Abstract
LincRNAs are long non-coding transcripts (>200 nt) from the intergenic regions of annotated protein-coding genes. One of the most highly induced lincRNAs in macrophages upon TLR ligation is lincRNA-Cox2, which has recently been shown to mediate both the activation and repression of distinct classes of immune genes in innate immune cells. We report here that lincRNA-Cox2 located at chromosome 1 proximal to the prostaglandin-endoperoxide synthase 2 (Ptgs2/Cox2) gene is an early-primary inflammatory gene controlled by NF-κB signaling in murine macrophages. Functionally, lincRNA-Cox2 is required for the transcription of NF-κB-regulated late-primary inflammatory response genes stimulated by bacterial lipopolysaccharide. Specifically, lincRNA-Cox2 is assembled into the SWI/SNF (SWItch/Sucrose NonFermentable) complex in cells after lipopolysaccharide stimulation. This resulting lincRNA-Cox2/SWI/SNF complex can modulate the assembly of NF-κB subunits to the SWI/SNF complex, and ultimately, SWI/SNF-associated chromatin remodeling and transactivation of the late-primary inflammatory response genes in macrophages in response to microbial challenge. Therefore, our data indicate a new regulatory role of NF-κB-induced lincRNA-Cox2 to act as a co-activator of NF-κB for the transcription of late-primary response genes in innate immune cells through modulation of epigenetic chromatin remodeling.
Keywords: LincRNAs, LincRNA-Cox2, Macrophages, NF-κB, SWI/SNF complex, Ccl5, Saa3, Inflammation, Histone modifications
INTRODUCTION
The transcription factor nuclear factor-κB (NF-κB) is an essential regulator of inflammatory and immune responses (1). Dysregulation of NF-κB signaling has been linked to cancer, inflammatory and autoimmune diseases, infectious diseases, and improper immune development (2,3). NF-κB is composed of homo- or hetero-dimeric complexes of NF-κB subunits, which include RelA (p65), RelB, c-Rel, p50, and p52 in humans (1). NF-κB signaling can be activated via two distinct NF-κB signal transduction pathways, the so-called canonical pathway (as stimulated by lipopolysaccharide [LPS] and tumor necrosis factor-α [TNFα]) and non-canonical pathway (e.g., stimulated by lymphotoxin B) (1). In addition to its initial cytoplasmic activation, both the recruitment of NF-κB to target genes in the nuclei and NF-κB-induced transcriptional events after recruitment are finely controlled events to ensure proper transactivation of NF-κB target genes (2).
Several waves of gene transcription, broadly categorized as the early-primary (e.g., Tnfa, Cxcl2, Ptgs2, Il1b), the late-primary (e.g., Ccl5, Saa3, Ifnb1), and the secondary response genes (e.g., Il6, Il12b, Nos2, Marco), have been demonstrated in macrophages following LPS stimulation (4-6). The early-primary response genes have promoters constitutively permissive for transcription (4,7). The molecular mechanisms underlying the transcription of late-primary and secondary response genes are unclear and may require synthesis of additional molecules and/or chromatin-remodeling triggered by NF-κB activation (4,8). Indeed, transcription of the secondary response genes, but not the late-primary genes, has been shown to require new protein synthesis (4,6). Promoter recruitment of the ATP-dependent SWItch/Sucrose NonFermentable (SWI/SNF) complex has been demonstrated in the transcription of late-primary and secondary response genes following NF-κB activation (5,9). The SWI/SNF complex is a nucleosome remodeling complex composed of several proteins encoded by the SWI and SNF genes (e.g., SWIs, Brg1, or Brm) (9,10). The SWI/SNF complex has DNA-stimulated ATPase activity and can destabilize histone-DNA interactions in reconstituted nucleosomes in an ATP-dependent manner (9,11). How NF-κB signaling controls dynamic gene transcription through SWI/SNF-mediated chromatin remodeling remains elusive.
Long intergenic ncRNAs (lincRNAs) are long non-coding transcripts (>200 nt) from the intergenic regions of annotated protein-coding genes (12). Several thousand lincRNAs have been identified in the mouse genome (13,14). These lincRNAs are functional but have less evolutionary conservation than protein-coding genes (15). LincRNAs may regulate gene transcription in cis by recruiting protein complexes to the site of transcription and creating a locus-specific address (16). It has been speculated that lincRNAs may also regulate distantly located genes in trans (17). The recent discovery of lincRNAs in association with specific chromatin modification complexes, including the SWI/SNF complex (18,19), suggests a role for lincRNAs in managing chromatin states in a gene-specific fashion. Additionally, lincRNAs have been associated with human inflammatory diseases, neurological disorders, and tumorigenesis (19-22).
LincRNAs may be critical mediators of NF-κB-regulated gene transcription and participate in the pathogenesis of various inflammatory diseases and, thus, be targets for therapeutic interventions. Increasing evidence indicates that lincRNAs can be induced in innate immune cells and may act as key regulators of the inflammatory response (13,23-26). LincRNAs are differentially regulated in virus-infected cells (27) and in dendritic cells or macrophages following stimulation by ligands for TLR4 (LPS) and TLR2 (Pam3CSK4) (23,24,26). LincRNA-Cox2 is one of the most highly induced lincRNAs in studies examining the innate immune response (23,24,26). The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus (28). Pattern recognition receptors such as the Toll-like receptors (TLRs) induce the expression of lincRNA-Cox2 (23). Intriguingly, lincRNA-Cox2 has been shown to mediate both the activation and repression of distinct classes of immune genes (23,26). Functionally, lincRNA-Cox2 has been demonstrated to interact with other RNA-binding proteins, including heterogeneous ribonucleoprotein (hnRNP) A/B and hnRNA-A2/B1 (23). Transcriptional repression of target genes is dependent on interactions of lincRNA-Cox2 with hnRNP-A/B and hnRNP-A2/B1 (23). We recently reported that lincRNA-Cox2 suppresses TNF-α-induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi-2/NuRD-mediated epigenetic histone modifications (29). How lincRNA-Cox2 may mediate transcriptional activation of target genes in innate immune cells upon TLR ligation is still unclear.
In the work described here, we demonstrated that lincRNA-Cox2 is an early-primary gene controlled by the NF-κB signaling in macrophages and microglia. LincRNA-Cox2 transcript is assembled into the SWI/SNF complex in both macrophages and microglia in response to LPS stimulation. This resulting lincRNA-Cox2/SWI/SNF complex can modulate SWI/SNF-associated chromatin remodeling and, consequently, transcription of late-primary response genes in cells following LPS stimulation or microbial challenge. Therefore, our data indicate a new regulatory role of lincRNA-Cox2 for the NF-κB-controlled transcription of late-primary response genes in innate immune cells.
MATERIALS AND METHODS
Cells and cell culture
The BV2 mouse microglia cells and RAW264.7 mouse macrophage cells were obtained from ATCC. Culture media were supplied with 10% FBS (Ambion) and antibiotics (100 IU/ml of penicillin and 100 μg/ml of streptomycin).
Nuclear and cytoplasmic extracts and Western blot
Cells were trypsinized with trypsin-EDTA (Sigma), washed with PBS, and the cell pellet was resuspended in 1 ml of cold buffer A (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT). Nuclear pellets were isolated from the whole cell protein by centrifugation at 14,000 rpm for 1 min at 4°C and resuspended in two-thirds packed cell volume of cold buffer B (20 mM HEPES, 1.5 mM MgCl2, 25% glycerol, 420 mM NaCl, 0.2 mM EDTA, 1 mM DTT) with vigorous agitation in the cold room for 30 min. The supernatant containing nuclear proteins was collected for Western blot. The following antibodies were used for blotting: anti-RelA (Santa Cruz), anti-IκBα (Cell Signaling), anti-PARP (Cell Signaling), anti-Actin (Sigma-Aldrich), and anti-p50 (Santa Cruz).
