Abstract
Treatment options for older patients with acute myeloid leukemia (AML) range from supportive care alone to full-dose chemotherapy. Identifying factors that predict response to therapy may help increase efficacy and avoid toxicity. The phase II SWOG S0703 study investigated the use of hydroxyurea and azacitidine with gemtuzumab ozogamicin in the elderly AML population and found survival rates similar to those expected with standard AML regimens, with less toxicity. As part of this study, global DNA methylation along with promoter DNA methylation and expression analysis of six candidate genes (CDKN2A, CDKN2B, HIC1, RARB, CDH1 and APAF1) were determined before and during therapy to investigate whether very early changes are prognostic for clinical response. Global DNA methylation was not associated with a clinical response. Samples after 3 or 4 days of treatment with azacitidine showed significantly decreased CDKN2A promoter DNA methylation in patients achieving complete remission (CR) compared to those who did not. Samples from day 7 of treatment showed significantly decreased RARB, CDKN2B and CDH1 promoter DNA methylation in responders compared to nonresponders. Gene-specific DNA methylation analysis of peripheral blood samples may help early identification of those older AML patients most likely to benefit from demethylating agent therapy.
Keywords: AML, Decitabine, DNA methylation, Elderly, Prognostic
1. Introduction
Treatment of AML in older patients remains a therapeutic challenge. While outcomes for younger patients with AML have improved over time, this trend has not been seen in patients over the age of 60. This population has a higher frequency of medical comorbidities and is more likely to have a suboptimal performance status. Additionally, there is a higher frequency of secondary AML (due to transformation from another hematologic malignancy or from prior therapies), adverse karyotypes and drug resistance in older patients which all contribute to inferior outcomes. There is no accepted standard of care in these patients and current therapeutic options range from supportive care alone to low intensity regimens such as demethylating agents and low dose cytarabine, or full dose chemotherapy [1–5]. While better therapies are badly needed, there would also be an advantage in being able to select from available therapies those to which a given patient would most likely respond.
Gene silencing via methylation of promoter CpG islands appears to play a significant role in the pathogenesis of hematologic malignancies such as myelodysplastic syndromes (MDS) and AML. Studies in MDS have shown epigenetic silencing of tumor suppressor genes such as CDKN2B (pi 5, INK4b), aberrant DNA methylation, and clinical activity of agents that affect DNA methylation, such as 5-azacytidine (azacitine) and 5-azadeoxycytidine (decitabine) [6–12]. In AML, DNA methylation inhibitors have also shown activity [13,14]. Distinct DNA cytosine methylation patterns distinguish AML subgroups [15]. DNA promoter regions of critical genes are inactivated through hypermethylation in AML [16,17]. CDKN2B and E-cadherin (CDH1) are independently associated with poor prognosis in AML when methylated. Patients with both CDKN2B and CDH1 methylation had worse prognosis compared to those with either gene methylated alone [18]. Reactivation of the tumor suppressor p73 by demethylation of its promoter region has been shown to occur after treatment of AML cells with decitabine [19]. This leads to p21WAF1 (CDKN1A) expression which correlated with AML cell cycle arrest [19]. APAF1 is another tumor suppressor inactivated in some cases of AML that can be re-expressed after treatment with a hypomethylating agent [20]. CDH1, HIC1, CDKN2A and CDKN2B genes can be hypermethylated in patients with AML [21–23]. The degree of DNA methylation has been shown to decrease after treatment with decitabine [14]. Another study measured global methylation status of bone marrow specimens from older patients with AML before and after treatment with decitabine; although post-treatment specimens showed significantly less methylation, it did not correlate with the percentage of bone marrow blasts [24]. Comparison of demethylation status before and after treatment and whether this correlates with clinical outcome has not been firmly established in acute leukemia.
The monoclonal antibody-antineoplastic conjugate Gemtuzumab ozogamicin (GO), which targets CD33, a myeloid antigen expressed on most AML leukemic cells, has been shown to reduce relapse rate and increase survival when added to induction chemotherapy in older adults [25]. Similarly, a recent Phase 3 trial showed that GO added during induction and consolidation may improve outcome of AML patients aged 50–70 years [26]. This drug was withdrawn from the U.S. market in 2010 when a confirmatory trial showed no improvement in survival and a higher fatality rate in the group treated with GO [27]. More recently, mutation analysis of patients enrolled on a Phase 3 clinical trial found that cytogenetically normal (CN)-AML patients had a preferential benefit from GO treatment as compared to AML patients with abnormal cytogenetics [28]. Two recent meta-analyses examined data from five randomized trials and concluded that GO improves overall survival (OS) and reduces relapse [29,30]; it was also shown to reduce the development of disease resistance [29].
The phase 2 Southwest Oncology Group (SWOG S0703) study by Nand et al. investigated the use of hydroxyurea and azacitidine with GO in the elderly AML population [31]. This regimen was shown to be at least as effective as standard therapy but with lower toxicity in poor risk patients (70 years and older, Zubrod performance status [PS] of 2–3). Similar outcomes were seen in the good risk group (60–69 years old or PS 0–1) although data in this subset of patients did not reach predefined significance goals [31].
Here we report the laboratory findings of samples accrued as part of the S0703 study. Global DNA methylation, promoter DNA methylation of six candidate genes chosen because of their previous association with DNA methylation in AML, and expression analysis of the same genes were determined at several time points before and during therapy. Goals of this study were to investigate DNA methylation or gene expression as indicators of clinical response.
