in hundreds of thousands and can occupy more than half of the genome. The increasing number of reference genomes from nonmodel species has begun to outpace efforts to identify and annotate TE content and methods that are used vary significantly between projects. Here, we demonstrate variation that arises in TE annotations when less than optimal methods are used. We found that across a variety of taxa, the ability to accurately identify TEs based solely on homology decreased as the phylogenetic distance between the queried genome and a reference increased. Next we annotated repeats using homology alone, as is often the case in new genome analyses, and a combination of homology and de novo methods as well as an additional manual curation step. Reannotation using these methods identified a substantial number of new TE subfamilies in previously characterized genomes, recognized a higher proportion of the genome as repetitive, and decreased the average genetic distance within TE families, implying recent TE accumulation. Finally, these finding—increased recognition of younger TEs—were confirmed via an analysis of the postman butterfly (Heliconius melpomene). These observations imply that complete TE annotation relies on a combination of homology and de novo–based repeat identification, manual curation, and classification and that relying on simple, homology-based methods is insufficient to accurately describe the TE landscape of a newly sequenced genome.
Keywords: transposable elements, genome annotation, Heterocephalus glaber, Microtus ochrogaster, Heliconius melpomene
Introduction
Transposable elements (TEs) are DNA sequences that occupy large portions of eukaryotic genomes (de Koning et al. 2011) and may have significant impacts on organismal biology and evolution (Kazazian 2004). As such, a thorough annotation of TEs in newly sequenced genomes is of utmost importance. However, because most sequencing projects are interested in genomic components more commonly associated with evolution of phenotypic characters, the repetitive portion of the genome is often either ignored or given only minimal attention.
The accurate identification of repetitive sequences is computationally challenging. Two computational methodologies exist for TE discovery in newly sequenced genomes: Homology-based searches and de novo identification. TE annotations via homology are, by definition, only able to identify TEs similar to those already described. Often homology-based annotations will miss lineage-specific subfamilies or TEs deposited via horizontal transfer. De novo methods identify TEs using structural features associated with TEs (e.g., LTR_finder; Xu and Wang 2007) or use k-mer counting methods (e.g., RepeatScout; Price et al. 2005) to identify overrepresented sequences. TEs lacking canonical structural features or that are present in low copy number would not be identifiable using de novo methods. In both cases, identification of heavily mutated TEs is difficult.
Most genome projects identify and annotate TEs using homology and de novo methods (Hoen et al. 2015), but the most accurate assessment of TE landscapes is currently only possible through a combination of de novo and homology-based repeat identification in conjunction with an additional manual curation step (Flutre et al. 2012). One reason for this is that longer elements are often recovered as multiple smaller fragments by both types of searches. Because elements are subsequently classified using homology and/or sequence hallmarks, including target site duplications, terminal inverted repeats, and the presence/length of open reading frames to provide a preliminary classification (Abrusán et al. 2009; Feschotte et al. 2009), these classifications are heavily dependent on the library of elements being fully and accurately represented.
One can verify that an element is fully and accurately represented via a manual curation process (Alfoldi et al. 2011; Platt and Ray 2012; Ge et al. 2013; Lavoie et al. 2013; Green et al. 2014). This usually consists of some variation on the following process: First elements are queried against the subject genome using BLAST (Altschul et al. 1997). The best hits to each element are extracted along with flanking sequence and aligned. If either the 5′ or 3′ end of the alignment terminates within the repetitive region, the BLAST/extension process is reiterated until both the 5′ and 3′ ends of the alignment show evidence of single-copy DNA. A majority-rule consensus sequences is then generated to approximate the source element (Kapitonov and Jurka 1999, 2003). Once the manual curation step is completed, sequence hallmarks along the entire consensus repeat can be used for TE classification. Unfortunately, this manual curation and validation process is often bypassed during genome sequencing and assembly efforts.
Recently, an effort to develop a set of standardized benchmarks to judge TE annotation quality was announced (Hoen et al. 2015). Here we illustrate the necessity of a coherent annotation strategy when analyzing genome drafts for TEs. To do so, we compare the results of TE annotations using minimal versus comprehensive analyses and describe the potential pitfalls of the former strategy. The data include examples from mammalian and insect genomes and adds to the growing recognition described by Hoen et al. (2015) that a need exists for a coherent set of TE annotation and benchmarking strategies.