Mice with LPS injection and Theiler’s murine encephalomyelitis virus (TMEV) infection
Male C57BL/6J mice (4-6 weeks old, The Jackson Laboratory, Bar Harbor, ME) were injected with LPS (10 mg/kg body weight, intraperitoneal) and peritoneal macrophages collected and cultured as previously reported (30). Cells isolated from mice following PBS administration were used as the control. For TMEV infection studies, female FVB/n mice (4-6 weeks old, The Jackson Laboratory) were anesthetized with isofluorane and injected with 2 × 105 plaque forming units of TMEV (DA strain) in a total volume of 10 μl via an intracerebral (i.c.) injection. Mice were monitored daily until time of sacrifice with an overdose of sodium pentobarbital (Sleepaway, Kalamazoo, MI). At 7 and 42 days after TMEV infection, the brains and spinal cords from 3-5 mice were collected for biochemical analysis or morphological observation. Control mice were injected with 10 μl HBSS (virus diluent) i.c. All animal experiments were done in accordance with procedures approved by the Institutional Animal Care and Use Committee of Creighton University.
siRNAs and plasmids
Two siRNAs targeted separate sequence of lincRNA-Cox2 were synthesized by Exiqon and used to knockdown lincRNA-Cox2 in cells: lincRNA-Cox2-siRNA-A (Sense sequence: GCCCUAAUAAGUGGGUUGUUU) and lincRNA-Cox2-siRNA-B (Sense sequence: AAGAGUAAGAUUCUGAAGAUCUU). LincRNA-Cox2-siRNA-A targets the 1008-1027 nt of lincRNA-Cox2 sequence (NR_110420.1). LincRNA-Cox2-siRNA-B targets the 558-580 nt of lincRNA-Cox2 as in the study by the Fitzgerald group (23). Sense sequence of siRNA for MyBBP1A is CCGGAGUGTAUUUGGUCAUAUCUUU and non-specific scrambled sequence UUCUCCGAACGUGUCACGUUU synthesized by Exiqon as for the control. The lincRNA-Cox2 expression vector was generated by RT-PCR amplification of lincRNA-Cox2 cDNA, using RNA from BV2 cells and cloned into the pcDNA3.3-TOPO vector (Life Technologies). Plasmids for luciferase reporter assay were generated by PCR amplification of DNA from BV2 cells using primers with built in Mlu I and Xho I restriction sites, allowing directional cloning into the pGL3-CMV reporter construct (Promega). The genomic coordinates of the luciferase reporter inserts are listed in Table S1.
PCR and RACE PCR
For quantitative analysis of mRNA and lincRNA expression, comparative real-time PCR was performed using the SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA). 5′ and 3’-RACE PCR was utilized to identify the 5′ and 3’ end of lincRNA-Cox2 to localize the transcriptional start site. The SMART RACE cDNA Amplification Kit (Clontech) was used. The sequences for all used primers are listed in Table S1.
Microarray
The LS Science Agilent SurePrint G3 Mouse Gene Expression Microarray and service to process the samples were used for genome-wide analysis. Briefly, BV2 cells were grown to 80% confluence and exposed to LPS (1 μg/ml) for 4h. Total RNA was isolated with the RNeasy Mini kit (Qiagen) according to the manufacturer’s instruction (Ambion). The quality of isolated RNAs was verified by an Agilent 2100 Bioanalyzer profile.
Luciferase reporter assay
The promoter of lincRNA-Cox2 was amplified by PCR from mouse genomic DNA, using primers as listed in Table S1. Cells were transfected with each reporter construct for 24h and then exposed to LPS for 8h in the presence or absence of SC-514 (100 μm), followed by assessment of luciferase activity as previously reported (31).
RNA Stability
Cells were transfected with lincRNA-Cox2-siRNA-A or the control siNRA for 24h and exposed to LPS (1 μg/ml) for 4h. The relative abundance of each mRNA was measure by real-time PCR, calculated using the ΔΔCt method and normalized to GAPDH and the half-lives of the RNAs calculated as previously reported (32).
Restriction enzyme accessibility assay (REAA)
Experiments were performed as described previously (5,33,34). Briefly, purified DNA from cells was digested using the restriction enzyme EcoN I for Ccl5 and PstI for Saa3. The digested DNA was ligated to 2.5 μL of 40 μM double-stranded linker oligonucleotide (See Table S1). The diluted ligation reaction was amplified with a primer to the linker and a second primer upstream or downstream of the anticipated cleavage site (P1a and P2). PCR reactions were performed with the linker primer and specific primer (P1b and P2) (Table S1). The amount of cleavage at each promoter site in the control cells was set to 1.
in situ hybridization
Frozen tissue sections were treated with 10 μg/ml proteinase K (Roche) at 37°C for 10 min. After washing with PBS, slides were incubated with the hybridization buffer (50% formamide, 100 μg/ml salmon sperm DNA, 200 μg/ml yeast tRNA, 600 mM NaCL, 1×Denhardt's solution, 0.25% SDS, 1 mM EDTA) at 42°C for 1 h. Slides were then hybridized with 20 nM DIG-labeled lincRNA-Cox2 probe (Exiqon) diluted in the hybridization buffer at 42°C overnight. Slides were incubated with anti-DIG-POD Fab fragments (Roche) at 4°C overnight, and lincRNA-Cox2 was visualized in a staining reaction with Renaissance Tyramide Signal Amplification Fluorescence Systems (PerkinElmer). A negative control (i.e., staining without lincRNA-Cox2 probe) was included.
Immunoprecipitation (IP) and Co-IP
Cell pellets were resuspended in 1 ml lysis buffer (10 mM Tris-HCl pH7.4, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40 and cocktail protease inhibitor) and nuclei were collected by centrifugation (500 g, 5 min) with the lysis buffer devoid of NP-40. A total of 600 μg nuclear protein was incubated with the primary antibodies at 4°C overnight to immunoprecipitate the protein complexes in the presence or absence of 100 micrograms per ml ethidium bromide. Immune complexes were collected by direct binding to protein A Agarose. The following antibodies were used for IP/Co-IP analysis: anti-RelA (Santa Cruz), anti-MyBBP1A (Santa Cruz), anti-BAF170 (Cell Signaling), anti-Brg1 (Santa Cruz), and anti-RelB (Santa Cruz).
RNA immunoprecipitation (RIP), chromatin immunoprecipitation (ChIP), and chromatin isolation by RNA purification (ChIRP) analyses
The formaldehyde crosslinking RIP was performed as described (35). Briefly, cells in culture were treated with formaldehyde at a final concentration of 0.3 % (v/v) at room temperature for 10 min. Crosslinking reactions were quenched by the addition of glycine (pH7.0) to a final concentration of 0.25 M. The cells were then harvested and nuclei pellets were collected. The crosslinked complexes were then incubated with the specific antibody–coated beads. Formaldehyde cross-links were reversed by incubation at 65 °C with rotation for 4h. Presence of RNA was measured by quantitative, strand-specific RT-PCR using the iCycler iQ Real-time detection system (BioRad). Gene-specific PCR primer pairs are listed in Table S1. The following antibodies were used for RIP analysis: anti-RelA (Santa Cruz), anti-p105/p50 (Abcam), anti-p100/p52 (Abcam), anti-p50 (Millipore), anti-Brg1 (Santa Cruz), anti-MyBBP1A (Santa Cruz).
ChIP analysis was performed with a commercially available ChIP Assay Kit (Upstate Biotechnologies) in accordance with the manufacturer's instructions. In brief, the chromatin fraction was immunoprecipitated overnight at 4°C using the antibodies. PCR amplification was performed in a total volume of 25 μl with specific primers. The percent input method was used to normalize the ChIP data. Signals obtained from the ChIP were divided by signals obtained from an input (1% of starting chromatin was as input). The forward and reverse primers used for each gene are listed in Table S1. The following antibodies were used for ChIP analysis: anti-RelA (Millipore), anti-Brg1 (Santa Cruz), anti-MyBBP1A (Santa Cruz), anti-H3K36me3 (Abcam), and anti-H3K4me3 (Abcam).
