Abstract
NK cell maturation is critical for normal effector function and the innate immune response to tumors and pathogens. However, the molecular pathways that control NK cell maturation remain largely undefined. Here, we investigate the role of SPPL3, an intramembrane aspartyl protease, in murine NK cell biology. We find that deletion of SPPL3 in the hematopoietic system reduces numbers of peripheral NK cells, clearance of MHC Class I-deficient tumors in vivo, and cytotoxicity against tumor cells in vitro. This phenotype is concomitant with reduced numbers of CD27+CD11b+ and CD27−CD11b+ NK cells, indicating a requirement for SPPL3 in efficient NK cell maturation. NK cell-specific deletion of SPPL3 results in the same deficiencies, revealing a cell-autonomous role for SPPL3 in these processes. CRISPR/Cas9 genomic editing in murine zygotes was used to generate knock-in mice with a catalytically compromised SPPL3 D271A allele. Mice engineered to express only SPPL3 D271A in NK cells phenocopy mice deleted for SPPL3, indicating a requirement for SPPL3 protease activity in NK cell biology. Our results identify SPPL3 as a cell-autonomous molecular determinant of NK cell maturation and expand the role of intramembrane aspartyl proteases in innate immunity.
INTRODUCTION
As founding members of the innate lymphoid cell family, natural killer (NK) cells are critically important for tumor surveillance and clearance of virally-infected cells by the innate immune system (1). Robust NK cell effector function depends upon normal NK cell development and maturation, which are governed by cell-intrinsic transcription factors as well as cytokines, including IL-15 (2–4). The major source of NK cell precursors and the site of their development in mice is the bone marrow (5). After development, NK cells migrate into the periphery and populate all the organs of the body. During the final stages of development, NK cells undergo an ordered maturation program defined by the expression of the tumor necrosis family (TNF) member CD27 and the integrin CD11b. NK cells progress from an immature CD27+CD11b− stage to an intermediate CD27+CD11b+ stage, and finally to a CD27−CD11b+ stage (6–9). Progression through these stages is required for optimal NK cell activity, and it occurs at the same time NK cells acquire the full complement of activating and inhibitory receptors that control their response to targets. Although the maturation steps have been defined by expression of these markers, the molecular details that govern these transitions are poorly understood.
The intramembrane aspartyl protease family is a class of multi-pass transmembrane proteins with diverse proteolytic roles in biology (10, 11). The family includes presenilin-1 and −2, and the signal peptide peptidase (SPP)-like subfamily members SPP, SPPL2a, SPPL2b, SPPL2c, and SPPL3. All intramembrane aspartyl proteases share YD and GXGD motifs that function in catalysis. Presenilins cleave Type I transmembrane proteins and function as the catalytic subunits of the γ-secretase complex. The SPP-like subfamily members cleave Type II transmembrane proteins due to their opposite orientation in the lipid bilayer, and function autonomously.
Recent studies have highlighted emerging roles in innate and adaptive immunity for SPP-like subfamily members. SPP cleaves the signal peptide from MHC class I proteins for presentation on HLA-E, which regulates NK cell function (12, 13), and also generates peptides for presentation by MHC class I, which impacts T cell activity (14). SPPL2a controls B cell and dendritic cell (DC) survival through cleavage of the N-terminal fragment of invariant chain, CD74, the build-up of which is toxic to these cells (15–17). SPPL2a can also cleave Fas ligand to generate an intracellular domain that inhibits B and T cell activation downstream of antigen receptor engagement (18). SPPL2a and SPPL2b are also reported to cleave TNF-α, releasing an intracellular domain that elicits IL-12 production in DCs (19).
SPPL3 is perhaps the least understood member of this family. Both protease-dependent and protease-independent functions have very recently been described for SPPL3. In a protease-independent manner, SPPL3 functions in the endoplasmic reticulum (ER) in T cells to promote store-operated calcium entry in response to T cell receptor engagement by enhancing the interaction between STIM1 and Orai1 (20). SPPL3 has also been shown to function as a protease in cell lines and murine embryonic fibroblasts (MEFs) to regulate the extent of constitutive complex glycosylation by targeting several glycosylation enzymes for cleavage and shedding from the ER or Golgi (21, 22).
In order to study SPPL3 function in the immune system, we generated and evaluated mice with a conditional SPPL3 allele that allowed targeted, tissue-specific SPPL3 deletion. Here we report an obligate, cell-autonomous, protease-dependent role for SPPL3 in NK cell maturation and cytotoxicity.
MATERIALS AND METHODS
Mice
Mice containing two LoxP sites flanking the third exon of SPPL3 on chromosome five were generated by conventional embryonic stem (ES) cell homologous recombination by Ingenious Targeting Limited in the C57Bl/6N × 129/SvEv strain background. These mice were back-crossed to C57Bl/6J (The Jackson Laboratory) mice for at least eight generations before crossing to Cre-containing strains. Sox2-Cre- (23) and Vav1-iCre-expressing mice (24) were purchased from The Jackson Laboratory. NKp46-iCre mice (8) were a gift from G. Trinchieri (NCI, Bethesda), with permission from E. Vivier (INSERM, France).
PCR was used to confirm the genotype of all mice. Males and females at least 7 weeks old were used with age- and gender-matched littermate controls. All mice were maintained in accordance with the Johns Hopkins University Institutional Animal Care and Use Committee. The SPPL3 D271A allele was generated by CRISPR/Cas9 genome engineering (25). An SPPL3-specific, sgRNA-encoding sequence, 5’-atcggggacattgtgatgcc-3’, was cloned into the BbsI site of pX330 (Addgene), amplified from pX330 with a leading T7 promoter by PCR, in vitro transcribed using the HiScribe T7 in vitro transcription kit (New England Biolabs), purified using the MEGAclear Kit (Ambion) and resuspended in water. A T7 promoter was cloned into pX330 directly upstream of Cas9 at the AgeI site, to create pX330+T7. Cas9 mRNA was in vitro transcribed using NotI-linearized pX330+T7 and the mMESSAGE mMACHINE T7 Ultra kit (Ambion), purified by LiCl precipitation, and resuspended in water. The sequence of the DNA oligo for homology-directed repair (HDR) was 5’- TTAACGTGTGCCCTTGTGTTTCAGCTCCACTGGCAGTCACTTCTCTATGCTGGGCATCGGGGcgATcGTGATGCCCGGCC TCCTGTTATGCTTTGTTCTTCGCTATGACAACTACAAGAAACAAG-3’ (lower case letters indicate mutations). An endogenous MslI restriction site was destroyed by the mutation, and a novel PvuI site was engineered to aid genotyping. The HDR oligo was purchased from IDT (4nM Ultramer) and resuspended in water. 25 ng/ml Cas9 mRNA, 12.5 ng/ml sgRNA and 25ng/ml HDR DNA oligo were injected into C57Bl/6J embryos generated by The Transgenic Core Laboratory at the Johns Hopkins University School of Medicine. Three founders were obtained from a cohort of 23 live pups and crossed to C57Bl/6J mice to demonstrate germline transmission. PCR followed by overnight PvuI digestion was used to confirm the presence of the mutant allele at each generation. Heterozygous pups from the N1 generation were used for survival curves and further breeding. Survival curves were completed with at least 100 pups from each founder. The relevant SPPL3 locus from one founder line was sequenced and this line was used in all other experiments. The top two off-target sites predicted by the server at CRISPR.mit.edu were sequenced and showed no evidence of Cas9 activity.