Materials and Methods
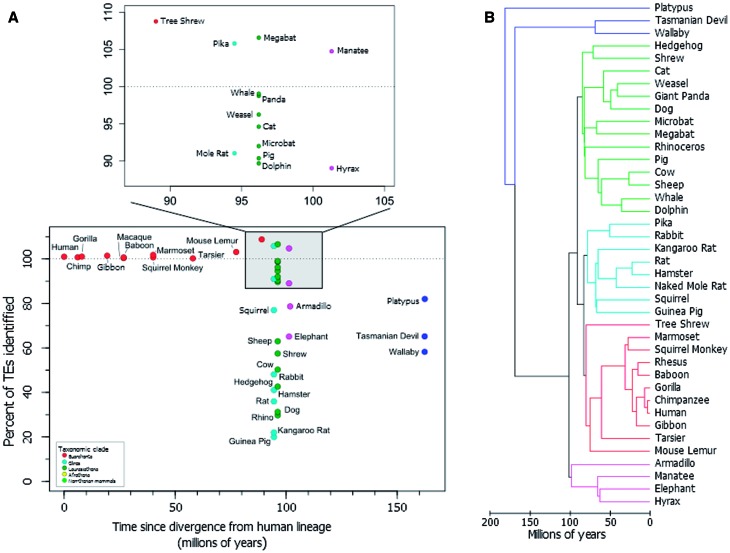
TEs were quantified in mammalian genomes available through the UCSC Genome Browser using the complete human TE library and sensitive (-s) RepeatMasker (Smit and Hubley 2014) searches that excluded low complexity regions (-nolow). All regions homologous to TEs were retained regardless of size or percent identity to a consensus TE. The sum of all TE content from these human-specific runs was then compared with species-appropriate analyses available at RepeatMasker.org (http://repeatmasker.org/genomicDatasets/RMGenomicDatasets.html, last accessed August 10, 2014) which are assumed to be the most accurate representations available for most species. RepeatMasker.org searches were conducted using the “-s” (sensitive settings) and each species were generated using libraries specific to that species. For example, the Rattus norvegicus genome was queried using R. norvegicus repeats (-species “R. norvegicus; Hubley R, personal communication). Only taxa available through the UCSC genome browser and RepeatMasker.org website were included. Human TE content of each genome was quantified as a percentage of the expected species-specific TE content. Time since divergence for each taxa was recorded as the mean divergence time provided at TimeTree.org (accessed 14 August 2014). These were then plotted to show the relationship between time since divergence and the ability to identify TEs using homology-dependent searches (fig. 1).
Fig. 1.—
Homology-based TE annotations using human TEs. (A) TEs in several mammalian genomes were identified and quantified using human TEs. The percentage of TEs identified using human TEs is given as a percentage of the known repeat content. Time since divergence from the human lineage for each taxa was taken from TimeTree.org. Taxonomically related species are grouped by color. The dotted line represents 100% recognition. (B) A phylogram depicting the radiation of the mammals, modified from Murphy et al. (2007).
To identify lineage-specific TEs, the naked mole rat (hetGla2; accession number AHKG00000000.1) and vole genomes (micOch1; accession number AHZW00000000.1) were masked using the standard Rodent libraries available from Repbase as implemented in RepeatMasker (-lib “Rodentia”). At the time, no known repeats were present in Repbase for either the prariie vole or the mole rat. The masked genomes were then used for de novo repeat identification using RepeatModeler. Those elements identified by RepeatModeler but not masked by the initial RepeatMasker run are expected to be specific to the mole rat or vole lineages. The BLAST, extract, extend process was used to guarantee capture of lineage-specific elements across their entire length. The Perl script used to automate these steps is available at www.github.com/nealplatt/bioinfo (extractAlignTEs.pl; last accessed 23 February 2015). There tend to be fewer TE families in mammalian genomes than other vertebrates (e.g., fish), but these TE families reach very high copy numbers (Furano et al. 2004). With this observation in mind, consensus elements were generated during each iteration from a minimum of 20 elements. Query sequences with fewer than 20 hits in any iteration were culled. After verifying the full length capture of each lineage-specific element, consensus elements were classified based on overall structure (size, poly-A tails, target site duplications) and similarities to previously identified TEs in Repbase. Elements were compared with each other using cd-hit-est (Huang et al. 2010). Those meeting the 80-80-80 rule was subsumed within a larger TE (sub)family (Wicker et al. 2007).