ChIRP analysis was performed as previously reported (36). Briefly, a pool of tiling oligonucleotide probes with affinity specific to the lincRNA-Cox2 sequence was used and glutaraldehyde cross-linked for chromatin isolation. The sequences for each probe are listed in Table S1; probe 1, 3 and 5 are mixed as the probe pool A and probe 2, 4 and 6 as the probe pool B. The DNA sequences of the chromatin immunoprecipitates were confirmed and quantified by real-time PCR using the same primer sets covering the gene promoter regions of interest as for ChIP analysis. A pool of oligo probes and primers for LacZ were served as controls. The percent input method was used to normalize the ChIRP data.
Statistical analysis
All values are given as mean ± S.E. Means of groups were from at least three independent experiments and compared with Student’s t test (unpaired) or the ANOVA test when appropriate. p values < 0.05 were considered statistically significant.
RESULTS
Expression of LincRNAs in Microglia and Macrophages in Response to LPS Stimulation
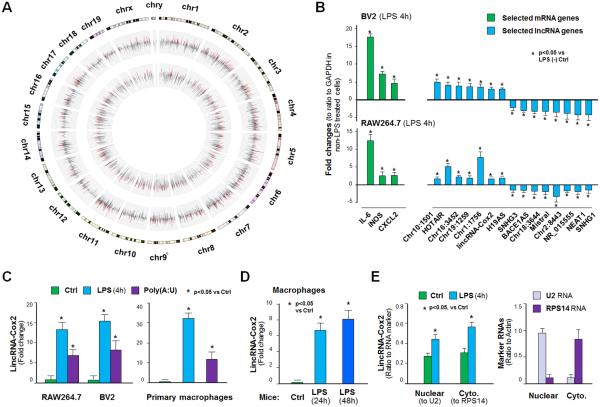
To identify lincRNAs transcribed during the innate immune response in macrophages, we conducted whole-transcriptome analysis of mouse microglia (BV2 cells) stimulated with a TLR4 ligand (LPS, 1 μg/ml) for 4h. The Agilent SurePrint G3 Mouse Gene Expression Microarray (G4852A) was used for the analysis, which provides full coverage of genes and transcripts with the most up-to-date content, including mRNAs and lincRNAs (http://www.chem.agilent.com/store/en_US/Prod-G4852A/G4852A). All array data were described in accordance with MIAME guidelines and deposited at ArrayExpress (database accession number under https://www.ebi.ac.uk/arrayexpress/ E-MTAB-3450). A total of 577 protein-coding genes were upregulated and 86 protein-coding genes downregulated in cells following LPS stimulation (Figure 1A and database E-MTAB-3450). A total of 423 lincRNAs were upregulated and 33 lincRNAs downregulated in the LPS-treated BV2 cells (Figure 1A and database E-MTAB-3450). LPS-induced alteration of selected protein-coding and lincRNA genes was further confirmed by quantitative real-time PCR in BV2 and RAW264.7 cells (Figure 1B). Consistent with previous studies (23,24,26), lincRNA-Cox2 was upregulated following incubation with LPS (Figure 1B and database E-MTAB-3450). Induction of lincRNA-Cox2 by LPS or a TLR3 ligand (PolyA:U, 20 μg/ml) was further demonstrated by real-time PCR of RAW264.7 macrophages and BV2 cells, as well as primary mouse peritoneal macrophages (PMPM) (Figure 1C). When LPS (15 mg/kg body weight) was injected to the abdominal cavity of mice, a significant increase in lincRNA-Cox2 level was detected in the PMPM isolated at 24h and 48h after LPS administration (Figure 1D). To assess the intracellular distribution of lincRNA-Cox2 following LPS stimulation, we exposed RAW264.7 cells to LPS for 4h and measured the lincRNA-Cox2 levels in the nuclear and cytoplasmic extracts. While an increased level of lincRNA-Cox2 content was detected in the nuclear and cytoplasmic extracts in LPS-treated cells, the distribution ratios in the nuclear and cytoplasmic extracts were similar in the non-LPS and LPS-treated cells (Figure 1E).
Figure 1.
Expression of LincRNAs in BV2 Cells or Macrophages in Response to LPS Stimulation. (A) The Circos plot shows genome-wide differential expression between untreated and LPS-stimulated (4h) BV2 cells. The outer track shows the log2 relative changes for protein-coding genes. The inner track shows the log2 relative changes of lincRNAs. These upregulated and downregulated genes are shown in red or green, respectively, whereas unchanged genes are shown in grey. (B) Expression of select protein-coding mRNA genes and lincRNA genes in BV2 and RAW264.7 cells following LPS stimulation (4h) by PCR. (C) LincRNA-Cox2 expression in BV2 and RAW264.7 and primary mouse peritoneal macrophages by LPS and PolyA:U (20 μg/ml, 4h). (D) Induction of lincRNA-Cox2 in primary peritoneal macrophages isolated from mice after LPS peritoneal injection (24h and 48h). (E) Nuclear versus cytoplasmic distribution of lincRNA-Cox2 in RAW264.7 cells following LPS stimulation. The U2 RNA (a nuclear RNA) and the RPS14 (a cytoplasmic RNA) were used as the control RNAs and their expression levels in unstimulated cells are shown. Data represent mean ± SE from three independent experiments. Cyto. = Cytoplasmic.
LincRNA-Cox2 is an NF-κB Early-primary Response Gene
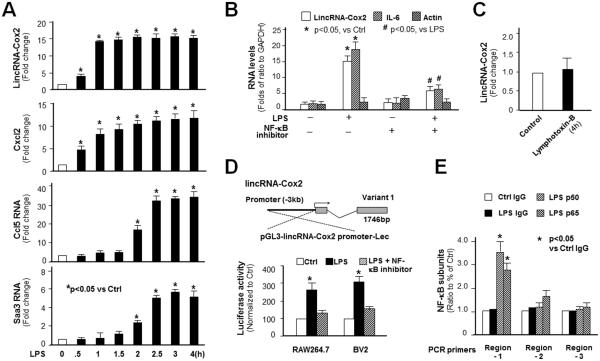
We performed real-time PCR analysis of lincRNA-Cox2 expression in macrophages after exposure to LPS for various periods of time. Data from the time-course analysis revealed that lincRNA-Cox2 is an early-primary response gene. Similar to Cxcl2, an early-primary response gene (5), levels of lincRNA-Cox2 began to increase at 30 min and reached a high level at 60 min in RAW264.7 cells after LPS stimulation, compared with Ccl5 and Saa3 induction, two typical late-primary response genes (5) (Figure 2A). Inhibition of the NF-κB pathway by a NF-κB inhibitor (SC-514) attenuated LPS-induced lincRNA-Cox2 expression (Figure 2B). NF-κB activation through the non-canonical pathway by lymphotoxin B did not alter lincRNA-Cox2 expression (Figure 2C). Consistent with previous data (23), we identified three splice variants of lincRNA-Cox2 by RACE PCR. Our variant 1 transcript from RAW264.7 and BV2 cells revealed a 1762 nt transcript, with extra 17 (176) nt at the 3’end compared with previous results (23). Consistently, variant 1 was the most abundant lincRNA-Cox2 transcript in cells following LPS stimulation (data not shown). Promoter analysis using the NF-κB inhibitor and a luciferase reporter vector encompassing the −3 ĸb upstream region of the transcription start site of the gene further confirmed that lincRNA-Cox2 variant 1 induction is controlled by NF-κB (Figure 2D). Moreover, ChIP analysis detected an enrichment of NF-κB p65 and p50 to the putative promoter region-1 (Chr1:150161930-150162133) of lincRNA-Cox2 gene locus in RAW264.7 cells following LPS stimulation (Figure 2E).