2. Materials and methods
Patients
All patients were enrolled on SWOG phase 2 clinical trial S0703. Patients with a newly diagnosed non-M3 AML under the WHO classification who had reached their 60th birthday and had a performance status of 0–3 were eligible for entry in the study. A total of 142 patients were enrolled in the study, with 83 and 59 in the good-risk and poor-risk cohorts, respectively. Good risk (age between 60 and 69 years or performance status of 0–1) and poor risk (age 70 years or older and performance status of 2 or more) based on prior experience in SWOG with older patients, were as previously defined for this clinical trial [31]. The patients were given hydroxyurea to bring the WBC count to less than <10,000 × 109/L and started on azacitidine 75 mg/m2 subcutaneously or intravenously daily for 7 days. Gemtuzumab ozogamicin 3 mg/m2 intravenously was administered on day 8. Those achieving CR or CRi received an identical treatment as consolidation therapy followed by 4 cycles of azacitidine therapy. The study design and the clinical findings from the study were recently published [31]. For statistical analysis, patients were grouped based on their clinical response. Complete Response (CR) was defined as <5% marrow blasts by morphology, no Auer rods, absolute neutrophil count (ANC) 1,000 × 109/L or higher, platelet count 100,000 × 109/L or higher and no evidence of extra-medullary disease. CR with incomplete recovery of blood counts (CRi) was the same as CR but ANC may be <1000 × 109/L and/or platelet count <100,000 × 109/L. Relapse from CR or CRi was defined as reappearance of leukemic blasts 5% or higher in peripheral blood or bone marrow or appearance/reappearance of extra-medullary disease. Written informed consent for treatment and for correlative studies was obtained from all patients in accordance with the Declaration of Helsinki. The study was approved by the institutional review boards of participating institutions.
2.2. Samples
Peripheral blood samples for laboratory correlative studies were collected pre-study, on Day 3 or 4 (range 2–5) and on Day 7 (range 6–9) of induction treatment. Samples were also submitted around Day 30 (range 27–35), after achievement of complete remission, completion of all required therapy and at the time of relapse. All specimens were sent to SWOG central laboratories for DNA and RNA isolation using standard methods. Peripheral blood samples analyzed required a prestudy sample, plus either day 3 or 4, or day 7 samples of sufficient quality from the same patient. DNA methylation and RNA expression were analyzed on peripheral blood samples from 84 and 82 patients, respectively, with a range of 3–8 DNA or RNA samples per patient analyzed. The samples analyzed for DNA methylation and RNA expression had a similar distribution outcome (CR, CRi treatment distribution) as the complete clinical trial cohort (Table 1).
Table 1.
Study samples analyzed and promoter methylation changes of APAF1 and HIC1 genes.
| Samples for analysis | Total | CR(%) | CRI(%) | Remission failure (%) |
|---|---|---|---|---|
| Total enrolled in S0703 | 133 | 35 (26) | 19 (14) | 79 (59) |
| DNA methylation analysisa | 84 | 25 (30) | 12 (14) | 47 (60) |
| RNA expression analysisa | 82 | 24 (29) | 12 (15) | 46 (56) |
| APAF1 promoter methylation complete cohort | ||||
| TIme points | N | Median change (IQR) | P-value | |
| Prestudy and day 3–4 | 84 | 7.46 (0.34, 6.35) | <0.01 | |
| Prestudy and day 7 | 83 | 4.22 (0.18, 2.53) | 0.095 | |
| HIC1 promoter methylation changes at day 3–4 | ||||
| Good risk group (N = 51) | Poor risk group (N = 31) | P-value | ||
| 1.56 (0.82, 2.83) | 0.96 (0.34, 1.79) | 0.021 |
Those analyzed required prestudy plus either day 3 or 4, or day 7 samples of sufficient quality available for analysis.
2.3. DNA methylation
Global DNA methylation was determined using a 5-mC-specific antibody capture ELISA-based method (MethylFlash Methylated DNA Quantification Kit, P-1034, Epigentek). DNA (100 ng) was analyzed in duplicate following the manufacturer’s protocol. Synthetic DNA containing known percentages of cytosine and 5-methylcytosine were used as controls. Methylated DNA status was quantified as a percentage of 5-methylcytosine in total DNA.
For gene-specific promoter DNA methylation, DNA was bisulfite converted using the EpiTect Bisulfite Conversion Kit (59104, Qiagen) following the manufacturer’s protocol. Methylation status of APAF1, CDH1, CDKN2A, CDKN2B, HIC1, and RARB promoter regions of bisulfite converted DNA was quantified using SYBR Green qPCR and normalized to COL2A1 using a primer probe combination with TaqMan Universal qPCR mix (4440040, Applied Biosystems). Briefly, 25–50 ng of bisulfite converted DNA was combined with methylation-specific primers (500 nM) and SYBR Green mix or with probe (100 nM) and TaqMan mix in a final volume of 20 µL Primer sequences (Supplemental Table 1) were previously published (citations in Supplemental Table 1). Reactions were performed in duplicate using a 7300 Real Time PCR thermocycler (Applied Biosystems). The mean Ct value for each promoter was normalized to COL2A1 (dCt). Fold change in promoter methylation was calculated using the 2−ΔΔCt method [32]. To ensure that the primers used were methylation-specific, DNA from a healthy donor was subjected to whole genome amplification (WGA) using the REPLI-g Mini kit (150023, Qiagen) which results in non-methylated DNA. A portion of this WGA DNA was then in vitro methylated by incubation with the CpG methyltransferase M.SssI (M0226, New England Biolabs) in the presence of S-adenosylmethionine resulting in DNA in which all CpG cytosine residues are methylated. The WGA and in vitro methylated WGA DNA were then bisulfite converted and assessed with qPCR as above.