Reagents
Antibodies were purchased recognizing mouse CD3 (145–2C11, BD), CD19 (1D3, BD), Ter119 (TER119, BD), Gr-1 (RB6–8C5, BD), CD122 (TM-β1, Biolegend), CD49b (DX5, BD and Biolegend), CD11b (Mac-1, BD), Ki67 (16A8, Biolegend), B220 (RA3–6B2, BD), CD8-α (53–6.7, BD), NK1.1 (PK136), NKG2D (CX5), NKp46 (29A1.4, Biolegend), CD27 (LG.7F9, eBioscience), phospho-S6 (235/6) (D57.2.2E, Cell Signaling), CXCR4 (2B11, eBioscience), Eomesodermin (Dan11Mag, eBioscience), KLRG1 (2F1/klrg1, Biolegend), CD51 (RMV-7, eBiosicence), Ly49H (3D10, eBioscience), CD69 (H1.2F3, BD), NKG2A/C/E (20D5, BD), Ly49G2 (eBio4D11, eBioscience), CD127 (A7R34, Biolegend), CXCR3 (CXCR3–173, BD), CD98 (RL388, Biolegend), Ly49D (4E5, Biolegend), phospho-STAT5 (47/Stat5(pY694), BD), GAPDH (D16H11, Cell Signaling), Tubulin (AA12.1, Developmental Studies Hybridoma Bank), and MGAT5 (clone 706824, R&D Systems). The SPPL3 antibody was previously described (20).
Concanavalin A, CellTrace CFSE, and CellTrace Violet (CTV) proliferation kits were purchased from Molecular Probes. Annexin V was purchased from Biolegend. Murine recombinant IL-15 was purchased from Peprotech. PHA-L was purchased from Life Technologies. IC fixation buffer, and FoxP3 fixation and permeabilization buffer and concentrate, and 10x permeabilization buffer were all purchased from eBioscience. Cytofix and Permeabilization buffer IV were purchased from BD Biosicence.
Flow cytometry
Organs were harvested into media (RPMI, 5% FBS, 1% P/S, 1% L-glutamine) and dissociated using frosted glass slides. Single cell suspensions were obtained by passing the cells over a 70 µm filter. Liver cells were spun over a 35% Percoll (Sigma) solution to separate lymphocytes (pelleted) from hepatocytes (top layer). Red blood cells (RBCs) were lysed using Ack lysing buffer (Quality Biologics). The final cell pellets were resuspended in PBS and counted using trypan blue exclusion. Negative isolation was performed according to manufacturer’s directions (Miltenyi) and enriched over LS columns.
Surface staining was carried out in FACS buffer (PBS, pH 7.4, 0.5% BSA, 2mM EDTA, 0.02% sodium azide) on ice for 30–60 minutes.
For intracellular staining of eomesodermin, the eBioscience Foxp3 fixation/permeabilization kit was used.
Annexin V staining was performed in 1x Annexin V binding buffer (10mM HEPES, pH 7.4, 140mM NaCl, 2.5mM CaCl2) for 15 minutes after surface staining. Additional Annexin V binding buffer was added and samples were run immediately.
Lineage markers used in all figures are CD3, CD19, Ter119, and Gr-1.
Data was collected on an LSR II flow cytometer (BD) and analyzed using FlowJo software (TreeStar).
For sorted cells, after the final wash, cells were passed through a 35 µm filter and sorted on a FACS Aria (BD) at the Johns Hopkins Ross Flow Cytometry Core.
RMA/s killing assay
RMA and RMA/s cells were a gift from J. Sun (MSKCC, New York). RMA/s cells were loaded with 2.5µM CFSE and RMA cells with 5µM CTV according to manufacturer’s directions. Cells were mixed in even ratios and injected (5×105 cells each) intraperitoneally into mice. 48 hours later, peritoneal lavage was performed by injecting 4ml PBS using a 27G needle into the cavity and massaging. The fluid was collected with a 22G needle. The ratio of RMA/s cells recovered is reported as a fraction of the total CFSE+ and CTV+ cells collected.
YAC-1 lysis assay
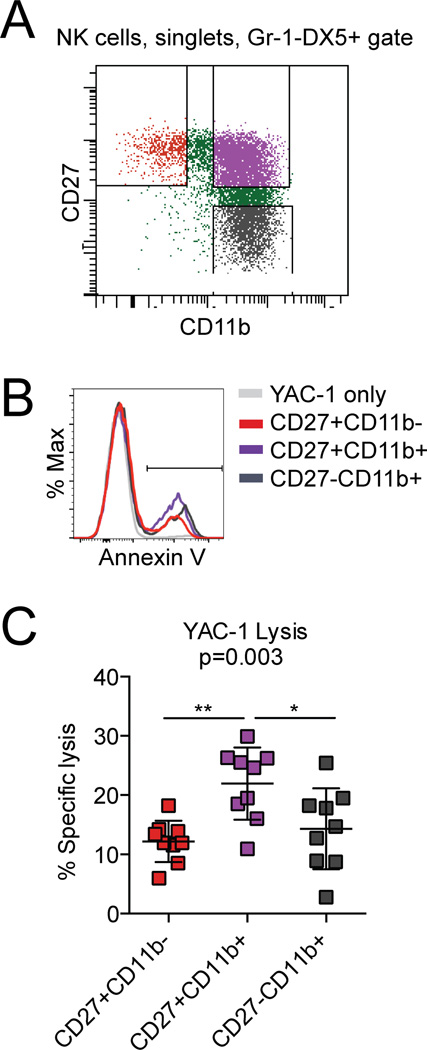
Splenic NK cells were isolated using the NK Negative Isolation II Kit (Miltenyi), then stained for CD49b (DX5) expression. YAC-1 cells were loaded with 500nM CFSE according to manufacturer’s directions. Equivalent numbers of CD49b+ cells were mixed in 96-well plates with YAC-1 cells in the indicated effector: target ratios. After 4–5 hour culture, cells were stained with Annexin V. YAC-1 lysis was calculated as the fraction of Annexin V+ cells as a percentage of CFSE+ cells, and is reported as the percent specific lysis in each experiment. For Figure 5, spleens from three C57Bl/6J mice were pooled. After isolation, cells were stained and sorted for Gr-1−DX5+ cells and CD27 and CD11b expression. Cells were counted and co-cultured with CFSE-loaded YAC-1 cells for 3.5 hours at a 2:1 E:T ratio before Annexin V staining.
Figure 5.
Maturation stage correlates with cytolytic function. (A) Representative flow panel showing the CD27 and CD11b gates used for sorting (Previously gated for single cells that were Gr-1−DX5+). (B) Representative flow panel of the CFSE+ gate after co-culture. (C) Percent specific YAC-1 lysis after 3.5-hour co-culture with splenic NK cells sorted on the indicated receptors at a 2:1 E:T ratio. Each data point represents three pooled mice. Pooled data from three independent experiments with n=9 mice. p-value calculated using one-way ANOVA (with Tukey’s multiple comparisons test for significance **q>5, *2<q<5).
Western Blots
For Figure 1a, C57Bl/6J spleens were harvested and sorted on the indicated markers. Splenocytes were isolated using the CD4+ T cell isolation kit, CD8a+ T cell isolation kit, Pan-B cell isolation kit II, or NK isolation kit II (Miltenyi). For NKp46-iCre strains, cells were further sorted for Gr-1−DX5+ cells. Isolated cells were lysed in equal volumes IP lysis buffer (150 mM NaCl, 50 mM HEPES pH 7.9, 1 mM EDTA, 10% glycerol, 1% igepal) for 1 hour at room temperature. Sorted cells were lysed in equivalent IP lysis buffer volume per cell. After clearing cell debris, total protein of isolated cells was analyzed by Bradford assay (Bio-Rad). Buffer D (150 mM Tris-HCl at pH 6.8, 15% SDS, 12.5% (v/v) 2-mercaptoethanol, 25% (v/v) glycerol, and 0.02% (w/v) bromphenol blue) was added and lysates were separated on a 12% SDS-PAGE gel then transferred to PVDF membranes. Membranes were stained with Ponceau S (0.1% Ponceau S w/v, 5% acetic acid v/v) to confirm equivalent loading, then blocked in 5% milk in TBST 1 hour to overnight. Anti-SPPL3 (1:20,000 dilution) and anti-MGAT5 (1:250 dilution) antibodies were incubated overnight. Anti-GAPDH (1:2000–5000 dilution) and anti-Tubulin (1:20,000 dilution) antibodies were incubated 1 hour to overnight. HRP-conjugated secondary antibodies (Santa Cruz) were incubated at 1:2000 dilution for 1 hour and HRP was detected by Clarity ECL Western substrate (SPPL3 and MGAT5, Bio-Rad) or ECL Western blotting substrate (GAPDH and Tubulin, Pierce) on Amersham Hyperfilm ECL (GE).