Figure 2.
LincRNA-Cox2 is an Early-primary Responsive Gene Controlled by NF-κB in RAW264.7 Cells. (A) Time course of LPS-induced expression of lincRNA-Cox2 and two known late-primary responsive genes (Ccl5 and Saa3). (B) Inhibition of LPS-induced lincRNA-Cox2 expression in cells by an IKK2/NF-ĸB inhibitor, SC-514. LincRNA-Cox2 levels were quantified by real-time PCR and presented as fold changes to ratio to GAPDH. Expression of IL-6 (a NF-ĸB-controlled gene) and Actin (not controlled by NF-ĸB) were also measured for control. (C) No induction of lincRNA-Cox2 in cells in response to lymphotoxin B (10 μg/ml for 4h). (D) LincRNA-Cox2 promoter luciferase reporter assay. The −3ĸb upstream of lincRNA-Cox2 TSS was cloned and inserted into the Lec-luciferase construct. SC-514 attenuated LPS-triggered luciferase activity in cells transfected with the luc-construct. (E) ChIP analysis of recruitment of NF-κB p65 and p50 subunits to the lincRNA-Cox2 promoter region in cells following LPS stimulation (for 2h). Data represent mean ± SE from three independent experiments.
Knockdown of LincRNA-Cox2 Attenuates the Transcription of Late-primary Genes Triggered by LPS
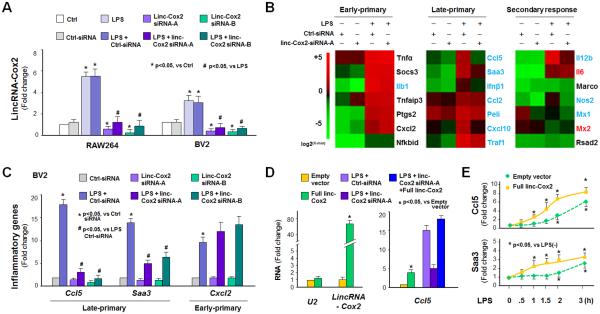
Using an RNAi approach, we measured the effects of lincRNA-Cox2 siRNA on LPS-induced gene transcription in BV2 cells by whole-transcriptome analysis. Two separate siRNAs were designed to target lincRNA-Cox2 and a scrambled non-specific siRNA was used as the control. While the control siRNA showed no effect on lincRNA-Cox2 expression, both siRNAs to lincRNA-Cox2 significantly decreased lincRNA-Cox2 expression levels in the non-stimulated and LPS-stimulated RAW264.7 and BV2 cells (Figure 3A). Silencing of lincRNA-Cox2 with the lincRNA-Cox2 siRNA-A resulted in significant alterations of protein-coding gene expression in non-stimulated BV2 cells (Figure 3B and database E-MTAB-3450). Silencing of lincRNA-Cox2 also attenuated LPS-triggered expression of numerous protein-coding genes, including many inflammatory genes (Figure 3B). Interestingly, LPS-triggered expression of the late-primary response genes, including Ccl2, Ccl5, Cxcl10, Peli1, Traf1, Saa3, and Ifnβ1, was globally suppressed by lincRNA-Cox2 siRNA (Figure 3B). Notably, we found an inhibitory effect of lincRNA-Cox2 siRNA on LPS-induced Ccl5 expression in both BV2 and RAW264.7 cell lines, different from the results reported in mouse bone marrow–derived macrophages (23). Moreover, knockdown of lincRNA-Cox2 attenuated LPS-induced expression of a few early-primary genes and several secondary response genes, as determined by whole-transcriptome analysis (database E-MTAB-3450). This suppressive effect on the late-primary response genes by lincRNA-Cox2 siRNA was further confirmed by real-time PCR using two separate siRNAs to lincRNA-Cox2 in BV2 cells (Figure 3C) and in RAW264.7 cells (data not shown). Suppression of these genes by lincRNA-Cox2 siRNA is at the transcriptional level, as their mRNA stability was not altered by lincRNA-Cox2 siRNA (data not shown). Moreover, the lincRNA-Cox2 siRNA did not have a significant effect on the cytoplasmic activation of the NF-κB pathway triggered by LPS, as there was no significant difference in the cytoplasmic degradation of IκBα and nuclear translocation of NF-κB RelA in the siRNA-treated RAW264.7 cells (Figure S1). When RAW264.7 cells were transfected with full-length lincRNA-Cox2 for 24h, a modest induction of Ccl5 and Saa3 genes was detected (Figure 3D and Figure S2). Transfection of full-length lincRNA-Cox2 attenuated the inhibitory effects of lincRNA-Cox2 siRNA on LPS-induced expression of Ccl5 (Figure 3D) and Saa3 (Figure S2). When cells were transfected with full-length lincRNA-Cox2 for 24h following by LPS stimulation for up to 4h, induction of Saa3 and Ccl5 genes in RAW264.7 cells expressing lincRNA-Cox2 in response to LPS stimulation was shifted to an earlier time point (Figure 3E).
Figure 3.
LincRNA-Cox2 Modulates Transcription of Genes in Response to LPS Stimulation in Macrophages. (A) Quantitative PCR analysis of lincRNA-Cox2 induction in RAW264.7 and BV2 cells treated by two separate siRNAs to lincRNA-Cox2. (B) Heatmaps of representative early-primary, late-primary, and secondary responsive genes in BV2 cells in response to LPS stimulation (4h). The siRNA-A was used to silence lincRNA-Cox2 in cells. (C) LincRNA-Cox2 silencing on LPS-induced expression of Ccl5, Saa3, and Cxcl2 genes in BV2 cells as measured by real-time PCR (LPS for 4h). (D) Overexpression of lincRNA-Cox2 induces Ccl5 expression and attenuates the inhibitory effect of lincRNA-Cox2 siRNA on LPS-induced Ccl5 expression in BV2 cells. Cells were transfected with the full-length of lincRNA-Cox2 or siRNA-A to lincRNA-Cox2 for 24h, exposed to LPS stimulation for 4h and followed by real-time PCR. U2 RNA was measured as a control to rule out non-specific effect of full-length lincRNA-Cox2 transfection. (E) Shift of Saa3 and Ccl5 as early-responsive genes in RAW264.7 cells expressing lincRNA-Cox2. Cells were transfected with the full-length of lincRNA-Cox2 for 24h followed by LPS stimulation for up to 4h. Expression levels of Saa3 and Ccl5 genes were measured by PCR. Data represent mean ± SE from three independent experiments.