2.4. Gene expression
RNA was converted to cDNA using the High Capacity cDNA Synthesis Kit (4368813, Life Technologies) following the manufacturer’s protocol. TaqMan Gene Expression Assays (APAF1: Hs00559441_m1, CDH1: Hs01023894_m1, CDKN2A: Hs00923894_m1, CDKN2B: Hs00793225_m1, HIC1: Hs00359611_s1, RARB: Hs00977140_m1, GAPDH: Hs02758991_g1, Applied Biosystems) and TaqMan Universal qPCR Master Mix (4440040, Applied Biosystems) were used to determine gene expression. All assays were performed in triplicate. Relative expression was calculated using the 2−ΔΔCt method [32].
2.5. Statistical methods
Wilcoxon rank sum tests were used to evaluate changes in global DNA methylation from prestudy, DNA methylation fold-change, and gene expression fold-change. Kruskal-Wallis tests were used to compare changes in global DNA methylation from prestudy, DNA methylation fold-change, and gene expression fold-change across categories. Spearman’s rank correlation was used to quantify associations between DNA methylation fold-change and gene expression fold-change. Two-sided p-values are reported and the 0.05 significance level was used. For the data presented in boxplots, thick horizontal black lines indicates the median values, horizontal lines above and below the median indicate the 25% and 75% values. The interquartile range (IQR) is the difference between the 75% and 25% values. The upper “whisker” is drawn at the largest data point not more than 75% +IQR. Data points more 75% +IQR is marked with a circle. The lower “whisker” is drawn at the smallest value data point not less than 25%−IQR. Any data point less than 25% + IQR is marked with a circle. The range (minimum, maximum) are printed below the boxplots to indicate the full scale of the data.
3. Results
3.1. Global DNA methylation
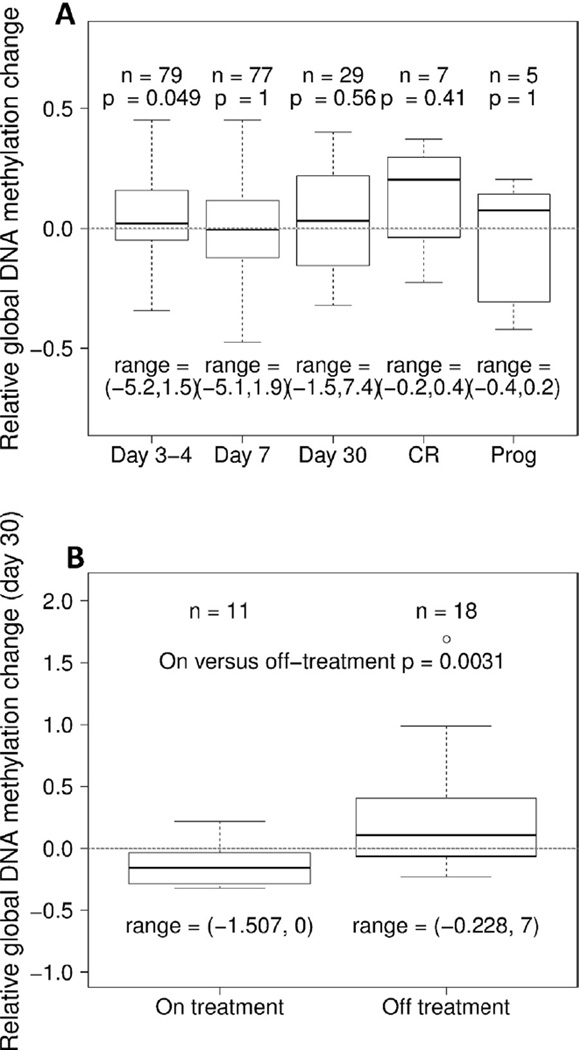
Total DNA methylation status was compared between DNA isolated from pre- versus post-treatment samples. The average global DNA methylation for the entire cohort was not significantly decreased at any time point following treatment initiation (Fig. 1A). A slight increase in global methylation was observed when comparing day 3–4 of treatment to prestudy samples that just reached statistical significance (p = 0.049). In addition, significantly decreased global DNA methylation was found in samples analyzed around one month after initiation of treatment (range 27–35 days) in patients on treatment as compared to those off treatment (Fig. 1B). This was not simply a reflection of those patients in CR at this time point; among those on treatment only 43% were in CR at the time of specimen collection, and those off treatment included patients who were in CR/CRi, those who later achieved CR/CRi, as well as those who never achieved a CR/CRi.
Fig. 1.
Global DNA methylation changes compared to prestudy samples. A Samples from the entire cohort. B Samples from around day 30, showing decreased global DNA methylation comparing those on treatment versus those off treatment. Box-plots show 25% and 75% values, and thick horizontal black lines indicate the median. Upper and lower whisker indicate the largest and smallest value data points not more or less than 75% + IQR and 25% − IQR, respectively. The range are printed below the boxplots to indicate the full scale of the data.