The unmasked version of the naked mole rat and vole genomes was reannotated using RepeatMasker and two different TE libraries—a rodent library containing the rodent TEs from Repbase and the fully curated library containing the rodent TEs plus the lineage-specific elements. After RepeatMasker annotations, Kimura 2-parameter (Kimura 1980) divergence values between the library elements and those found in the naked mole rat and vole genomes were calculated using the calcDivergenceFromAlign.pl utility packaged with RepeatMasker. Highly mutable C-phosphate-G (CpG) sites were excluded from distance analyses (Hodgkinson and Eyre-Walker 2011). Element accumulation over time was calculated by binning elements based on their Kimura two-paramter distance value from the consensus library TEs.
The Heliconius melpomene genome was analyzed in a way similar to the mole rat and vole with few exceptions. First, Lavoie et al. (2013) recently completed a complete annotation of the H. melpomene genome using the protocols recommended here in. To compare different annotation strategies a fully curated library was created by using all known arthropod elements from Repbase. Becasue this already included all the H. melpomene repeats from Lavoie et al. (2013), this served as the fully curated library. An arthropod-only repeat library was created by removing elements deposited by Lavoie et al. (2013). CpG sites were included in calculating the age distribution of TE accumulation.
Analysis of Mammalian Genomes
TE annotations that rely on homology alone are likely to result in biologically unrealistic annotations. In extreme cases of homology-based annotations, TE libraries from related species or groups serve as the sole source for TE identification within newly assembled genomes. Because most TEs, especially the retrotransposons, are inherited and accumulate in a vertical manner, the more distantly related two taxa are, the more divergent the TE component of the genomes are expected to be. As the phylogenetic distance between the genome under study and the most closely related organism with available TE libraries increases, the more problematic the analysis becomes.
To illustrate this problem we analyzed the TE fraction of 40 mammalian genomes using a library of human TEs. Each genome was subjected to standard RepeatMasker searches and the recovered TE content was quantified as a percentage of the known TE content. Known TE content here is defined as that identified in the current RepeatMasker tables (Smit and Hubley 2014). Known TE content does not always equate actual TE content, due to varying levels of knowledge per taxon (discussed below). To determine the effect of increased genetic distance on our ability to identify TEs using homology-based searches, the percentage of identified TEs was arranged temporally using the mean divergence times between each species and the human lineage (fig. 1A). One might expect rates of TE identification to exhibit an inverse relationship with genetic distance, particularly within the primates. Instead, we see a more inverted L-shaped pattern with high rates of TE identification in primates but no discernable pattern in all other mammals examined. Because most major groups of mammals diverged over a very brief period ∼90–100 Ma (fig. 1B), the nonprimate taxa accumulate in essentially a single column with TE identification rates varying from <20% (Guinea Pig) to more than 109% (Tree Shrew) of known TEs in various taxa. Thus, there appears to be no relationship between phylogenetic distance and ability to identify TEs solely based on homology. It is possible that TE identification rates are impacted by low-quality genome assemblies; however, regressing percent TE content identified against basic genome assembly statistics (contig N50 and number of scaffolds) fails to support this hypothesis (supplementary fig. S1, Supplementary Material online). In some taxa more than 100% of the known TE content is captured using human-specific repeats. Examples include the megabat, manatee, and pika. This is likely driven by several factors including unique TE biology (Cantrell et al. 2008), stochasticity in individual RepeatMasker analyses, or variation in the quality of TE annotations across mammalian genomes (see discussion below) along with combined effects of rates of mutation in each lineage, and the nature of TE (especially retrotransposon) accumulation.