LincRNA-Cox2 is Assembled to the SWI/SNF Complex in Macrophages Following LPS Stimulation
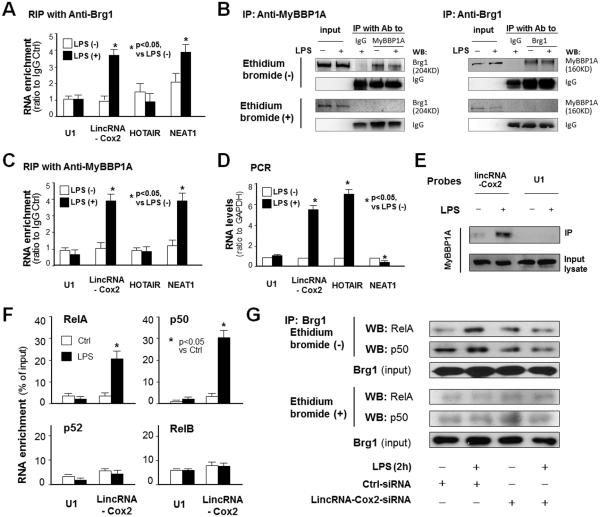
As the SWI/SNF complex is required for late-primary gene transcription (5), we asked whether this complex is involved in lincRNA-Cox2-modulated expression of late-primary gene in macrophages in response to LPS stimulation. We first conducted formaldehyde crosslinking RIP analysis to determine whether there is a direct physical association between lincRNA-Cox2 and the SWI/SNF complex. NEAT1, a recently identified SWI/SNF-associated lincRNA (18), U1 RNA and lincRNA HOTAIR were used as the RNA controls and a non-specific anti-IgG as the antibody control. We detected no obvious presence of U1 and HOTAIR in the immunoprecipitates pulled down from unstimulated and LPS-stimulated RAW264.7 cells by anti-Brg1, a known component of the SWI/SNF complex (37). Immunoprecipitates pulled down from unstimulated cells by anti-Brg1 showed presence of NEAT1, but not lincRNA-Cox2. However, a significant level of lincRNA-Cox2 and NEAT1 was detected in immunoprecipitates from LPS-treated cells (Figure 4A). MyBBP1A is a known RNA-binding protein which can be assembled into the SWI/SNF complex (38,39). Using Co-IP with a buffer without ethidium bromide, we detected a physical association between MyBBP1A and the SWI/SNF complex in both unstimulated and LPS-stimulated macrophages (Figure 4B). In contrast, this physical association was not observed when the co-IP was performed in the presence of ethidium bromide (Figure 4B), suggesting that interactions between MyBBP1A and the SWI/SNF complex may be mediated by nucleic acids. Moreover, immunoprecipitation of LPS-stimulated cells with anti-MyBBP1A revealed the presence of lincRNA-Cox2 and NEAT1, but not the control U1 and HOTAIR (Figure 4C). Presence of specific RNAs in the SWI/SNF complex from LPS-stimulated cells is not correlated to the RNA expression levels (Figure 4D). Our RNA pull-down experiment, using a biotinylated probe to lincRNA-Cox2 followed by Western blotting for MyBBP1A, also detected the presence of MyBBP1A (Figure 4E). Interestingly, knockdown of MyBBP1A by a siRNA attenuated LPS-triggered assembly of lincRNA-Cox2 to the SWI/SNF complex (Figure S3).
Figure 4.
LincRNA-Cox2 Interacts with MyBBP1A and Required for the assembly of NF-κB Subunits to the SWI/SNF complex in Macrophages Following LPS Stimulation. (A) RNA immunoprecipitation (RIP) analysis of lincRNA-Cox2 in the SWI/SNF complex in RAW264.7 cells in response to LPS stimulation using anti-Brg1. Cells were exposed to LPS for 2h, following by formaldehyde crosslinking RIP. (B) Co-IP analysis of MyBBP1A and Brg1. RAW264.7 cells were exposed to LPS for 2h and whole cell lysates were precipitated by anti-Brg1 or anti-MyBBP1A, followed by blotting in the presence or absence of ethidium bromide using anti-MyBBP1A or anti-Brg1, respectively. (C) RIP analysis of lincRNA-Cox2 in the SWI/SNF complex in RAW264.7 cells in response to LPS stimulation using anti-MyBBP1A. (D) Expression levels of associated genes in RAW264.7 cells in response to LPS stimulation. (E) Pull-down analysis of lincRNA-Cox2 assembly into the SWI/SNF complex. RAW264.7 cells were exposed to LPS for 2h and whole cell lysates were precipitated using biotinylated probe pool to lincRNA-Cox2, followed by blotting for MyBBP1A. (F) Increased presence of lincRNA-Cox2 in the immunoprecipitates from LPS-stimulated RAW264.7 cells using antibodies to RelA and p50, but not antibodies to p52 and RelB. Cells were exposed to LPS for 2h, followed by RIP detection of lincRNA-Cox2. (G) Co-IP analysis of NF-ĸB subunits (RelA and p50) and Brg1. RAW264.7 cells were transfected with the lincRNA-Cox2-siRNA for 24h and exposed to LPS for 4h and whole cell lysates were precipitated by anti-Brg1, followed by blotting in the presence or absence of ethidium bromide using anti-RelA or anti-p50, respectively. Experiments were performed in triplicate and representative blots are shown.
LincRNA-Cox2 Promotes the Assembly of NF-κB Subunits to the SWI/SNF Complex in Macrophages Following LPS Stimulation
Previous studies have demonstrated the assembly of “non-core components”, such as NF-κB subunits, to the SWI/SNF complex in cells following inflammatory stimulation (40). To explore the possibility that lincRNA-Cox2 may promote the assembly of NF-κB subunits to the SWI/SNF complex in cells following LPS stimulation, we performed RIP analysis for lincRNA-Cox2 assembly using antibodies to various NF-κB subunits in RAW264.7 cells following LPS stimulation. We detected a significant increase of lincRNA-Cox2 in the immunoprecipitates from LPS-stimulated cells using antibodies to RelA and p50, but not p52 and RelB (Figure 4F). Using Co-IP with a buffer without ethidium bromide, we detected a physical association between NF-κB subunits (RelA and p50) and the SWI/SNF complex in LPS-stimulated macrophages (Figure 4G). In contrast, this physical association was not observed when the co-IP was performed in the presence of ethidium bromide (Figure 4G), suggesting that interactions between NF-κB subunits and the SWI/SNF complex may be mediated by nucleic acids. Indeed, knockdown of lincRNA-Cox2 decreased the association between NF-κB subunits (RelA and p50) and the SWI/SNF complex (Figure 4G).
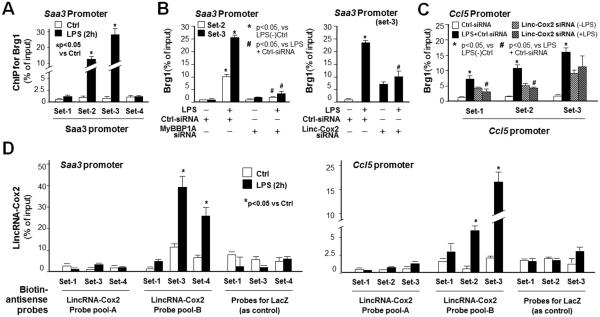
The SWI/SNF Complex and LincRNA-Cox2 Are Recruited to the Promoter/Enhancer Region of Saa3 and Ccl5 Genes Following LPS Stimulation
ChIP was used to assess the recruitment of the SWI/SNF complex to the Saa3 gene promoter region in RAW264.7 cells following LPS stimulation for 2h. Anti-Brg1 was used to precipitate the DNA:SWI/SNF complex. PCR was then performed using primer sets that covered ~3kb of the Saa3 promoter (Figure 5A). A significant increase in SWI/SNF recruitment was detected at the Saa3 promoter in LPS-treated cells (+168 ~ −18 and −361 ~ −544 of TSS, encompassed by primer set-2 and set-3, respectively) (Figure 5A). Knockdown of MyBBP1A or lincRNA-Cox2 by siRNAs attenuated the recruitment of the SWI/SNF complex to according Saa3 promoter induced by LPS (Figure 5B). Recruitment of the SWI/SNF complex to set 1-3 regions of the Ccl5 promoter and inhibition of its recruitment to set 1 and set 2 regions (but not the set 3 region) by lincRNA-Cox2 siRNA-A was detected in LPS-treated cells (Figure 5C). In contrast, a slight increase in SWI/SNF recruitment to Saa3 and Ccl5 promoters was detected in cells treated with the lincRNA-Cox2 siRNA-A in the absence of LPS (Figure 5B and 5C), probably due to the broad effects of lincRNA-Cox2 knockdown in the non-stimulated cells, as shown in Figure 3B. To test whether lincRNA-Cox2 is recruited to the Saa3 promoter, we adapted the ChIRP technique (36), using a pool of tiling oligonucleotide probes with affinity specific to the lincRNA-Cox2 sequence. The DNA sequences of the chromatin immunoprecitates were confirmed by real-time PCR using the same primer sets covering the Saa3 promoter as for the ChIP analysis for SWI/SNF recruitment. We identified a significant increase in occupancy of lincRNA-Cox2 to the Saa3 promoter with the probe pool-B for lincRNA-Cox2 (Figure 5D). Intriguingly, it appears that lincRNA-Cox2 and the SWI/SNF complex occupy the same region of Saa3 promoter encompassed by primer set-3 (Figure 5D). In addition, a significant increase in occupancy of lincRNA-Cox2 to the Ccl5 promoter with the probe pool-B for lincRNA-Cox2 was also detected by ChIRP analysis (Figure 5D).