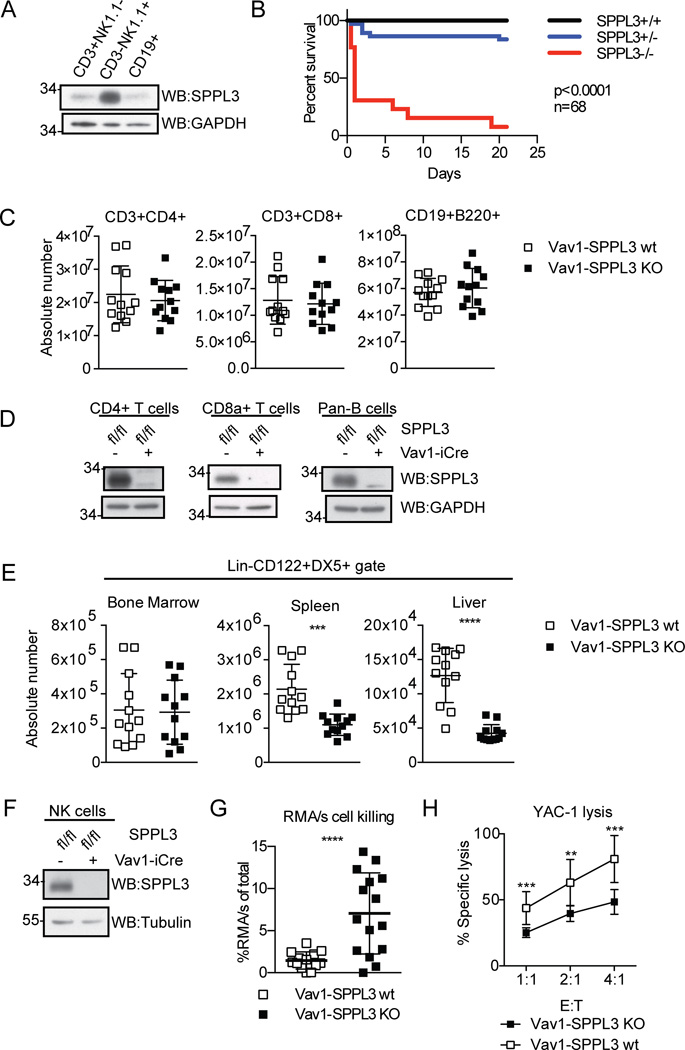
Figure 1.
SPPL3 is required in the immune system for normal NK cells. (A) Expression of SPPL3 in C57Bl/6J splenocytes sorted on the indicated markers. Representative panel from three independent experiments with one mouse per experiment. (B) Kaplan-Meier survival curve of nonconditional SPPL3 knock-out mice (n=68 pups, p value was calculated by Mantel-Cox test). (C) Absolute number of lymphocytes in the spleen. Pooled data from three experiments with n=3–4 mice per genotype. (D) SPPL3 expression in splenocytes isolated using the indicated Miltenyi negative isolation kits. Representative images from two independent experiments with n=1–2 mice per genotype. (E) Absolute number of NK cells (Lin−CD122+DX5+) in the indicated organs. Pooled data from four independent experiments with n=3–4 mice per genotype. (F) Western blot of splenic NK cells after negative isolation. Representative blot from four independent experiments with n=2–3 mice per genotype. (G) Percent RMA/s cells (of total RMA + RMA/s cells collected) remaining 48 hours after intraperitoneal injection. Pooled data from three independent experiments with n=2–8 mice per genotype. (H) Percent specific YAC-1 lysis after four-hour co-culture with isolated splenic NK cells (equivalent DX5+ cell number) at the indicated effector: target (E:T) ratios. Pooled data from three independent experiments with n=3 mice per genotype. For experiments in panels C, E, and G, each data point represents one mouse. **p<0.01, ***p<0.001, ****p<0.0001 (two-tailed unpaired Student’s t-test with Welch’s correction). For experiments in panel H, each data point represents the mean of three pooled experiments. The data was analyzed by unpaired Student’s t test, and statistical significance determined using the Holm-Sidak method, with alpha=5.000%. Each ratio was analyzed individually, without assuming a consistent SD.
IL-15 stimulation
Splenic or bone marrow lymphocytes were harvested, RBCs were lysed, and cells were aliquotted into 96-well U-bottom plates. 2x IL-15 or media alone was added to the cultures and incubated at 37°C for 40 minutes. Cells were washed once with PBS and stained for surface proteins. After another PBS wash, cells were fixed in pre-warmed BD Cytofix for 10 minutes at 37°C. Cells were washed with PBS, then resuspended in 1x permeabilization buffer IV (BD) for 15 minutes. After a spin, cells were resuspended in FACS buffer with the phospho-S6 and phospho-STAT5 antibodies for 45 minutes at 4°C.
NK cell proliferation
Splenic NK cells were isolated and loaded with 10µM CFSE for 15 minutes according to manufacturer’s instructions. 2.5×104 cells were plated in 96-well U-bottom plates and 2x IL-15 or media alone was added. Cells were cultured for 3 days. After a wash, cells were stained for DX5 followed by AnnexinV staining for proliferation analysis.
PCR
For tail cells, tissue was digested in proteinase K (25U/ml) 2 hours to overnight in Laird’s buffer (100 mM Tris pH 8.5, 5 mM EDTA, 0.2% SDS 400 mM NaCl). DNA was extracted by ethanol precipitation and the pellet was resuspended in 0.1X TE.
DNA from sorted NK cells was isolated using the QIAamp DNA mini kit (QIAGEN).
RESULTS
SPPL3 is necessary for normal numbers of peripheral NK cells
To explore a role for SPPL3 in the immune system, we assayed its expression in B cells, T cells, and NK cells sorted from murine splenocytes (Fig. 1 A). All cells assayed expressed SPPL3, but NK cells exhibited the highest levels, suggesting a potential role for SPPL3 in NK cell biology.
To determine an in vivo role for SPPL3, we generated mice with an allele of SPPL3 in which the third exon is flanked by loxP sites, SPPL3fl/+. We bred SPPL3fl/+ mice to Sox2-Cre mice to generate a constitutive null allele, SPPL3−/+, then bred these mice to homozygosity to evaluate the effects of global SPPL3 deletion. All genotypes were born in Mendelian ratios, but SPPL3−/− mice were smaller than their littermates and died of undetermined cause shortly after birth (Fig. 1 B, and data not shown).
We next bred SPPL3fl/fl mice to Vav1-iCre mice to investigate hematopoietic cell-specific effects of SPPL3 deletion. SPPL3fl/fl /Vav1-iCre (Vav1-SPPL3 KO) mice were born at Mendelian ratios and were overtly healthy. SPPL3fl/+/Vav1-iCre, and SPPL3fl/fl mice were used as controls (Vav1-SPPL3 WT). Vav1-SPPL3 KO mice displayed normal numbers of T (CD3+CD4+, CD3+CD8+) and B cells (CD19+B220+) in the spleen (Fig. 1 C). Western blot analysis confirmed that SPPL3 was completely excised in each cell type, excluding the possibility that Vav1-iCre is ineffective in these cell types (Fig. 1 D). Thus, SPPL3 is dispensable for normal T and B cell numbers. In contrast, the number of NK cells (Lin−CD122+DX5+) in Vav1-SPPL3 KO mice was reduced about twofold in the spleen, and threefold in the liver, but was normal in the bone marrow (Fig. 1 E, Supplemental Fig. 1). The remaining NK cells in the spleen showed complete excision of SPPL3, indicating there was no selection for non-recombined cells (Fig. 1 F). The data indicate that SPPL3 is selectively required for normal numbers of peripheral NK cell populations.