3.2. Promoter DNA methylation
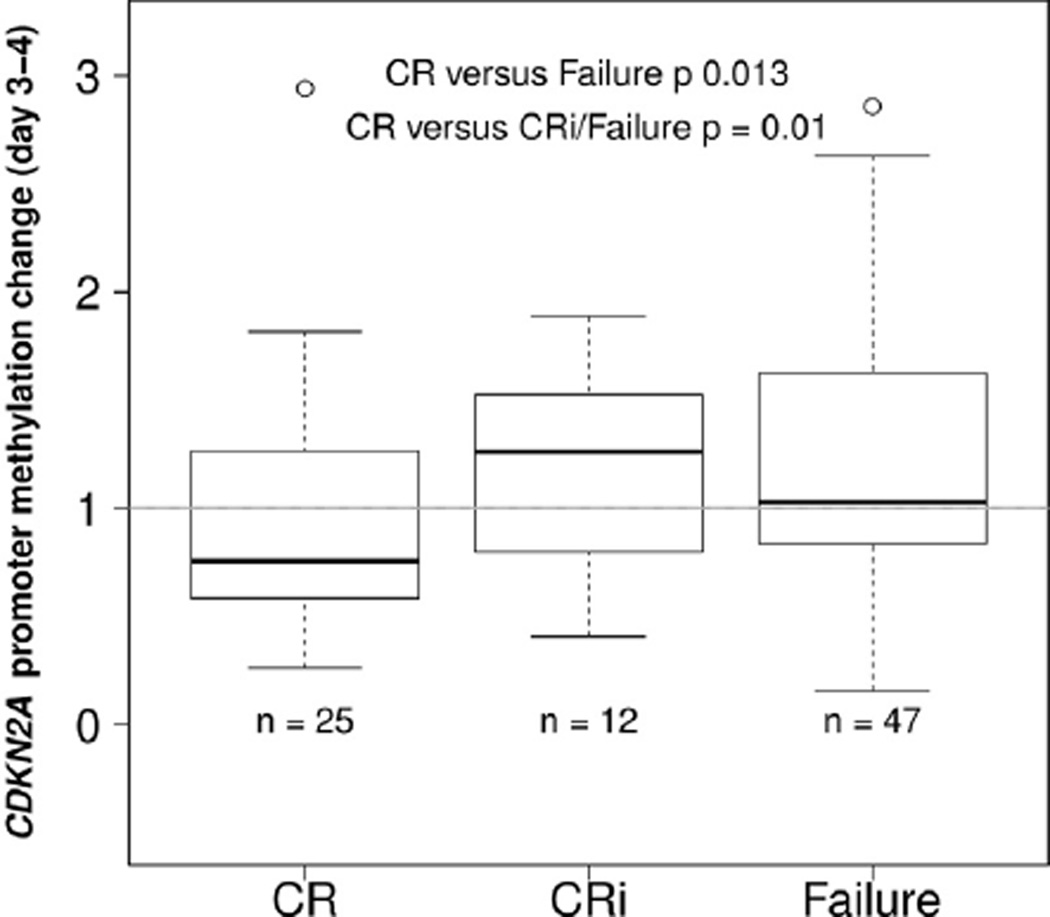
DNA methylation changes in the promoter regions of six genes, CDKN2A, CDKN2B, HIC1, RARB, CDH1 and APAF1 were determined relative to prestudy levels. A significant decrease in APAF1 methylation was found in day 3–4 samples regardless of patient response (Table 1). However, more selective changes in promoter DNA methylation were found for most of the other analyzed genes. Samples from days 3–4 after start of treatment showed significantly decreased CDKN2A promoter methylation in patients achieving CR as compared to those with failure to achieve CR (Fig. 2). In patients achieving CR the average CDKN2A promoter methylation was decreased compared to prestudy (median fold-change = 0.76), while in patients achieving CRi the average CDKN2A promoter methylation was increased compared to prestudy (median fold-change = 1.26). Among patients who were remission failures, the average CDKN2A promoter methylation was similar compared to prestudy (median fold-change = 1.03) (Fig. 2).
Fig. 2.
CDKN2A promoter methylation changes at Day 3–4 comparing patients who later achieved CR, CRi, or who had treatment failure. Patients who later achieved CR had significantly decreased CDKN2A promoter methylation by Day 3–4. Boxplots show 25% and 75% values, and thick horizontal black lines indicate the median. Upper and lower whisker indicate the largest and smallest value data points not more or less than 75% + IQR and 25% − IQR, respectively. The range are printed below the boxplots to indicate the full scale of the data.
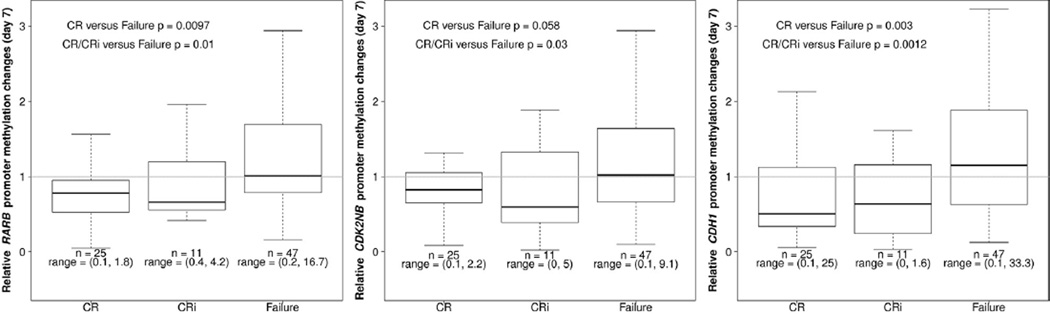
Samples from day 7 after start of treatment showed significantly decreased RARB promoter methylation in patients achieving CR and/or CRi as compared to those with failure to achieve CR (Fig. 3). Similarly, day 7 samples showed significantly decreased CDKN2B promoter methylation in patients achieving CR and/or CRi as compared to those who failed to achieve CR (Fig. 3). For the CDH1 gene, patients who later achieved CR and/or CRi also showed decreased promoter methylation by day 7 (Fig. 3). There were no significant findings for HIC1 promoter methylation (data not shown).
Fig. 3.
Gene-specific promoter methylation changes at Day 7, comparing patients who later achieved CR; CRi; or who had treatment failure for A RARB, B CDKN2B, and C CDH1. Each of these promoters demonstrated significantly decreased promoter methylation by Day 7 in patients who later achieved CR or CRi. Boxplots show 25% and 75% values, and thick horizontal black lines indicate the median. Upper and lower whisker indicate the largest and smallest value data points not more or less than 75% + IQR and 25% − IQR, respectively. The range are printed below the boxplots to indicate the full scale of the data.