Figure 5.
The lincRNA-Cox2/SWI/SNF complex is recruited to the promoter regions of Saa3 and Ccl5 loci in cells in response to LPS stimulation. The lincRNA-Cox2/SWI/SNF complex is recruited to the promoter regions of Saa3 and Ccl5 loci in cells in response to LPS stimulation. (A) ChIP analysis of occupancy of SWI/SNF complex to the promoter region of Saa3 gene in RAW264.7 cells in response to LPS stimulation. Cells were exposed to LPS (1 μg/ml) for 2h followed by ChIP analysis for the occupancy of SWI/SNF complex using anti-Brg1. (B) Inhibition of SWI/SNF occupancy to the Saa3 promoter in response to LPS stimulation by siRNAs to MyBBP1A or lincRNA-Cox2. Cells were transfected with the siRNAs to MyBBP1A or lincRNA-Cox2 for 24h, exposed to LPS for 2h, followed by ChIP analysis for Brg1. (C) Occupancy of SWI/SNF complex to the promoter region of Ccl5 gene and its inhibition by lincRNA-Cox2 knockdown in RAW264.7 cells in response to LPS stimulation. Cells were transfected with the siRNAs to lincRNA-Cox2 for 24h, exposed to LPS for 2h, followed by ChIP analysis for Brg1. (D) ChIRP analysis of occupancy of lincRNA-Cox2 to the promoter region of Saa3 and Ccl5 gene promoters in RAW264.7 cells in response to LPS stimulation. Cells were exposed to LPS (1 μg/ml) for 2h followed by ChIRP analysis for the occupancy of lincRNA-Cox2. Data represent mean ± SE from three independent experiments.
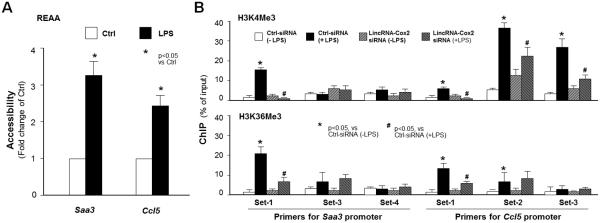
LincRNA-Cox2 Promotes Histone H3 Methylations with Transcriptional Activation at the Promoter Regions of Saa3 and Ccl5 Genes Following LPS Stimulation
It has been speculated that the chromatin of late-primary genes is packed tightly compared with the chromatin of early-primary genes (5). The recruitment of SWI/SNF complex to late-primary genes will trigger chromatin remodeling, resulting in access of κB sites for NF-κB binding and finally, gene transcription (4,5). Consistent with previous findings (5) using REAA, we found an increase in the restriction enzyme accessibility to the promoter regions of the Saa3 and Ccl5 genes in RAW264.7 cells following LPS stimulation (Figure 6A). ChIP analysis demonstrated that an increase in H3K4Me3 and H3K36Me3 to the Saa3 promoter was detected in cells following LPS stimulation (Figure 6B). Moreover, lincRNA-Cox2 siRNA attenuated LPS-triggered enrichment of H3K4Me3 and H3K36Me3 in the Saa3 promoter (Figure 6B). LPS-induced enrichment of H3K4Me3 and H3K36Me3 markers and the inhibitory effects by lincRNA-Cox2 siRNA were also observed in the Ccl5 gene (Figure 6B).
Figure 6.
LincRNA-Cox2 Promotes Histone H3 Methylations with Transcriptional Activation at the Promoter Regions of Saa3 and Ccl5 Genes Following LPS Stimulation. (A) LPS stimulation increases the accessibility of the promoter regions of the Saa3 and Ccl5 genes. RAW264.7 cells were exposed to LPS for 2h, followed by the restriction enzyme accessibility assay (see also materials). (B) Enrichment of H3K4me3 and H3K36me3 to the promoter regions of Saa3 and Ccl5 genes induced by LPS and its inhibition by lincRNA-Cox2-siRNA. RAW264.7 cells were transfected with the siRNA to lincRNA-Cox2 for 24h, exposed to LPS for 2h, followed by ChIP analysis for H3K4me3 and H3K36me3, respectively, using PCR primers to the corresponding regions of gene promoters. Data represent mean ± SE from three independent experiments.
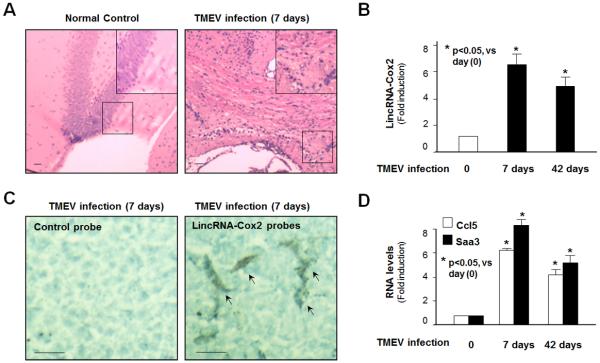
Expression of LincRNA-Cox2 and Late-primary Genes in the Central Nervous System (CNS) in Mice Following TMEV Infection
To determine the relevance of our findings in vivo, we used a well-studied mouse model of CNS inflammation and demyelination induced by TMEV (41). Female FVB/n mice were intracerebrally infected with the DA strain of TMEV, and lincRNA-Cox2 levels in infected and uninfected brain tissues were measured. TMEV-induced CNS inflammation was evident by HE staining of the brain tissues around the hippocampus region at 7 days postinfection (p.i.) with TMEV (Figure 7A). A significant increase in lincRNA-Cox2 levels was found in the whole mouse brains at 7 days p.i., a peak time point in the inflammatory response and a time when viral titers are high (42). At day 42 p.i., a time when the chronic demyelinating phase is underway, a modest increase in lincRNA-Cox2 levels was detected in the brains of infected mice (Figure 7B). Microglia may be the major source of lincRNA-Cox2 expression, as evidenced by in situ hybridization analysis of the hippocampus region of the brain tissues (Figure 7C). Interestingly, increased expression of Ccl5 and Saa3 was also identified in the whole brain tissues from mice at 7 days p.i. (Figure 7D).
Figure 7.
Induction of LincRNA-Cox2 and Late-primary Genes in Microglia in Mice Following TMEV Infection. (A) Hematoxylin and eosin staining of the brain tissues around the hippocampus region from mice at 7 days postinfection with TMEV demonstrating CNS inflammation. A higher power image is shown in the boxed region in the upper right of each image. (B) TMEV infection increased lincRNA-Cox2 expression in the brains of infected mice. Animals were infected with TMEV, and levels of lincRNA-Cox2 in the brain tissues at 7 and 42 days postinfection were quantified by PCR. (C) In situ hybridization of lincRNA-Cox2 around the hippocampus region of the brain tissues from mice at 7 days post-TMEV infection. While the control probe detected no signal, the probe to lincRNA-Cox2 detected intense positive signal in cells with characteristics of microglia (indicated by arrows). (D) TMEV infection increased expression of Ccl5 and Saa3 in the CNS of infected mice. Animals were infected with TMEV, and the mRNA levels of Ccl5 and Saa3 in the whole brain tissues at 7 and 42 days postinfection were quantified by PCR.