To test whether the reduction in NK cell numbers in Vav1-SPPL3 KO mice resulted in a functional defect, we injected mice with equal numbers of the MHC class I-deficient tumor cell line, RMA/s, and the MHC class I-sufficient parental line, RMA, and compared relative cell recovery after 48 hours. NK cell killing in control mice resulted in 1.5% recovery of RMA/s cells, while the recovery in Vav1-SPPL3 KO mice was 7%, indicating reduced NK cell function (Fig. 1 G). To exclude total NK cell number and migration in vivo as factors contributing to the cytotoxic defect, we tested equal numbers of splenic NK cells (DX5+) from Vav1-SPPL3 KO and control mice in a YAC-1 target lysis assay in vitro. NK cells from Vav1-SPPL3 KO mice had approximately 60% of the cytotoxic activity seen in control mice across a range of effector to target ratios (Fig, 1 H), indicating an important role for SPPL3 in NK cell cytolytic function.
SPPL3 regulates NK cell maturation
We next addressed whether the reduced NK cell frequency and activity in Vav1-SPPL3 KO mice was associated with altered receptor expression. NK cells showed changes in all of the receptors tested (Fig. 2 A). At the population level, the percent of NK cells that were NKG2D+ in the spleen was moderately reduced from 97% to 88%. NK1.1+ NK cells were also moderately reduced from 99% to 89%. The loss of NKp46+ NK cells was somewhat larger, from 97% to 72%. The most significant change was in CD11b+ NK cells, from 81% to 38%, a twofold loss. The same fold changes in these populations were seen in the bone marrow (Fig. 2 B).
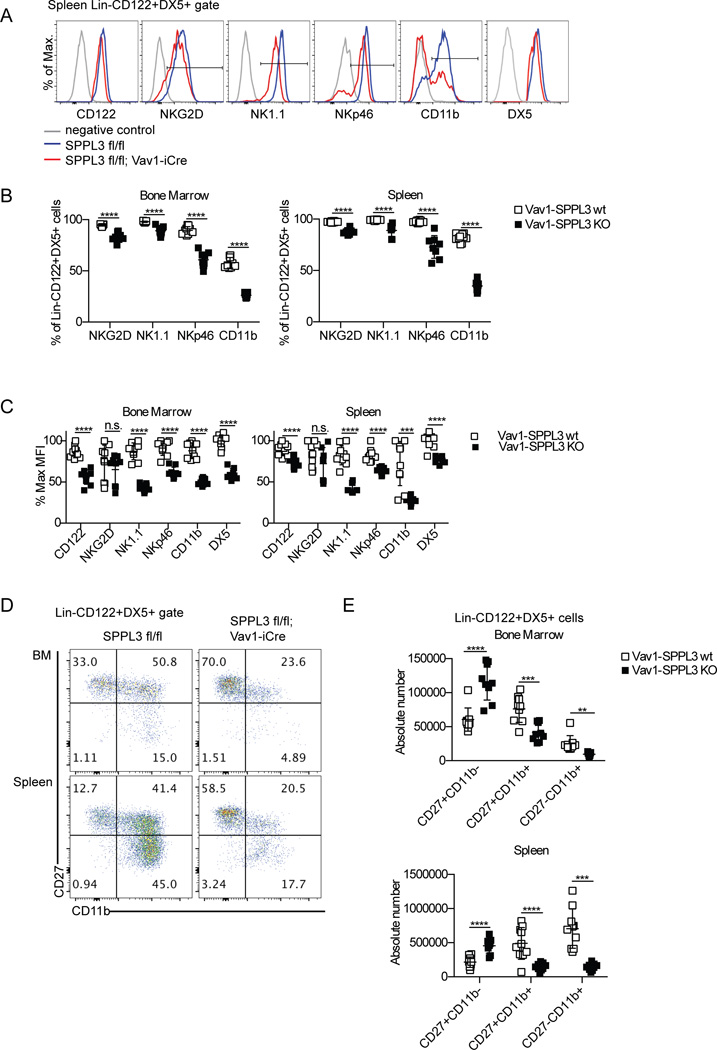
Figure 2.
SPPL3 is required for normal receptor expression and development of NK cells. (A) Representative flow panels of the NK cell gate (Lin−CD122+DX5+) showing the profile of the indicated receptors in the spleen. NK1.1 and DX5 negative controls are unstained cells, others are isotype controls. (B) The percentage of the NK cell gate that is positive for the indicated receptors as depicted in (A). (C) The median fluorescence intensity of the positive gates in (A) of indicated receptors on NK cells, calculated as a percent of the maximum for each receptor. (D) Representative flow panels of the NK cell gate in the indicated organs. (E) The absolute number of each maturation stage in NK cells. Pooled data from three independent experiments with n=3 mice per genotype. Each data point represents one mouse. **p<0.01, ***p<0.001, ****p<0.0001 (unpaired Student’s t-test, statistical significance determined using the Holm-Sidak method, with alpha=5.000%. Each row was analyzed individually, without assuming a consistent SD).
On a per cell basis, NK1.1 expression on splenic NK cells in Vav1-SPPL3 KO mice was reduced from 82% of the maximum fluorescence intensity to only 42% on positively staining cells. NKp46 was moderately reduced, from 82% to 63% of the maximum, and CD122 expression was reduced from 89% to 72%. No significant change in NKG2D levels was observed. DX5 expression on Vav1-SPPL3 KO cells was reduced to 76% of control mice (Fig. 2 C). Interestingly, similar fold reductions in these receptors were observed in the bone marrow of Vav1-SPPL3 KO mice, where NK cell numbers were normal. The most dramatic difference in expression levels between Vav1-SPPL3 KO and control mice was CD11b, an integrin that marks terminal maturation in NK cells (6), which was reduced 2.7-fold, from 72% to 27%, on splenic NK cells and 88% to 50% on bone marrow NK cells in Vav1-SPPL3 KO mice. This prompted us to evaluate NK cell maturation in Vav1-SPPL3 KO and control mice.
NK cells mature sequentially from CD27+CD11b− to CD27+CD11b+ to CD27−CD11b+ cells. NK cells in Vav1-SPPL3 KO mice displayed a twofold increase in the total number of immature, CD27+CD11b− cells in the spleen and bone marrow. Vav1-SPPL3 KO mice showed a threefold loss of CD27+CD11b+ in the spleen and a twofold loss in the bone marrow (Fig. 2, D and E), consistent with a partial block in maturation at the CD27+CD11b− stage. Vav1-SPPL3 KO mice exhibited a 2.6-fold reduction in the most mature CD27−CD11b+ NK cells in the bone marrow, and a 4.9-fold reduction of CD27−CD11b+ cells in the spleen. Thus, the absence of SPPL3 in the hematopoietic system resulted in the reduced expression of several NK cell surface receptors and a dramatic defect in NK cell maturation.