3.3. Gene expression
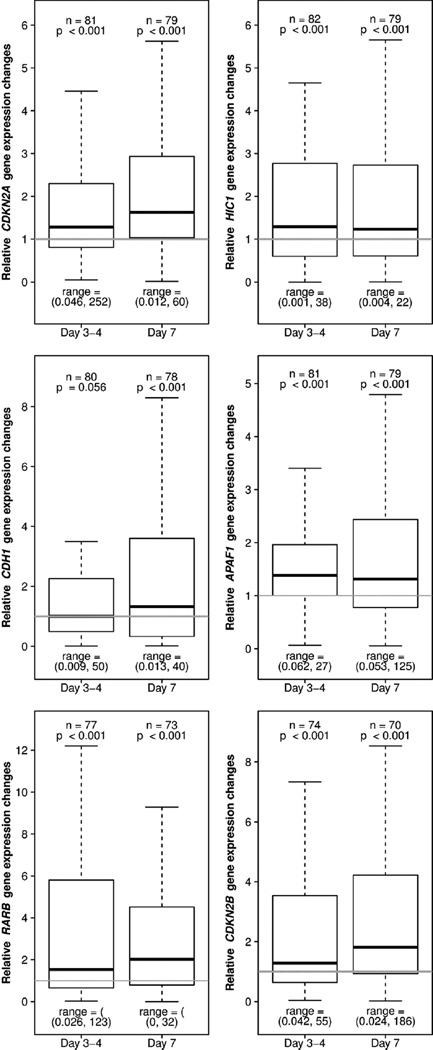
The same genes analyzed for DNA promoter methylation were also assessed for RNA expression using Taqman-based quantitative RT-PCR. CDKN2A, CDKN2B, RARB, HIC1 and APAF1 expression were significantly increased in the samples from days 3–4 and from day 7 after start of therapy in the entire patient cohort analyzed (Fig. 4), but did not distinguish patients’ response to therapy. Similarly, CDH1 expression was increased in samples from all patients, but only achieved statistical significance at day 7. However, a significant increase in HIC1 expression at day 3–4 was found in good risk patients as compared to poor risk patients (Table 1).
Fig. 4.
Gene expression changes at days 3–4 and at day 7 compared to prestudy for (A) CDKN2A, CDKN2B, RARE, APAF1, CDH1 and HIC1 in the entire cohort. Boxplots show 25% and 75% values, and thick horizontal black lines indicate the median. Upper and lower whisker indicate the largest and smallest value data points not more or less than 75% + IQR and 25% − IQR, respectively. The range are printed below the boxplots to indicate the full scale of the data.
4. Discussion
Here we report DNA methylation and gene expression analyses in peripheral blood samples from a phase 2 trial of older patients with AML treated with azacitine and GO therapy. We demonstrate that as early as day 3–4 following DNA methylation inhibitor treatment, decreases in gene-specific DNA methylation can be detected. Consistent with other studies [24,33], this suggests that early epigenetic changes are found after initiation of DNA demethylation therapy. However, in the current study, early changes are in the order of days as compared to weeks in most previous studies. Furthermore, peripheral blood is analyzed in the current study, without need for additional bone marrow sampling. Of the genes analyzed, there were several DNA methylation changes that distinguished patients who would later respond to therapy from those who would not as early as day 3–4 (for CDKN2A) or day 7 (for RARB, CDKN2B and CDH1). These data are consistent with those of Claus et al., who analyzed very early DNA methylation changes of candidate gene promoters in peripheral blood blasts isolated from eight AML patients treated with decitabine [33]. They found increased CDH1 and CDKN2B methylation in AML samples prior to treatment compared to normal hematopoietic precursor cells. Although they did not observe statistically significant decreases in these two genes post-decitabine, perhaps due to the smaller number of patients’ samples analyzed, they did observe other significant gene-specific methylation changes by one week of treatment. In our study using peripheral blood samples without cell fractionation, we found very early gene-specific DNA methylation changes which are prognostic for response. Thus, gene-specific promoter methylation analyses on a limited number of genes with comparison to the same patient’s prestudy sample, could be performed quickly and easily from peripheral blood samples, which could be of benefit to the patient.
Our findings also appear to possibly distinguish between patients with CR and CRi. In the case of CDKN2A, the promoter DNA methylation seen in CRi was similar to non-responders. Often, reports of clinical results combine CR and CRi. Our results indicate that these two categories of response may indeed be different and should continue to be reported separately.
In this trial, unseparated samples were used for DNA and RNA analyses. Thus, the exact cell populations studied may have differed from sample to sample. This same caveat is true for many other large studies, and it is possible that very profound changes in cell populations within a very short time-frame (3–7 days) are reflected by significant methylation or expression changes. Although the current results leave unanswered the question of whether these changes were happening in specific lineages, they do not diminish the importance of the findings. It is precisely the less invasive and relatively simple sample preparation prior to analysis which suggests that critical predictive information may be relatively straightforward. Here, we have identified specific genes whose promoter methylation may be useful as early prognostic indicators of response. Future comparison of the current genes to other established outcome predictors and across other clinical trials will determine broad applicability of these specific genes. More importantly, however, our study provides proof-of-principle that information obtained from peripheral blood samples a few days after treatment initiation may be impactful.