Interestingly, the data (fig. 1) can also be used to make generalizations about TE identification in individual mammalian genomes. We expected that the repeat library from an outgroup species would identify elements at a similar rate in two sister taxa. If true, closely related species in figure 1 should cluster more closely together than to distantly related taxa. For example, a human repeat library should identify similar numbers of TEs in all rodents and similar numbers in all carnivores, and so on, but in reality, the percentage of identified TEs across phylogroups varies drastically (fig. 1A, rodents 19.9–94.5%, carnivores 31.3–98.7%). In the pika (Ochotona princeps), the human-based RepeatMasker analysis identified 200.8 Mb of TEs accounting for more than 100% of TEs currently annotated in the pika genome. For comparison, the most closely related species to the pika in our analysis was the rabbit (Oryctolagus cuniculus). These taxa diverged from each other ∼50 Ma, compared with the ∼90 Myr divergence between the pika–rabbit common ancestor and primates (Murphy et al. 2007). Despite the relatively close relationship between the rabbit and pika, the human libraries were much less successful at identifying known TEs in the rabbit genome with <530 Mb of ∼1,100 Mb identified in the current RepeatMasker tables (Smit and Hubley 2014).
When compared with the TE component of other glirean taxa (e.g., rat 1,035 Mb; guinea pig 672 Mb; hamster 560 Mb; squirrel 801 Mb), it becomes clear that the 200.8 Mb repetitive content in the pika genome is low and an outlier. Both the rabbit and pika genomes have undergone some level of de novo TE identification (Jurka 2009a, 2009b). These seemingly conflicting results could be driven by multiple scenarios including the cessation of recent activity or the active removal of TE insertions. Both these processes are considered rare (van de Lagemaat et al. 2005). A more likely explanation for the apparent reduced TE load may be driven by differences in annotation quality. Further work on TEs in lagomorphs will be able to determine if this result is biological or methodological.
We attempted a more tractable comparison by using a library of all known rodent TEs in RepBase to identify repetitive DNAs in the naked mole rat genome (Heterocephalus glaber; Kim et al. 2011). Using a version of the genome masked for rodent repeats (rodent-only), we employed a de novo repeat identification using RepeatModeler, which yielded 388 potentially novel repetitive sequences. After manually verifying each element, combining similar hits, and extending fragments of longer elements as described above (Wicker et al. 2007), 66 putative TEs from the naked mole rat genome that were not present in Repbase remained.
TE content and accumulation in the naked mole rat genome was recalculated using this new library of fully curated elements in combination with the rodent-only TE library. From a genome content perspective, both analyses recovered similar results (table 1). The rodent-only library underestimated the TE content of the naked mole rat genome by 88.3 Mb or 3.8% of the total genome assembly (table 1) but is a 13% overall increase in the total TE content. A substantial difference between the two annotations was in increased recognition of mole rat–specific retrotransposons that would be mischaracterized or overlooked in a homology-only approach. Specifically, this included long interspersed element (LINE) 1s (31.8Mb), short interspersed elements (SINEs) (17.5 Mb), and solo long terminal repeats (LTRs) (52.5 Mb) in conjunction with a decrease in recognized ERV3 LTRs (−26.3 Mb). The decrease in ERV3s was due to reclassification of several ERV3s as solo LTRs.
Table 1.
Transposable Element Load in the Naked Mole Rat (Heterocephalus glaber) and the Prairie Vole (Microtus ochrogaster) using Rodent-Specific and De Novo Repeat Transposable Element Libraries
| Naked Mole Rat |
Prarie vole |
|||||
|---|---|---|---|---|---|---|