SPPL3 is required in a cell autonomous manner for NK cell development and function
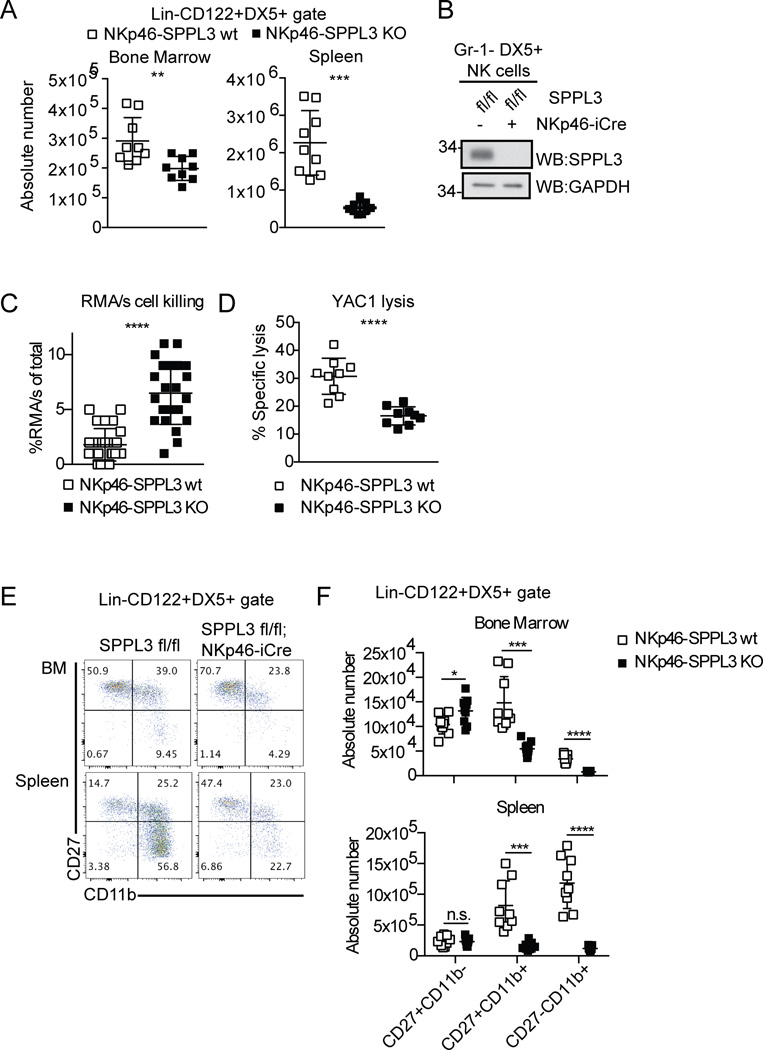
Previous studies have determined that other cells in the hematopoietic compartment can influence NK cell maturation and function (26, 27), leaving open the possibility that the effects on NK cells observed in Vav1-SPPL3 KO mice might be cell-nonautonomous. To test whether the requirement for SPPL3 in NK cell maturation and function was cell-autonomous, we bred the NK cell-specific NKp46-iCre (8) onto the SPPL3fl/fl background. SPPL3fl/fl, SPPL3fl/+, SPPL3 fl/+/NKp46-iCre mice were all used as controls (NKp46-SPPL3 wt). SPPL3fl/fl/NKp46-iCre mice (NKp46-SPPL3 KO) had a fourfold reduction in NK cell number in the spleen compared to control mice. In the bone marrow there was a 1.5-fold reduction in NKp46-SPPL3 KO mice (Fig. 3 A). Western blot analysis of NK cells sorted from the spleen confirmed efficient SPPL3 deletion in these cells, again ruling out selection for non-recombined cells (Fig. 3 B). RMA/s cells were not killed as efficiently in NKp46-SPPL3 KO mice, which had 6.5% recovery of the tumor cells, compared to control mice, which had 1.8% recovery (Fig. 3 C). Splenic NK cells isolated from NKp46-SPPL3 KO mice displayed 53% of control levels of YAC-1 killing in vitro (Fig. 3 D), indicating a cell-autonomous role for SPPL3 in NK cell frequency and cytolytic function.
Figure 3.
SPPL3 is required in a cell intrinsic manner for NK cell number and function. (A) Absolute number of NK cells in the spleen and bone marrow from NKp46-iCre mice. Pooled data from three experiments with n=3 mice per genotype. (B) Western blot showing SPPL3 expression in splenic NK cells sorted for Gr-1−DX5+ after NK cell isolation. Representative panel from two experiments with n=4–8 mice per genotype. (C) Percent RMA/s cells (of the total RMA + RMA/s cells collected) remaining 48 hours after injection. Pooled data from three independent experiments with n=2–10 mice per genotype. (D) Percent specific YAC-1 lysis after four-hour co-culture with isolated splenic NK cells (equivalent DX5+ cell number) at 2:1 E:T ratio. Pooled data from three independent experiments with n=3 mice per genotype. (E) Representative flow panels of the NK cell gate in the indicated organs. (F) The absolute number of cells in each maturation stage in NK cells. Pooled data from three independent experiments with n=3 mice per genotype. Each data point represents one mouse, except YAC-1 lysis, where each data point represents the mean. *p<0.05, ***p<0.001, ****p<0.0001 (two-tailed unpaired Student’s t-test with Welch’s correction). For experiments in panel F, statistical significance was determined with an unpaired Student’s t-test using the Holm-Sidak method, with alpha=5.000%. Each row was analyzed individually, without assuming a consistent SD.
NK cell-specific SPPL3 deletion resulted in a defect in NK cell maturation similar to that seen in Vav1-SPPL3 KO mice (Fig. 3, E and F). NKp46-SPPL3 KO mice had fivefold fewer CD27+CD11b+ NK cells and ninefold fewer CD27−CD11b+ NK cells in the spleen. In the bone marrow there was a 2.7-fold loss of CD27+CD11b+ cells and fourfold loss of CD27−CD11b+ cells. There was a small, 1.3-fold, but significant build-up of CD27+CD11b− cells in the bone marrow, with no change in the spleen. These results establish that SPPL3 is required in a cell-intrinsic manner for NK cell maturation.