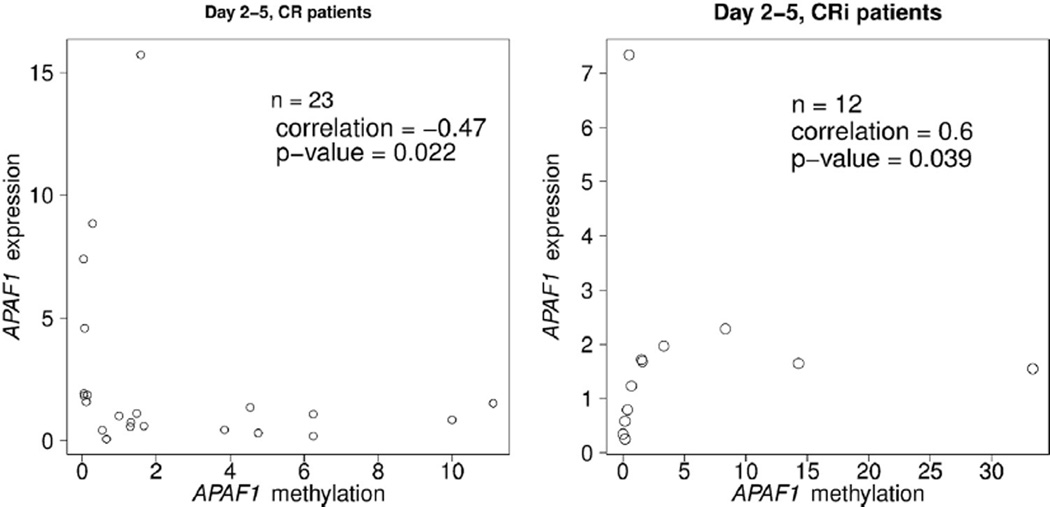
Promoter DNA methylation is often associated with gene silencing. Changes in promoter DNA methylation can result in gene expression changes; however, this is not universally true due to additional regulatory mechanisms that also influence gene expression. Of the genes assessed in this study, there was a significant negative correlation (r = −0.47) between APAF1 promoter methylation and APAF1 gene expression between pre-study and day 3 or 4 samples among patients who later went on to achieve CR, with decreased promoter methylation correlated with increased expression (Fig. 5). The genes analyzed in the current study were chosen based on previous association with DNA methylation in AML at the time the clinical trial was designed, not for RNA expression association. It remains to be determined whether DNA methylation or RNA expression analyses could be more useful in the routine clinical setting, but less invasive and simplified sample processing may facilitate earlier patient stratification.
Fig. 5.
Negative correlation between APAF1 promoter methylation and APAF1 expression between day 3–4 and prestudy among patients who later achieved CR. The scatterplots indicate each patient’s DNA methylation fold-change and gene expression fold-change. Each patient is represented by one circle.
Several large-scale DNA methylation analyses of AML patient samples have been reported recently. As part of The Cancer Genome Atlas (TCGA) Research Network, Ley et al. analyzed 192 samples selected from adults with de novo AML, using Methylation Bead-Chip profiling [34]. Methylation differences were found between different subtypes of AML as compared to normal CD34 + CD38-cells [34]. A large cohort of 623 cytogenetically normal de novo AML samples were analyzed for methylation of the CEBPA promoter and found no association between promoter methylation and prognosis [35]. Recently, two unappreciated subgroups were identified in samples from 344 AML patients analyzed for gene expression and DNA methylation using genome-wide methods in combination with determination of mutations in splicing factor genes [36]. Promoter methylation status of four genes, ALOX12, HS3ST2, GSTM1 and FZD9, was determined in 127 AML patients (median age: 59 years). Methylated GSTM1 was associated with worse overall survival and disease-free survival; and those with both GSTM1 and ALOX12 promoter methylation had worse outcomes compared to all AML tested [37]. It is not yet apparent whether these findings will be directly relevant to elderly AML patients.
Other recent studies have focused on the elderly AML patient population. Methylation status of the O6-methylguanine-DNA methyltransferase (MGMT) promoter, whose expression correlates with resistance to the alkylating agent temozolomide [38], was recently used to stratify the dose given to elderly AML patients [39]. Levels of the microRNA miR-29b, which targets multiple DNA methyltransferases [40], were assessed in pre-treatment samples from a Phase 2 single agent decitabine trial for older AML patients. Higher levels of miR-29b were significantly associated with clinical response in this elderly patient cohort [41]. However, compared to studies which analyze a single diagnostic sample, additional insight may be gained from analysis of sequential patient samples during therapy, as in the current study.
5. Conclusions
Treatment of acute myeloid leukemia (AML) in older patients remains a therapeutic challenge, in part due to relatively poor performance status and other medical comorbidities. The present study analyzed sequential patient samples acquired as part of a phase II SWOG clinical trial of older AML patients treated with azacitidine. We show that azacitidine therapy in older patients with AML induces early DNA demethylation changes in specific promoters; and that these changes are associated with clinical outcomes. Thus, relatively straightforward gene-specific DNA methylation analysis of peripheral blood samples may help very early identification of those older AML patients who are most likely to benefit from demethylating agent therapy. Such information may help decrease toxicity and improve efficacy of therapy in these patients.
Supplementary Material
Acknowledgments
This study was supported in part by the National Institutes of Health (NIH), National Cancer Institute (NCI), National Clinical Trials Network (NCTN) grants CA180888, CA180819, CA180801, CA180835, CA180828, CA180846; NIH/NCI SWOG Biorepository grant CA114748; NCI Community Oncology Research Program (NCORP): CA189848, CA045807, CA189856, CA189853, CA189808, CA189858, CA189860, CA189971, CA189954, CA189872; and in part by Celgene Corporation and Pfizer, Inc.
Footnotes
Authorship contributions
NJA was involved in experimental design, performed experiments, analyzed data and wrote the manuscript; KP performed experiments, analyzed data and wrote the manuscript; SZ performed experiments and analyzed data; MO and KC analyzed data and wrote the manuscript; JEG, FRA and JPR were involved in study design; HPE was involved in study design and analyzed data; SN and NJZ-L were involved in study design, analyzed data and wrote the manuscript.