DISCUSSION
The molecular basis underlying the delayed transcription of genes in macrophages in response to NF-κB activation following extracellular stimuli remains unclear. Transcription of the secondary response genes involves new protein synthesis, presumably the protein products of early responsive genes (7). A key difference between the secondary response genes and the late-primary response genes is that the late-primary genes are transcribed in a new protein synthesis-independent manner (5,7,8). One of the major findings of this study is that transcription of the late-primary response genes involves the induction of lincRNA-Cox2. Knockdown of lincRNA-Cox2 resulted in a global inhibition of transcription of the late primary genes in cells in response to LPS stimulation. Recruitment of lincRNA-Cox2 to the gene loci of selected late-primary response genes was confirmed by ChIRP analysis. Notably, the lincRNA-Cox2 gene appears to be an early-primary response gene. Overexpression of lincRNA-Cox2 shifts the late-primary genes to become early response genes in macrophages in response to LPS stimulation. Although knockdown of lincRNA-Cox2 also attenuated LPS-triggered expression of some early-primary genes and secondary genes, its main effects are the trans-suppression of the late-primary response genes. The involvement of an early-response lincRNA in the transcription of late-primary response genes may explain the “delayed but protein synthesis-independent” nature of these late-primary genes. Complementarily, lincRNAs have been implicated to function in trans to regulate distantly located genes (43).
It has been speculated that the late-primary response genes are tightly packed in the chromatin compared with the early-primary genes (5). Recruitment of the SWI/SNF complex to the late-primary genes may trigger chromatin remodeling, resulting in access of κB sites for NF-κB binding and subsequently, gene transcription (4,5). Nevertheless, only a modest increase in histone acetylation has been identified within the late-primary gene loci in cells following LPS stimulation (5), suggesting the involvement of other mechanisms to control gene transcription. Findings from this study support the hypothesis that lincRNA-Cox2 is involved in the recruitment of the SWI/SNF complex to the late-primary genes in macrophages in response to LPS stimulation. LincRNAs may interact with DNA molecules to form a triple-helical structure (15). As such, lincRNA-Cox2 may “guide” the initial recruitment of the SWI/SNF complex to the late-primary gene loci through direct binding to a specific DNA motif, presumably a common consensus sequence in the promoter regions of these late-primary genes. Nevertheless, recruitment of the SWI/SNF complex to the late-primary gene loci is not a consensus-specific procedure (37). Therefore, lincRNA-Cox2 is more likely to function as a scaffold molecule for the assembly of various SWI/SNF components to the late-primary gene loci in cells following LPS stimulation. Indeed, assembly of NF-ĸB subunits, RelA and p50, to the SWI/SNF complex in LPS-stimulated macrophages appears to require lincRNA-Cox2. It has been speculated that human lincRNAs may function as scaffold molecules to affect gene expression (44,45).
LincRNAs may function as scaffold molecules through their interactions with various RNA-binding proteins (15). MyBBP1A, a known RNA-binding protein, is associated with both the RNA polymerase I complex and the ribosome biogenesis machinery (46). Direct interactions between MyBBP1A and lincRNA-Cox2 were demonstrated by our RNA pull-down analysis. Although assembly of MyBBP1A into the SWI/SNF complex was previously demonstrated by Co-IP analysis (39), results from more systematic analyses do not support this protein as a “core” subunit for the human SWI/SNF complex (47). Data from our Co-IP analysis support the assembly of MyBBP1A to the SWI/SNF complex. However, the physical interactions between MyBBP1A and the SWI/SNF complex may be mediated by nucleic acids, as their physical associations were not observed when the co-IP analysis was performed in the presence of ethidium bromide. Whether lincRNA-Cox2 is required for the assembly of MyBBP1A to the SWI/SNF complex is unclear.
Our analyses revealed a transcriptional activation of Ccl5 through induction of lincRNA-Cox2, different from the findings from the Fitzgerald group showing an inhibitory effect of lincRNA-Cox2 on Ccl5 expression (23). Notably, one common siRNA sequence, the lincRNA-Cox2-siRNA-B, was used to knockdown the expression of lincRNA-Cox2. However, different stimuli/ligands and cell types were used in the two studies, which may account for the difference. In addition, lincRNA-Cox2 has been shown to interact with other RNA-binding proteins, including hnRNP-A/B and -A2/B1 (23). Because hnRNP-A/B and -A2/B1 function mainly to regulate the RNA stability and splicing of mRNA transcripts (48), such an interaction should not have a regulatory effect specific to the late-primary response genes.
Similar to the regulation of protein-coding genes, promoter binding of transcription factors may regulate lincRNA gene expression in macrophages following LPS stimulation. Specifically, we demonstrated that LPS stimulation induces binding of the NF-κB p65 and p50 subunits to the potential promoter region of the lincRNA-Cox2 gene, resulting in transcription of the early genes after LPS stimulation. Activation of the NF-κB signaling through the non-canonical pathway consistently failed to induce lincRNA-Cox2 expression, as the non-canonical NF-κB activation usually triggers the nuclear translocation of p52 and RelB subunits (49), implicating that lincRNA-Cox2 may not be a key regulator in the immune responses mediated by the non-canonical NF-κB signaling.
NF-κB-mediated lincRNA-Cox2 expression and its subsequent impact on transcription of late-primary response genes may be an important pathogenic factor to chronic inflammatory and infectious diseases. Production of late-primary genes in microglia, including Ifnβ1, Peli1, Ccl5, and Saa3, has been implicated in the pathogenesis of inflammatory response in the CNS following microbial infection (50-54). Activated microglia are an important source of inflammatory cytokines in the CNS following TMEV infection (55-56). In this study, we detected an increase in lincRNA-Cox2 expression, probably in microglia, and elevated levels of late-primary genes in the CNS from TMEV-infected mice. Although the pathogenic role of lincRNA-Cox2 in TMEV-mediated CNS inflammation requires further exploration using conditional lincRNA-Cox2 knockout mice, our data suggest, for the first time, that induction of lincRNA-Cox2 may be involved in the transcriptional regulation of late-primary genes in microglia in the CNS in mice following TMEV infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Barbara L. Bittner and Courtney E. Dolata for their assistance in writing the manuscript and Dr. Xin-Tian Zhang for stimulating discussion.
This work was partially supported by the National Institutes of Health (AI095532) and the Nebraska Cancer and Smoking Disease Research Program (LB595) to XMC and by Grant Number G20RR024001 from the National Center for Research Resources.
Abbreviations
- SWI/SNF
SWItch/Sucrose NonFermentable
- NF-κB
Nuclear factor-κB
- LincRNAs
Long intergenic ncRNAs
- hnRNA
heterogeneous ribonucleoprotein
- PMPM
Primary mouse peritoneal macrophages
- IP
Immunoprecipitation
- ChIP
Chromatin immunoprecipitation
- REAA
Restriction enzyme accessibility assay
- ChIRP
Chromatin isolation by RNA purification
- TMEV
Theiler’s murine encephalomyelitis virus
- CNS
Central Nervous System
Footnotes
Potential conflicts of interest: Nothing to report.
SUPPLEMENTAL DATA
Supplemental data include one supplemental table and three supplemental figures which are available online.