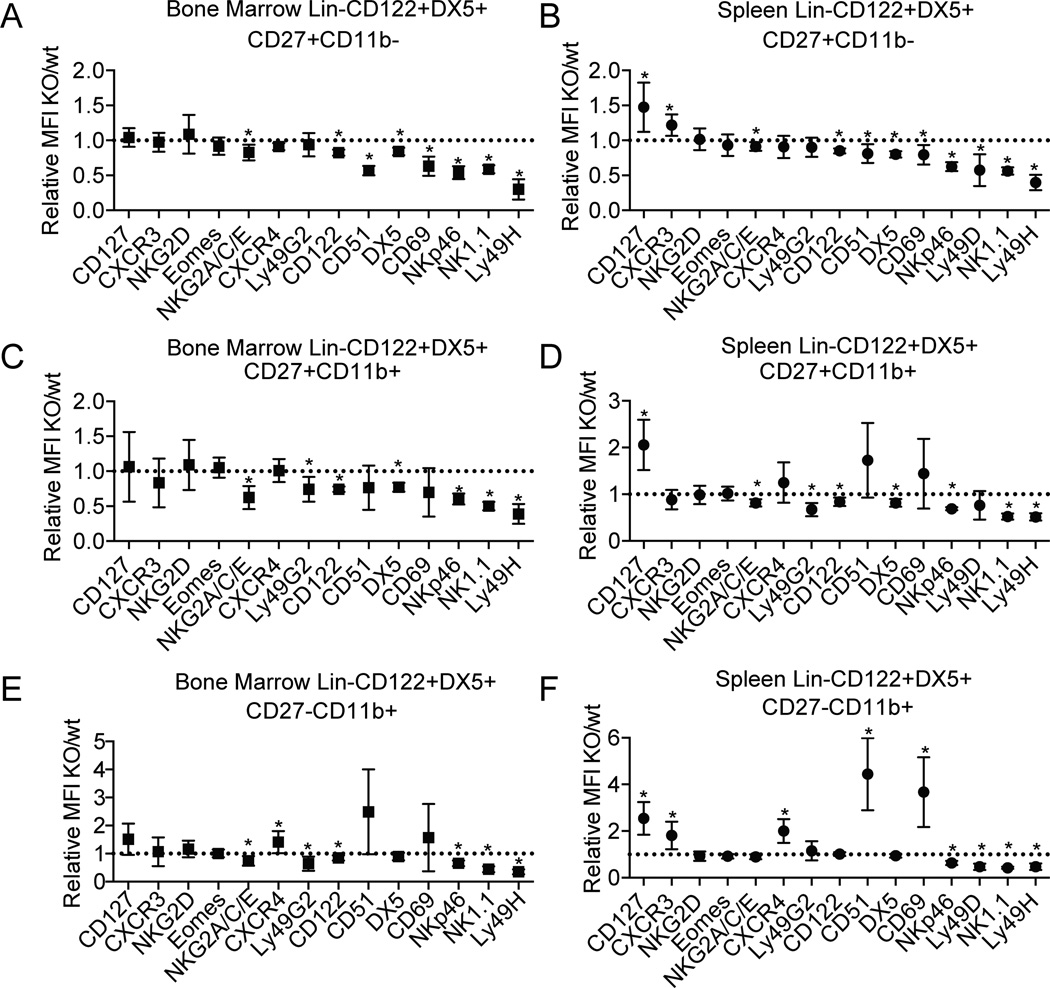
To probe for molecular defects that might parallel the specific loss of CD27+CD11b+ and CD27−CD11b+ NK cells in NKp46-SPPL3 KO mice, we assayed the different maturation stages for cell surface expression of activating and inhibitory Ly49 receptors, chemokine and cytokine receptors, adhesion molecules, as well as markers of NK cell differentiation that we had previously analyzed in the Vav1-SPPL3 KO model (Figure 4 and Supplemental Figure 2). Most of the molecules assayed displayed only subtle (less than twofold) changes in surface expression on NK cells from NKp46-SPPL3 KO mice without a selective change in CD11b+ stages that appeared to be most affected by SPPL3 deletion. The largest effects observed were on integrin alpha V (CD51) and CD69. CD51 was upregulated up to 4.4-fold in the spleen CD27-CD11b+ stage, and up to 2.4-fold in the same stage in the bone marrow (Fig. 4 E, F). CD69 was upregulated 3.7-fold in CD27-CD11b+ cells in the spleen, and 1.5-fold in the bone marrow. CD127 (the IL-7 receptor) was also upregulated by 1.5- to 2.5-fold on all maturation subsets in the spleen in NKp46-SPPL3 KO mice (Fig. 4 B, D, F). All subsets in both the spleen and bone marrow displayed two- to threefold reductions in the activating receptor Ly49H, but not in the activating receptor NKG2D or in the inhibitory receptor Ly49G2. Activating receptors NK1.1 and NKp46 were also reduced up to twofold in all three maturation stages in NKp46-SPPL3 KO mice. Notably, the transcription factor eomesodermin, which is required for NK cell progression to the CD11b–expressing stage as well as maintenance of the mature phenotype (4), was unaffected at any stage of maturation. The subtle changes in surface molecule expression that were measured in NKp46-SPPL3 KO mice did not obviously explain the cellular defects observed in NK cell maturation.
Figure 4.
Survey of expression of regulators in SPPL3-deficient NK cells. The relative mean fluorescence intensity of the indicated proteins are shown on NKp46-SPPL3 KO NK cells relative to NKp46-SPPL3 WT controls on CD27+CD11b− BM (A) and splenic NK cells (B), CD27+CD11b+ BM (C) and splenic NK cells (D), and CD27−CD11b+ BM (E) and splenic NK cells (F). Each data point represents the pooled mean of three independent experiments each with n=3–4 mice per genotype. *p<0.05 (unpaired Student’s t-test, statistical significance determined using the Holm-Sidak method, with alpha=5.000%. Each gene was analyzed individually, without assuming a consistent SD).
Alterations in maturation can influence cytotoxicity
The terminal maturation program coincides with the acquisition of the full complement of activating and inhibitory receptors on NK cells (7, 8). CD11blo NK cells from RAG1-deficient mice show reduced cytotoxic capacity against YAC-1 targets compared to CD11bhigh cells (9). Furthermore, CD27+CD11b+ NK cells from RAG1-deficient mice display increased YAC-1 cytotoxicity compared to the most mature, CD27−CD11b+, NK cells (7). To test whether the reduction in cytotoxicity of SPPL3-deficient NK cells could be at least partially attributed to their bulk population shift to a more immature status, we sorted wild type C57Bl/6J splenic NK cells into the three maturation stages (Fig. 5 A) and tested each in the YAC-1 lysis assay. CD27+CD11b− NK cells exhibited 55% of the cytotoxicity of the CD27+CD11b+ NK cells, which showed the maximum cytotoxic activity, and CD27−CD11b+ showed 65% of this activity (Fig. 5, B and C). This determination allowed us to conclude that the change in distribution among these maturation stages in NKp46-SPPL3 KO mice can account for the functional per-cell cytoxicity defect observed in vitro.
SPPL3 supports the proliferation and survival of NK cells but is not required for IL-15 signaling
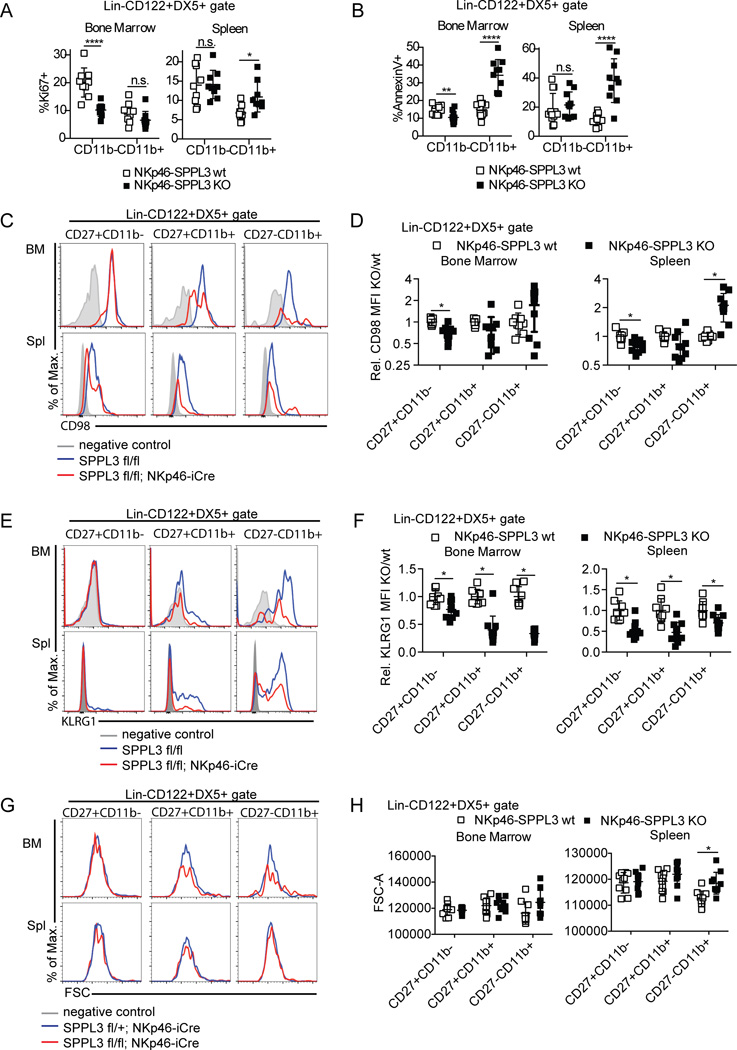
The effect of SPPL3 deletion on NK cell maturation could be explained by changes in NK cell proliferation or death. To assay proliferation, splenic and bone marrow NK cells were stained for the proliferation marker Ki67. NKp46-SPPL3 KO cells showed a twofold reduction in percent Ki67+CD11b− NK cells in the bone marrow, from 20% in control mice to 10% in NKp46-SPPL3 KO mice, with no change in the percent of Ki67+CD11b+ cells (Fig. 6 A). Splenic CD11b− NK cells showed no change in Ki67 staining, and even a small increase in the CD11b+ fraction (6.8% in controls and 10.8% in NKp46-SPPL3 KO). The reduced CD11b− precursor proliferation in the bone marrow likely affects output to the periphery, potentially contributing to lower peripheral NK cell numbers.