Conflict of interest
There are no conflicts of interest to disclose.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.Org/10.1016/j.leukres.2016.01.004.
References
- 1.Appelbaum FR, Gundacker H, Head DR, Slovak ML, Willman CL, Godwin JE, et al. Age and acute myeloid leukemia. Blood. 2006 May 9;107:3481–3485. doi: 10.1182/blood-2005-09-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wheatley K, Brookes CL, Howman AJ, Goldstone AH, Milligan DW, Prentice AG, et al. Prognostic factor analysis of the survival of elderly patients with AML in the MRC AML11 and LRF AML14 trials. Br. J. Haematol. 2009 Jun 5;145:598–605. doi: 10.1111/j.1365-2141.2009.07663.x. [DOI] [PubMed] [Google Scholar]
- 3.Lowenberg B, Ossenkoppele GJ, van Putten W, Schouten HC, Graux C, Ferrant A, et al. High-dose daunorubicin in older patients with acute myeloid leukemia. N. Engl. J. Med. 2009 Sep 13;361:1235–1248. doi: 10.1056/NEJMoa0901409. [DOI] [PubMed] [Google Scholar]
- 4.Rowe JM, Tallman MS. How I treat acute myeloid leukemia. Blood. 2010 Oct 17;116:3147–3156. doi: 10.1182/blood-2010-05-260117. [DOI] [PubMed] [Google Scholar]
- 5.Becker PS, Medeiros BC, Stein AS, Othus M, Appelbaum FR, Forman SJ, et al. G-CSF priming, clofarabine, and high dose cytarabine (GCLAC) for upfront treatment of acute myeloid leukemia, advanced myelodysplastic syndrome or advanced myeloproliferative neoplasm. Am. J. Hematol. 2014 Dec;:24. doi: 10.1002/ajh.23927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quesnel B, Guillerm G, Vereecque R, Wattel E, Preudhomme C, Bauters F, et al. Methylation of the p15(INK4b) gene in myelodysplastic syndromes is frequent and acquired during disease progression. Blood. 1998 Apr 8;91:2985–2990. [PubMed] [Google Scholar]
- 7.Figueroa ME, Skrabanek L, Jiemjit Li Y, Fandy A, Paietta TE, et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009 Oct 16;114:3448–3458. doi: 10.1182/blood-2009-01-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao X, Yang F, Li S, Liu M, Ying S, Jia X, et al. CpG island methylator phenotype of myelodysplastic syndrome identified through genome-wide profiling of DNA methylation and gene expression. Br. J. Haematol. 2014 Jun 5;165:649–658. doi: 10.1111/bjh.12811. [DOI] [PubMed] [Google Scholar]
- 9.Itzykson R, Fenaux P. Epigenetics of myelodysplastic syndromes. Leukemia. 2014 Mar 3;28:497–506. doi: 10.1038/leu.2013.343. [DOI] [PubMed] [Google Scholar]
- 10.Bravo GM, Lee E, Merchan B, Kantarjian HM, Garcia-Manero G. Integrating genetics and epigenetics in myelodysplastic syndromes: advances in pathogenesis and disease evolution. Br. J. Haematol. 2014 Sep 5;166:646–659. doi: 10.1111/bjh.12957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joeckel TE, Lubbert M. Clinical results with the DNA hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in patients with myelodysplastic syndromes: an update. Semin. Hematol. 2012 Oct 4;49:330–341. doi: 10.1053/j.seminhematol.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Zeidan AM, Linhares Y, Gore SD. Current therapy of myelodysplastic syndromes. Blood Rev. 2013 Sep 5;27:243–259. doi: 10.1016/j.blre.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006 Jun 12;66:6361–6369. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 14.Yang AS, Doshi KD, Choi SW, Mason JB, Mannari RK, Gharybian V, et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006 May 10;66:5495–5503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- 15.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010 Jan 1;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002 Aug;21:5400–5413. doi: 10.1038/sj.onc.1205651. (35) [DOI] [PubMed] [Google Scholar]
- 17.Costello J-F, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 2000 Febueary;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 18.Shimamoto T, Ohyashiki JH, Ohyashiki K. Methylation of p15(INK4b) and E-cadherin genes is independently correlated with poor prognosis in acute myeloid leukemia. Leuk. Res. 2005 Jun 6;29:653–659. doi: 10.1016/j.leukres.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 19.Schmelz K, Wagner M, Dorken B. Tamm 1.5-Aza-2′-deoxycytidine induces p21WAF expression by demethylation of p73 leading to p53-independent apoptosis in myeloid leukemia. Int. J. Cancer. 2005 May 5;114:683–695. doi: 10.1002/ijc.20797. [DOI] [PubMed] [Google Scholar]
- 20.Furukawa Y, Sutheesophon K, Wada T, Nishimura M, Saito Y, Ishii H, et al. Methylation silencing of the Apaf-1 gene in acute leukemia. Mol. Cancer Res. 2005 Jun 6;3:325–334. doi: 10.1158/1541-7786.MCR-04-0105. [DOI] [PubMed] [Google Scholar]
- 21.Melki JR, Vincent PC, Brown RD, Clark SJ. Hypermethylation of E-cadherin in leukemia. Blood. 2000 May 10;95:3208–3213. [PubMed] [Google Scholar]
- 22.Melki JR, Vincent PC, Clark SJ. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res. 1999 Aug 15;59:3730–3740. [PubMed] [Google Scholar]
- 23.Cechova H, Lassuthova P, Novakova L, Belickova M, Stemberkova R, Jencik J, et al. Monitoring of methylation changes in 9p21 region in patients with myelodysplastic syndromes and acute myeloid leukemia. Neoplasma. 2012;59(2):168–174. doi: 10.4149/neo_2012_022. [DOI] [PubMed] [Google Scholar]