| Rodent (Mb) | De novo (Mb) | Rodent (Mb) | De novo (Mb) | |||
| Class I retrotransposons | 594.79 | 661.63 | 646.21 | 790.68 | ||
| LTRs | 157.39 | 175.2 | 210.39 | 346.24 | ||
| ERV | 7.55 | 7.45 | 2.02 | 1.74 | ||
| ERV1 | 17.05 | 15.47 | 10.28 | 13.04 | ||
| ERV2 | 21.35 | 14.61 | 89.97 | 221.59 | ||
| ERV3 | 110.65 | 84.39 | 105.43 | 102.27 | ||
| Gypsy | 0.54 | 0.51 | 0.1 | 0.1 | ||
| LTR | 0.25 | 52.77 | 2.6 | 7.5 | ||
| LINEs | 368.83 | 400.35 | 213.68 | 230.1 | ||
| CR1 | 16.18 | 15.94 | 2.29 | 2.29 | ||
| L1 | 352.16 | 383.94 | 211.24 | 227.66 | ||
| L2 | 0.12 | 0.11 | 0.03 | 0.03 | ||
| Penelope | 0.01 | 0.01 | 0 | 0 | ||
| R4 | 0.01 | 0.01 | 0 | 0 | ||
| RTE | 0.02 | 0.02 | 0.01 | 0.01 | ||
| RTEX | 0.33 | 0.31 | 0.1 | 0.1 | ||
| Tx1 | 0.01 | 0.01 | 0 | 0 | ||
| SINEs | 68.5 | 86.03 | 222.12 | 214.31 | ||
| SINE1/7SL | 68.42 | 74.29 | 84.4 | 77.34 | ||
| SINE2 | 0 | 11.66 | 137.64 | 136.89 | ||
| SINE3/5S | 0.04 | 0.04 | 0.04 | 0.04 | ||
| Unk | 0.05 | 0.05 | 0.03 | 0.03 | ||
| Unclassified non-LTRs | 0.06 | 0.06 | 0.02 | 0.02 | ||
| Unclassified | 0.06 | 0.06 | 0.02 | 0.02 | ||
| Class II DNA transposons | 33.17 | 51.36 | 17.2 | 17.2 | ||
| PiggyBac | 0 | 1.73 | 0 | 0 | ||
| TcMariner | 14.45 | 30.07 | 4.88 | 4.88 | ||
| hAT | 15.33 | 16.42 | 9.18 | 9.17 | ||
| MuDR | 1.43 | 1.24 | 0.39 | 0.39 | ||
| Helitron | 0.13 | 0.13 | 0.03 | 0.03 | ||
| Kolobok | 0.02 | 0.02 | 0.01 | 0.01 | ||
| Unk | 1.8 | 1.76 | 2.71 | 2.71 | ||
| Unclassified tes | 5.61 | 8.89 | 26.63 | 13.51 | ||
| Unclassified | 5.61 | 8.89 | 26.63 | 13.51 | ||
| Total | 633.57 | 721.88 | 690.04 | 821.39 | ||
Note.—Rodent-specific libraries were taken from Repbase (August 2014). De novo libraries were combined with the rodent-specific libraries in an effort to generate the complete annotations.
Although similar numbers of elements were identified overall, the major difference between the two analyses lies in the area of sequence diversity, which was heavily affected when the different libraries were used. Analyses with the rodent-only library suggest that the vast majority of TE loci have divergences ≥10% from their respective consensus. Specifically, TEs exhibiting between 0% and 10% divergences from their consensuses make up only 0.6% (3.6 Mb) of TEs, implying that there has been no appreciable accumulation in the recent past for this taxon. By applying a mammalian neutral mutation rate of 2.2 × 10—9 per base per million years (Kumar and Subramanian 2002), figure 2A (rodent-only library) suggests that TE accumulation declined substantially between 40 and 45 Ma. Not coincidentally, this corresponds with the estimated divergence time between the naked mole rat and guinea pig lineages (Huchon et al. 2007), guinea pig being the most closely related taxon with available TE annotations. However, when the same analysis is performed using a properly curated library, 11.3% (81.23 Mb) of the overall TE content in the mole rat is < 10% divergent from the consensus and, by extension, indicates significant amounts of recent TE accumulation (fig. 2D).
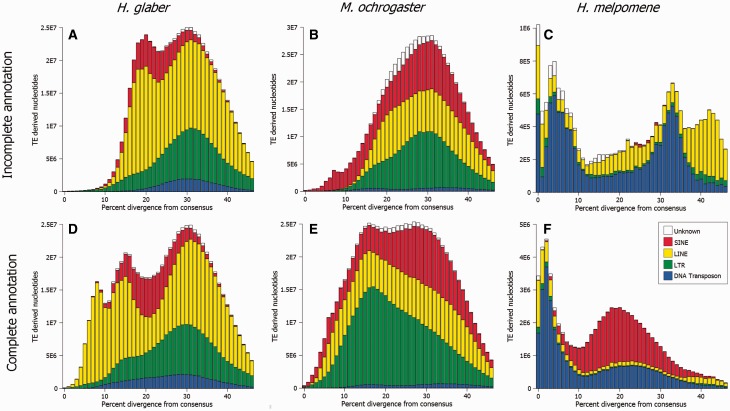
Fig. 2.—
Differences in TE accumulation histories of the (A, D) naked mole rat (Heterocephalus glaber), (B, E) prairie vole (Michrotus ochrogaster), and (C, F) postman butterfly (Heliconius melpomene) before and after de novo TE identification and curation. RepeatMasker searches against the (A) mole rat and (C) prairie vole used all known mammal TEs and all known arthropod TEs were used against the (E) postman butterfly genome to identify all known TEs based on homology only. De novo identification and curation altered the content, quantity, and distribution of elements identified for the (B) mole rat, (D) prairie vole, and (F) postman butterfly genomes. Divergence from a consensus sequence from each element was calculated and binned to demonstrate the accumulation profile for each taxa. For the mole rate and prairie vole, highly mutable CpG sites were excluded from analyses.