REFERENCES
- 1.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 2.Natoli G, Saccani S, Bosisio D, Marazzi I. Interactions of NF-kappaB with chromatin: the art of being at the right place at the right time. Nat. Immunol. 2005;6:439–445. doi: 10.1038/ni1196. [DOI] [PubMed] [Google Scholar]
- 3.Haddad JJ, Abdel-Karim NE. NF-κB cellular and molecular regulatory mechanisms and pathways: therapeutic pattern or pseudoregulation? Cell Immunol. 2011;271:5–14. doi: 10.1016/j.cellimm.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Bhatt DM, Pandya-Jones A, Tong AJ, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramirez-Carrozzi V, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2b nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sen R, Smale ST. Selectivity of the NF-κB Response. Cold Spring Harb Perspect. Biol. 2009;2:a000257. doi: 10.1101/cshperspect.a000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fowler T, Sen R, Roy AL. Regulation of primary response genes. Mol. Cell. 2011;44:348–360. doi: 10.1016/j.molcel.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Natoli G. Late-genes control of NF-κB-dependent transcriptional responses by chromatin organization. Cold Spring Harb Perspect. Biol. 2009;1:a000224. doi: 10.1101/cshperspect.a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Euskirchen GM, Auerbach RK, Davidov E, Gianoulis TA, Zhong G, Rozowsky J, Bhardwaj N, Gerstein MB, Snyder M. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 2011;7:e1002008. doi: 10.1371/journal.pgen.1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan HY, Trotter KW, Archer TK, Kingston RE. Swapping function of two chromatin remodeling complexes. Mol. Cell. 2005;17:805–815. doi: 10.1016/j.molcel.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 11.Bouazoune K, Miranda TB, Jones PA, Kingston RE. Analysis of 12--individual remodeled nucleosomes reveals decreased histone-DNA contacts created by hSWI/SNF. Nucleic Acids Res. 2009;37:5279–5294. doi: 10.1093/nar/gkp524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dempsey LA. LncRNAs in immune cells. Nat. Immunol. 2013;14:1036. [Google Scholar]
- 14.Luo H, Sun S, Li P, Bu D, Cao H, Zhao Y. Comprehensive characterization of 10,571 mouse large intergenic noncoding RNAs from whole transcriptome sequencing. PLoS One. 2013;8:e70835. doi: 10.1371/journal.pone.0070835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40:1–10. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaguchi T, Tanigawa A, Naganuma T, Ohkawa Y, Souquere S, Pierron G, Hirose T. SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc. Natl. Acad. Sci. U S A. 2015;112:4304–4309. doi: 10.1073/pnas.1423819112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Y, Rowley MJ, Böhmdorfer G, Wierzbicki AT. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Mol. Cell. 2013;49:298–309. doi: 10.1016/j.molcel.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corey DR, Dimmeler S, Kornfeld J-W. Targeting noncoding RNAs in disease: challenges and opportunities. Science. 2013;341:1021. [Google Scholar]
- 21.Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 22.Johnson R. Long non-coding RNAs in Huntington's disease neurodegeneration. Neurobiol. Dis. 2012;46:245–254. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, Bradel-Tretheway BG, Korth MJ, Castle JC, Biery MC, et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010;1:e00206. doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rapicavoli NA, Qu K, Zhang J, Mikhail M, Laberge RM, Chang HY. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q, Chen CY, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio. 2013;29:e00596–12. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong Q, Gong AY, Zhang XT, Lin C, Ma S, Chen J, Hu G, Chen XM. LincRNA-Cox2 modulates TNF-α-induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi-2/NuRD-mediated epigenetic histone modifications. FASEB J. 2015:fj.15–279166. doi: 10.1096/fj.15-279166. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008 doi: 10.1002/0471142735.im1401s83. Chapter 14, Unit 14.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou R, Hu G, Gong AY, Chen XM. Binding of NF-kappaB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Res. 2010;38:3222–3232. doi: 10.1093/nar/gkq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramaniam D, Ramalingam S, May R, Dieckgraefe BK, Berg DE, Pothoulakis C, Houchen CW, Wang TC, Anant S. Gastrin-mediated interleukin-8 and cyclooxygenase-2 gene expression: differential transcriptional and posttranscriptional mechanisms. Gastroenterology. 2008;134:1070–1082. doi: 10.1053/j.gastro.2008.01.040. [DOI] [PubMed] [Google Scholar]
- 33.Ohkawa Y, Mallappa C, Vallaster CS, Imbalzano AN. An improved restriction enzyme accessibility assay for analyzing changes in chromatin structure in samples of limited cell number. Methods Mol. Biol. 2012;798:531–542. doi: 10.1007/978-1-61779-343-1_32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao S, Procko E, Shannon MF. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J. Immunol. 2001;167:4494–4503. doi: 10.4049/jimmunol.167.8.4494. [DOI] [PubMed] [Google Scholar]
- 35.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 36.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Euskirchen GM, Auerbach RK, Snyder M. SWI/SNF chromatin-remodeling factors: multiscale analyses and diverse functions. J. Biol. Chem. 2012;287:30897–30905. doi: 10.1074/jbc.R111.309302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 39.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U S A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishizaka A, Mizutani T, Kobayashi K, Tando T, Sakurai K, Fujiwara T, Iba H. Double plant homeodomain (PHD) finger proteins DPF3a and -3b are required as transcriptional co-activators in SWI/SNF complex-dependent activation of NF-kB RelA/p50 heterodimer. J. Biol. Chem. 2012;287:11924–11933. doi: 10.1074/jbc.M111.322792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipton HL. Theiler's virus infection in mice: an unusual biphasic disease process leading to demyelination. Infect. Immun. 1975;11:1147–1155. doi: 10.1128/iai.11.5.1147-1155.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pavelko KD, Pease LR, David CS, Rodriguez M. Genetic deletion of a single immunodominant T-cell response confers susceptibility to virus-induced demyelination. Brain Pathol. 2007;17:184–196. doi: 10.1111/j.1750-3639.2007.00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 44.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 46.Hochstatter J, Hölzel M, Rohrmoser M, Schermelleh L, Leonhardt H, Keough R, Gonda TJ, Imhof A, Eick D, Längst G, Németh A. Myb-binding protein 1a (Mybbp1a) regulates levels and processing of pre-ribosomal RNA. J. Biol. Chem. 2012;287:24365–24377. doi: 10.1074/jbc.M111.303719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Middeljans E, Wan X, Jansen PW, Sharma V, Stunnenberg HG, Logie C. SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes. PloS One. 2012;7:e33834. doi: 10.1371/journal.pone.0033834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodarzi H, Najafabadi HS, Oikonomou O, Greco TM, Fish L, Salavati R, Cristea IM, Tavazoie S. Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature. 2012;485:264–268. doi: 10.1038/nature11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun S. Non-canonical NF-κB signaling pathway. Cell Res. 2011;21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen CJ, Chen JH, Chen SY, Liao SL, Raung SL. Upregulation of RANTES gene expression in neuroglia by Japanese encephalitis virus infection. J. Virol. 2004;78:12107–12119. doi: 10.1128/JVI.78.22.12107-12119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mi W, Belyavskyi M, Johnson RR, Sieve AN, Storts R, Meagher MW, Welsh CJ. Alterations in chemokine expression following Theiler's virus infection and restraint stress. J. Neuroimmunol. 2004;151:103–115. doi: 10.1016/j.jneuroim.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 52.Palma JP, Kwon D, Clipstone NA, Kim BS. Infection with Theiler's murine encephalomyelitis virus directly induces proinflammatory cytokines in primary astrocytes via NF-kappaB activation: potential role for the initiation of demyelinating disease. J. Virol. 2003;77:6322–6331. doi: 10.1128/JVI.77.11.6322-6331.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ransohoff RM, Wei T, Pavelko KD, Lee JC, Murray PD, Rodriguez M. Chemokine expression in the central nervous system of mice with a viral disease resembling multiple sclerosis: roles of CD4+ and CD8+ T cells and viral persistence. J. Virol. 2002;76:2217–2224. doi: 10.1128/jvi.76.5.2217-2224.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao Y, Jin J, Chang M, Chang JH, Hu H, Zhou X, Brittain GC, Stansberg C, Torkildsen Ø, Wang X, et al. Peli promotes microglia-mediated CNS inflammation by regulating Traf3 degradation. Nat. Med. 2013;19:595–602. doi: 10.1038/nm.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jarry U, Jeannin P, Pineau L, Donnou S, Delneste Y, Couez D. Efficiently stimulated adult microglia cross-prime naive CD8+ T cells injected in the brain. Eur. J. Immunol. 2013;43:1173–1184. doi: 10.1002/eji.201243040. [DOI] [PubMed] [Google Scholar]
- 56.Ransohoff RM, Brown MA. Innate immunity in the central nervous system. J. Clin. Invest. 2012;122:1164–1171. doi: 10.1172/JCI58644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.