Figure 6.
SPPL3 regulates NK cell proliferation and survival in vivo. (A) Percent Ki67+ NK cells (Lin−CD122+DX5+) in the spleen and bone marrow. Pooled data from three experiments with n=3 mice per genotype. (B) Annexin V staining of splenic and bone marrow NK cells. Pooled data from three independent experiments with n=3–4 mice per genotype. (C, E, G) Representative flow panels showing the expression of CD98 (C), KLRG1 (E), and forward scatter (FSC, G) on the indicated NK cell gates in each organ. (D, F, H) Mean fluorescensce intensity of CD98 (relative, D), KLRG1 (relative, F), and FSC (absolute, H) on NKp46-SPPL3 KO NK cells in spleen and bone marrow, as compared to NKp46-SPPL3 wt controls. Pooled data from three independent experiments with n=3–4 mice per genotype. Each data point represents one mouse. *p<0.05, ****p<0.0001 (unpaired Student’s t-test, statistical significance determined using the Holm-Sidak method, with alpha=5.000%. Each subset was analyzed individually, without assuming a consistent SD).
To assess cell death, we stained splenocytes and bone marrow lymphocytes with Annexin V. NKp46-SPPL3 KO CD11b− NK cells in the bone marrow actually showed a moderate reduction in cell death compared to control cells (14% in controls compared to 10% in NKp46-SPPL3 KO), while no change in Annexin V staining was observed in splenic CD11b− NK cells. However, NKp46-SPPL3 KO CD11b+ NK cells showed a twofold increase in Annexin V staining in the bone marrow, from 15% to 34%, and a threefold increase in the spleen, from 11% to 38% (Fig. 6 B). These results suggest that SPPL3 is required both for proliferation of CD11b− NK cells and for the survival of CD11b+ NK cells that do make the transition from CD11b− precursors, which may account for the maturation defect seen in SPPL3 deficient animals.
IL-15 signaling to immature NK cells regulates homeostatic proliferation in vivo, in part via the activation of the mTOR pathway, which regulates many metabolic processes (28). mTOR deficiency in NK cells results in a loss of mature, CD11b+ NK cells similar to what we observed in NKp46-SPPL3 KO mice, and mTOR-deficient NK cells also show reduced proliferation in the bone marrow and reduced peripheral activation (29). We assayed the levels of three downstream markers of mTOR activity in freshly isolated NK cells: CD98, KLRG1, and cell size (Fig. 6 C–H). CD98 expression was only moderately reduced in the immature CD27+CD11b- bone marrow NK cells, to 70% of control levels, and in the spleen to 79% of control levels (Fig. 6 D). KLRG1 was reduced at all maturation stages, by as much as threefold in CD27-CD11b+ cells (Fig. 6 E–F), consistent with a defect in NK cell maturation that might or might not be mTOR-dependent. However, cell size was not reduced at all in any subtype in either bone marrow or spleen (Fig. 6 G–H), suggesting that mTOR activity in freshly isolated SPPL3-deficient NK cells is likely intact.
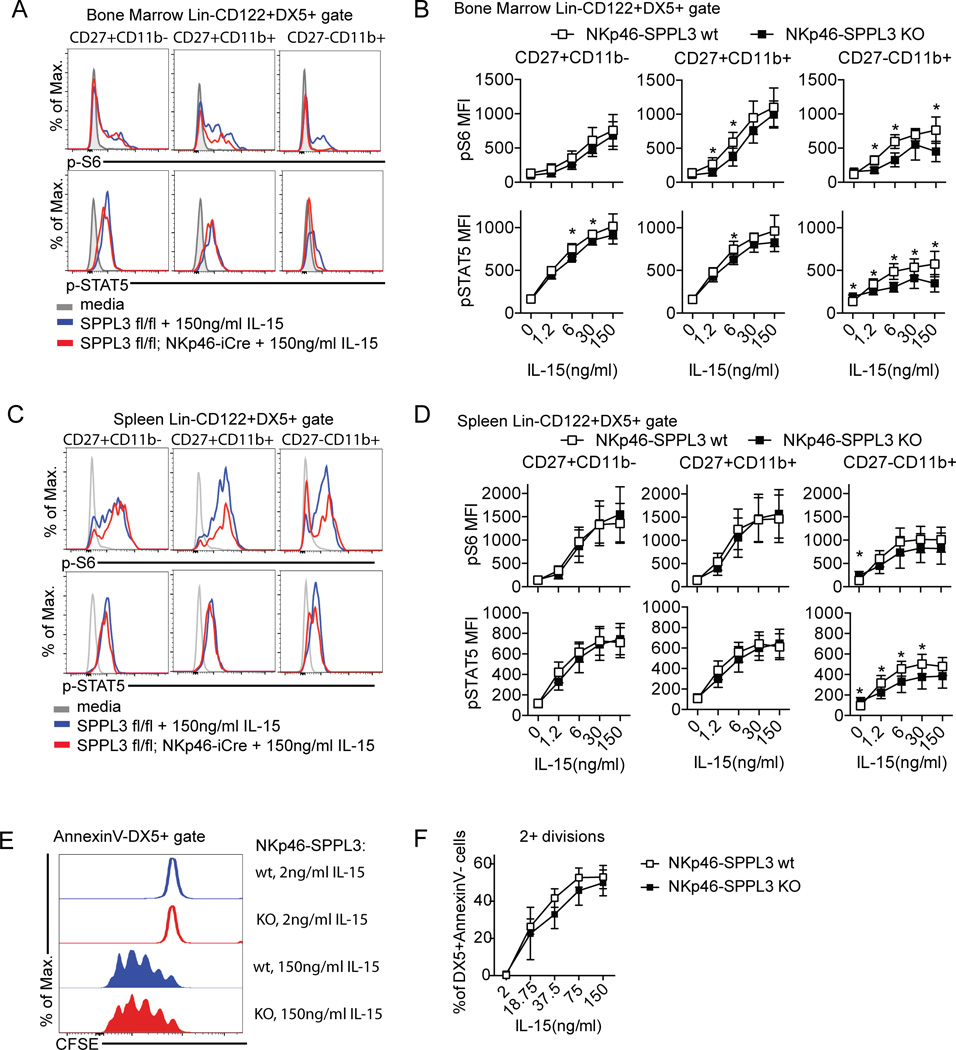
We next tested the response of NKp46-SPPL3 KO NK cells to IL-15 by assaying the induction of phospho-S6 kinase and phosho-STAT5, both markers of mTOR activation, after 40 minutes of IL-15 stimulation. SPPL3-deficient NK cells from the bone marrow and spleen showed robust responses to IL-15 in these assays (Fig. 7 A–D). Moreover, proliferation in response to any dose of IL-15 was normal in SPPL3-deficient NK cells (Fig. 7 E–F). The results suggest that the absence of SPPL3 in NK cells does not impact IL-15 signaling to mTOR in immature NK cells and likely affects NK cell maturation through a different pathway.
Figure 7.