- 24.Yan P, Frankhouser D, Murphy M, Tam HH, Rodriguez B, Curfman J, et al. Genome-wide methylation profiling in decitabine-treated patients with acute myeloid leukemia. Blood. 2012 Sep 12;120:2466–2474. doi: 10.1182/blood-2012-05-429175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burnett AK, Russell NH, Hills RK, Kell J, Freeman S, Kjeldsen L, et al. Addition of gemtuzumab ozogamicin to induction chemotherapy improves survival in older patients with acute myeloid leukemia. J. Clin. Oncol. 2012 Nov;30:3924–3931. doi: 10.1200/JCO.2012.42.2964. (32) [DOI] [PubMed] [Google Scholar]
- 26.Castaigne S, Pautas C, Terre C, Raffoux E, Bordessoule D, Bastie JN, et al. Effect of gemtuzumab ozogamicin on survival of adult patients with de-novo acute myeloid leukaemia (ALFA-0701): a randomised, open-label, phase 3 study. Lancet. 2012 Apr;379:1508–1516. doi: 10.1016/S0140-6736(12)60485-1. (9825) [DOI] [PubMed] [Google Scholar]
- 27.Petersdorf SH, Kopecky KJ, Slovak M, Willman C, Nevill T, Brandwein J, et al. A phase 3 study of gemtuzumab ozogamicin during induction and postconsolidation therapy in younger patients with acute myeloid leukemia. Blood. 2013 Jun 24;121:4854–4860. doi: 10.1182/blood-2013-01-466706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renneville A, Abdelali RB, Chevret S, Nibourel O, Cheok M, Pautas C, et al. Clinical impact of gene mutations and lesions detected by SNP-array karyotyping in acute myeloid leukemia patients in the context of gemtuzumab ozogamicin treatment: results of the ALFA-0701 trial. Oncotarget. 2014 Feburary;5:916–932. doi: 10.18632/oncotarget.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Xu SN, Oin DB, Tan Y, Gong Q, Chen JP. Effect of adding gemtuzumab ozogamicin to induction chemotherapy for newly diagnosed acute myeloid leukemia: a meta-analysis of prospective randomized phase III trials. Ann. Oncol. 2014 Feburary;25:455–461. doi: 10.1093/annonc/mdt566. [DOI] [PubMed] [Google Scholar]
- 30.Hills RK, Castaigne S, Appelbaum FR, Delaunay J, Petersdorf S, Othus M, et al. Addition of gemtuzumab ozogamicin to induction chemotherapy in adult patients with acute myeloid leukaemia: a meta-analysis of individual patient data from randomised controlled trials. Lancet Oncol. 2014 Aug 9;15:986–996. doi: 10.1016/S1470-2045(14)70281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nand S, Othus M, Godwin JE, Willman CL, Norwood TH, Howard DS, et al. A phase 2 trial of azacitidine and gemtuzumab ozogamicin therapy in older patients with acute myeloid leukemia. Blood. 2013 Nov 20;122:3432–3439. doi: 10.1182/blood-2013-06-506592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔct method. Methods. 2001 Dec 4;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Claus R, Pfeifer D, Almstedt M, Zucknick M, Hackanson B, Plass C, et al. Decitabine induces very early in vivo DNA methylation changes in blasts from patients with acute myeloid leukemia. Leuk. Res. 2013 Feburary;37:190–196. doi: 10.1016/j.leukres.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network, Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013 May 22;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fasan A, Alpermann T, Haferlach C, Grossmann V, Roller A, Kohlmann A, et al. Frequency and prognostic impact of CEBPA proximal, distal and core promoter methylation in normal karyotype AML: a study on 623 cases. PLoS One. 2013;8(2):e54365. doi: 10.1371/journal.pone.0054365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taskesen E, Havermans M, van Lom K, Sanders MA, van Norden Y, Bindels E, et al. Two splice-factor mutant leukemia subgroups uncovered at the boundaries of MDS and AML using combined gene expression and DNA-methylation profiling. Blood. 2014 May 21;123:3327–3335. doi: 10.1182/blood-2013-07-512855. [DOI] [PubMed] [Google Scholar]
- 37.Ohgami RS, Ma L, Ren L, Weinberg OK, Seetharam M, Gotlib JR, et al. DNA methylation analysis of ALOX12 and GSTM1 in acute myeloid leukaemia identifies prognostically significant groups. Br. J. Haematol. 2012 Oct 2;159:182–190. doi: 10.1111/bjh.12029. [DOI] [PubMed] [Google Scholar]
- 38.Baer JC, Freeman AA, Newlands ES, Watson AJ, Rafferty JA, Margison GP. Depletion of O6-alkylguanine-DNA alkyltransferase correlates with potentiation of temozolomide and CCNU toxicity in human tumour cells. Br. J. Cancer. 1993 Jun 6;67:1299–1302. doi: 10.1038/bjc.1993.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medeiros BC, Kohrt HE, Gotlib J, Coutre SE, Zhang B, Arber DA, et al. Tailored temozolomide therapy according to MGMT methylation status for elderly patients with acute myeloid leukemia. Am. J. Hematol. 2012 Jan 1;87:45–50. doi: 10.1002/ajh.22191. [DOI] [PubMed] [Google Scholar]
- 40.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009 Jun 25;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blum W, Garzon R, Klisovic RB, Schwind S, Walker A, Geyer S, et al. Clinical response and miR-29b predictive significance in older AML patients treated with a 10-day schedule of decitabine. Proc. Natl. Acad. Sci. U. S. A. 2010 Apr 16;107:7473–7478. doi: 10.1073/pnas.1002650107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.