The reason behind the discrepancy is easily understood given retrotransposon biology. At the moment of insertion, an element will be almost, if not completely, identical to the source element. Over time TE insertions accumulate mutations independently of each other. Because the rodent library contains TEs from taxa that diverged from the naked mole rat ≥40 Ma, overall genetic diversity among TE insertions and the TE library was much greater when the rodent-only library was used exclusively (fig. 2A vs. D). This is driven by the recognition of lineage-specific TEs as variants of older, ancestral families. The result is higher levels of sequence divergence that artificially skews the relative ages of elements.
The prairie vole (Microtus ochrogaster) genome provides an additional example of the need for proper repeat identification and curation. Analyses using homology-based searches with a rodent TE library and a fully curated library produce considerably different results (fig. 2B and E) despite sharing a common ancestor with Mus and Rattus ∼45 Ma (Adkins et al. 2003). Similar to the analyses in the naked mole rat, we masked the prairie vole genome for known rodent repeats before de novo repeat identification. Despite masking all previously known elements, 404 potentially novel elements were recovered. After manual curation, 121 TE families and subfamilies remained that were specific to the prairie vole lineage. As in the mole rat, reannotation of the prairie vole genome using a library that included the newly curated elements and known rodent TEs recovered similar overall TE content. However, substantial differences in the details of TE accumulation are seen. For example, ERV2 content more than doubled from 90 to 221.6 Mb. In addition, the skew in estimated divergence from each subfamily consensus was shifted substantially to the left, indicating a more recent accumulation of TEs in the genome than would be suggested by homology-based identification methods alone.
These two examples from Mammalia indicate a need to perform detailed analyses of the TE landscape in naive genome assemblies. In both instances, the overall TE content was similar when using fully curated versus ancestral rodent libraries, but the average sequence diversity was less in the fully curated analyses. A similar observation is also seen in the primates in figure 1A. Analysis using human TEs successfully identifies most TEs in other primates. This implies that clade-specific TE libraries are capable of identifying most TEs of relatively closely related species but the analysis of the prairie vole and mole rat imply that clade-specific libraries will skew estimates of sequence diversity, and by extensions perceived accumulation rates. Mammals are a particularly well-studied clade, with more than one hundred fully sequenced genomes. Indeed, they are overrepresented when compared with other, more species-rich clades. In a third example, we demonstrate similar biases in an invertebrate.
Analysis of the Heliconius melpomene Genome
We simulated a homology-only analysis of the H. melpomene genome using TEs from other insects (see Materials and Methods) to determine the likely results of a precuration approach. This analysis indicated that the repeat composition of this taxon is dominated by older Helitrons and that SINEs are not a significant presence (fig. 2C). Full curation of the H. melpomene genome increased the known repetitive portion of the genome from less than 20.4 to 75.3 Mb, an increase of almost 4-fold (Lavoie et al. 2013). Second, a previously unrecognized tRNA-derived SINE family, Metulj, was found to occupy 41.1% (31 Mb) of total repeat content and over 8% of the genome as a whole (fig. 2F). Finally, an additional 7.3 Mb of the total 14.4 Mb of Helitron content was identified. This is slightly less than the 17.1 Mb previously estimated in the H. melpomene genome (Han et al. 2013) although the accumulation pattern was considerably altered. Instead of the Helitron component comprising older, inactive elements, it becomes clear that these butterflies have been subjected to massive amounts of recent accumulation. Helitrons are known to be involved in exon shuffling, transduction in general, and the introduction of novel regulatory elements (Morgante et al. 2005; Pritham and Feschotte 2007; Thomas et al. 2014). Heliconius serves as model organisms for mimetic evolution and color variation (Brower 1996) and TEs have already been implicated in genomic regions associated with wing pattern variation in Heliconius (Papa et al. 2008). With the availability of a more accurately annotated genome, future research may focus on recent Helitron activity in these regions.
The striking change in our understanding of the TE landscape of H. melpomene when compared with the smaller (but still important) shifts in our picture of TE accumulation in mammals can be explained by our relative understanding of TEs in the two clades. In general, genomic data from insects are underrepresented when compared with mammals. This is compounded by the fact that Mammalia is relatively young (150 Myr; Murphy et al. 2007) compared with many other classes including Insecta (350 Myr; Gaunt and Miles 2002). Of the ∼8,500 elements known in Arthropoda, the majority of annotated TEs are from Aedes aegypti, Drosophila sp., and Locusta migratoria, all species more than 340 My diverged from Heliconius (Gaunt and Miles 2002). The most closely related taxon with a sequenced genome and well-annotated TEs is Bombyx mori, the silkworm moth, which diverged from the lineage leading to butterflies ∼145 Ma (Gaunt and Miles 2002). Thus, within insects the closest taxon with available genomic data may have diverged hundreds of millions of years prior. In the relatively data-rich mammals, gaps in our understanding of genomic TE content rarely span more than 50 My. Because genomic recourses in general and TE resources in particular are sparse in insects, the need for complete and accurate annotations is of utmost importance, as seen in H. melpomene.
Conclusions
TEs may contribute to structural rearrangements, rewire regulatory pathways, be exapted into protein-coding regions, and contribute to numerous other mechanisms of genome evolution (Kidwell and Lisch 2001; Suh 2015). Thus, a proper understanding of accumulation patterns is vital. Indeed, as demonstrated by the Heliconius example above, a proper understanding of TE accumulation in a genome may prompt ideas related to the mechanisms of phenotypic evolution within a clade. By improving TE annotations, the role of TEs in the development of lineage-specific characteristics, if any, can be better understood. Furthermore, we have not mentioned the possibility of missing horizontally transferred TEs when using a homology-only approach. Such events are known to occur (Pace et al. 2008) and could be significant players in the evolution of genome structure (Platt et al. 2014; Thomas et al. 2014). By limiting ourselves to minimal annotation strategies, especially approaches that utilize homology-based searches alone, many of these effects could be overlooked.
TEs may comprise up to 85% of eukaryotic genomes (Tenaillon et al. 2010), yet often receive significantly less attention than the protein-coding portions of the genome. We found that fully curated TE annotations are able to improve upon current or minimally curated annotations. TEs from closely related species can be used to identify repeats in other taxa in a relatively closely related clade as demonstrated here in the rodents. This strategy fails to accurately quantify levels of sequence diversity within TE families and by extension overestimates TE age and accumulation period. In other cases, like the H. melpomene example, homology-based searches fail to describe TE content at an acceptable level by any standards. These findings further support Hoen et al.’s (2015) call for a coherent annotation and benchmarking strategy that is applied across taxa. It is likely that this strategy will include combinations of homology and de novo TE identification methods with a manual curation step. By abiding to the principles outlined herein, our ability to understand the biology of TEs and genome evolution in general will be significantly impacted.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation (DEB-1354147, MCB-0841821, and DEB-1020865 to D.A.R.). Additional support was provided by College of Arts and Sciences at Texas Tech University.
Literature Cited
- Abrusán G, Grundmann N, DeMester L, Makalowski W. 2009. TEclass—a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 25:1329–1330. [DOI] [PubMed] [Google Scholar]
- Adkins RM, Walton AH, Honeycutt RL. 2003. Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Mol Phylogenet Evol. 26:409–420. [DOI] [PubMed] [Google Scholar]
- Alfoldi J, et al. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477:587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower AVZ. 1996. Parallel race formation and the evolution of mimicry in Heliconius butterflies: a phylogenetic hypothesis from mitochondrial DNA sequences. Evolution 50:195–221. [DOI] [PubMed] [Google Scholar]
- Cantrell MA, Scott L, Brown CJ, Martinez AR, Wichman HA. 2008. Loss of LINE-1 activity in the megabats. Genetics 178:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7:e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Keswani U, Ranganathan N, Guibotsy ML, Levine D. 2009. Exploring repetitive DNA landscapes using REPCLASS, a tool that automates the classification of transposable elements in eukaryotic genomes. Genome Biol Evol. 1:205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flutre T, Permal E, Quesneville H. 2012. Transposable element annotation in completely sequenced eukaryote genomes In: Grandbastien MA, Casacuberta JM, editors. Plant transposable elements. Berlin Heidelberg (Germany: ): Springer; p. 17–39. [Google Scholar]
- Furano AV, Duvernell DD, Boissinot S. 2004. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 20:9–14. [DOI] [PubMed] [Google Scholar]
- Gaunt MW, Miles MA. 2002. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 19:748–761. [DOI] [PubMed] [Google Scholar]
- Ge RL, et al. 2013. Draft genome sequence of the Tibetan antelope. Nat Commun. 4:1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, et al. 2014. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346:1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MJ, et al. 2013. Identification and evolution of the silkworm helitrons and their contribution to transcripts. DNA Res. 20:471–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson A, Eyre-Walker A. 2011. Variation in the mutation rate across mammalian genomes. Nat Rev Genet. 12:756–766. [DOI] [PubMed] [Google Scholar]
- Hoen DR, et al. 2015. A call for benchmarking transposable element annotation methods. Mobile DNA 6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Niu B, Gao Y, Fu L, Li W. 2010. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huchon D, et al. 2007. Multiple molecular evidences for a living mammalian fossil. Proc Natl Acad Sci U S A. 104:7495–7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. 2009a. Long terminal repeats from rabbit. Repbase Rep. 9:2900. [Google Scholar]
- Jurka J. 2009b. Long terminal repeats from the American Pika. Repbase Rep. 9:2205. [Google Scholar]
- Kapitonov V, Jurka J. 1999. Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica 107:27–37. [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. 2003. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci U S A. 100:6569–6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH. 2004. Mobile elements: drivers of genome evolution. Science 303:1626–1632. [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR. 2001. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55:1–24. [DOI] [PubMed] [Google Scholar]
- Kim EB, et al. 2011. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16:111–120. [DOI] [PubMed] [Google Scholar]
- Kumar S, Subramanian S. 2002. Mutation rates in mammalian genomes. Proc Natl Acad Sci U S A. 99:803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie C, Platt R, Novick P, Counterman B, Ray D. 2013. Transposable element evolution in Heliconius suggests genome diversity within Lepidoptera. Mobile DNA 4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M, et al. 2005. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet. 37:997–1002. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. 2007. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 17:413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace JK, Gilbert C, Clark MS, Feschotte C. 2008. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc Natl Acad Sci U S A. 105:17023–17028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa R, et al. 2008. Highly conserved gene order and numerous novel repetitive elements in genomic regions linked to wing pattern variation in Heliconius butterflies. BMC Genomics 9:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RN, II, Ray DA. 2012. A non-LTR retroelement extinction in Spermophilus tridecemlineatus. Gene 500:47–53. [DOI] [PubMed] [Google Scholar]
- Platt RN, II, et al. 2014. Large numbers of novel miRNAs originate from DNA transposons and are coincident with a large species radiation in bats. Mol Biol Evol. 31:1536–1545. [DOI] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA. 2005. De novo identification of repeat families in large genomes. Bioinformatics 21:i351–i358. [DOI] [PubMed] [Google Scholar]
- Pritham EJ, Feschotte C. 2007. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc Natl Acad Sci U S A. 104:1895–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF, Hubley RM. 2014. RepeatMasker Genomic Datasets. Available from: http://repeatmasker.org/genomicDatasets/RMGenomicDatasets.html.
- Suh A. 2015. The specific requirements for CR1 retrotransposition explain the scarcity of retrogenes in birds. J Mol Evol. 81:18–20. [DOI] [PubMed] [Google Scholar]
- Tenaillon MI, Hollister JD, Gaut BS. 2010. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 15:471–478. [DOI] [PubMed] [Google Scholar]
- Thomas J, Phillips CD, Baker RJ, Pritham EJ. 2014. Rolling-circle transposons catalyze genomic innovation in a mammalian lineage. Genome Biol Evol. 6:2595–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Lagemaat LN, Gagnier L, Medstrand P, Mager DL. 2005. Genomic deletions and precise removal of transposable elements mediated by short identical DNA segments in primates. Genome Res. 15:1243–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 8:973–982. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wang H. 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35:W265–W268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.