SPPL3 does not affect IL-15 signaling. (A, C) Representative flow panels showing the expression of phospho-S6 and phospho-STAT5 in response to graded doses of IL-15 in NK cells in the bone marrow (A) and spleen (C). (B, D) Mean fluorescence intensity of phospho-S6 and phospho-STAT5 in the indicated NK cell subsets in response to graded doses of IL-15 in the bone marrow (B) and spleen (D). (E) Representative flow panels showing CFSE dilution in NK cells (DX5+AnnexinV− gate) after three day culture in graded doses of IL-15 (F) Percent of NK cells that had two or more divisions by CFSE dilution after three day culture in IL-15. Pooled data from three independent experiments with n=3–4 mice per genotype. Each data point represents the pooled mean. Results were analyzed for significance with an unpaired Student’s t-test, statistical significance determined using the Holm-Sidak method, with alpha=5.000%. Each IL-15 dose was analyzed individually, without assuming a consistent SD).
SPPL3 protease activity is required for normal peripheral NK cell frequency
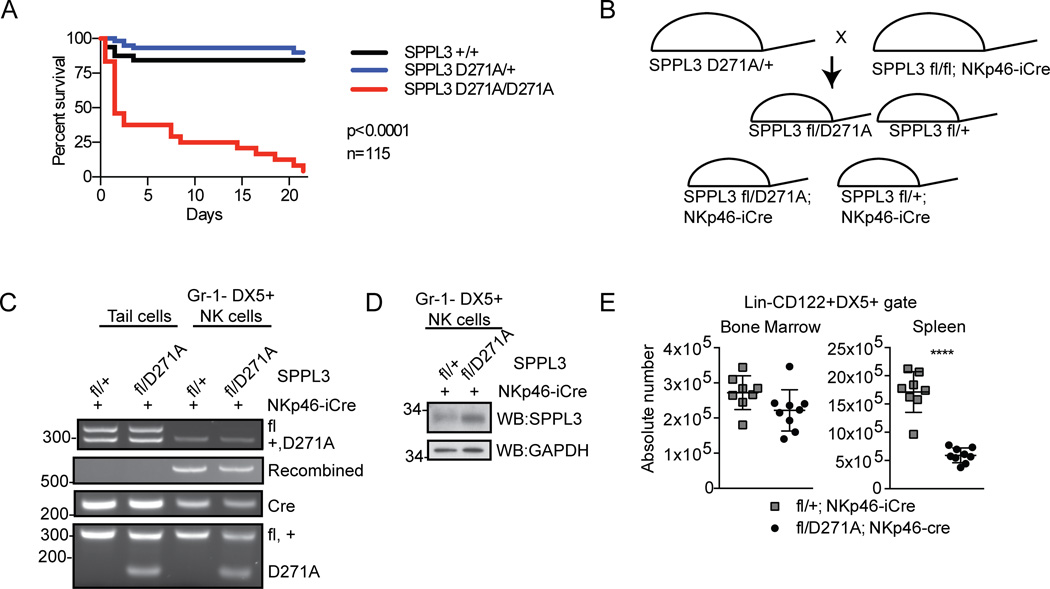
Both proteolytic and non-proteolytic functions of SPPL3 have recently been reported. To address whether SPPL3 protease activity is required for its role in NK cell biology, we used CRISPR/Cas9 genome editing in single cell zygotes to generate mice with a knock-in mutation of SPPL3 D271A (25). This residue is part of the YD and GXGD active site motif that is highly conserved among intramembrane aspartyl proteases and whose mutation to alanine has been shown to abrogate protease activity (30). SPPL3D271A/D271A mice displayed perinatal lethality similar to that observed with SPPL3−/− mice (Fig. 8 A). We next generated SPPL3fl/D271A/NKp46-iCre mice to engineer NK cell-specific expression of SPPL3 D271A in the absence of wild type SPPL3 (Fig. 8 B). These mice were obtained in Mendelian ratios and survived without overt phenotype. Genomic PCR and Western blot analysis of splenic NK cells sorted from SPPL3fl/D271A/ NKp46-iCre mice confirmed recombination of the floxed wild type SPPL3 allele and expression of SPPL3 D271A (Fig. 8, C and D). This demonstrated that the SPPL3 D271A mutant protein was expressed and that there was no selection for non-recombined NK cells. SPPL3fl/D271A/ NKp46-iCre mice exhibited no change in bone marrow NK cell number compared to control mice (SPPL3fl/+/NKp46-iCre), and a 2.9-fold reduction in NK cell number in the spleen (Fig. 8 E). The results establish that SPPL3 protease activity is required in a cell-autonomous manner for normal numbers of peripheral NK cells.
Figure 8.
SPPL3 protease activity is required in a cell-autonomous manner for normal NK cells. (A) Kaplan-Meier survival curve of SPPL3 D271A–expressing mice (n=115 pups, p value was calculated by Mantel-Cox test). (B) Schematic of breeding strategy to create conditional expression of SPPL3 D271A exclusively within NK cells. (C) PCR on bulk cells (tail biopsy) or sorted splenic NK cells (Gr-1−DX5+) after isolation. Panels from a single experiment with n=4–8 mice per genotype. (D) Western blot of sorted splenic NK cells (Gr-1−DX5+) after isolation from mice of the indicated genotypes. Representative panels from two experiments with n=4–8 mice per genotype. (E) Absolute number of NK cells (Lin−CD122+DX5+) in the bone marrow and spleen. Pooled data from four experiments with n=3–4 mice per genotype. Each data point represents one mouse. **p<0.01, ****p<0.0001 (two-tailed unpaired Student’s t-test with Welch’s correction).
SPPL3 protease activity is required for normal NK cell maturation
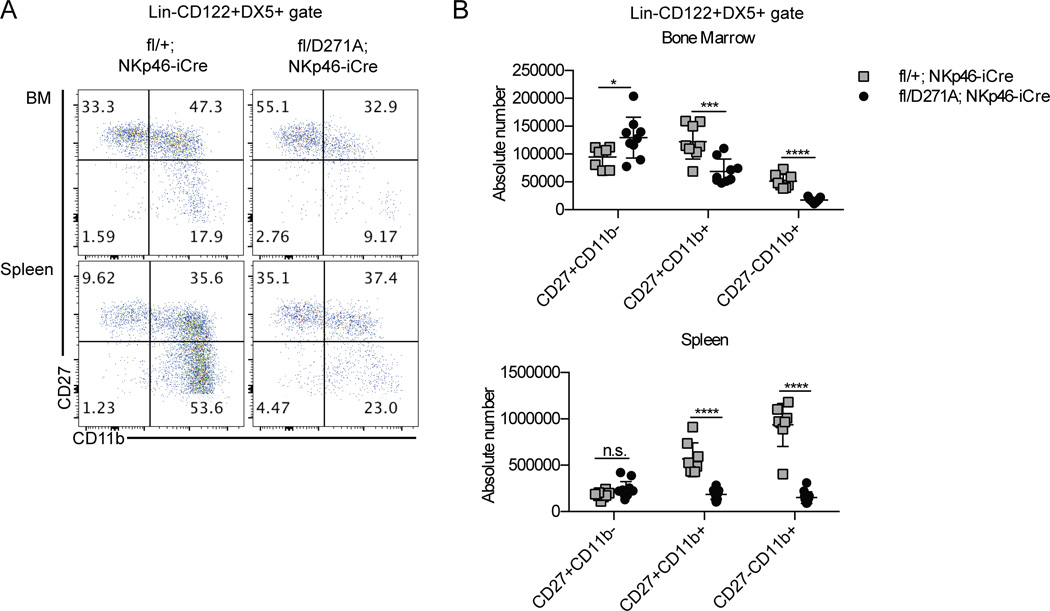
We next examined NK cell maturation in SPPL3fl/D271A/NKp46-iCre and control mice. SPPL3fl/D271A/NKp46-iCre mice exhibited a threefold reduction in CD27−CD11b+ NK cells in the bone marrow, and a sixfold reduction in the spleen (Fig. 9, A and B). There was a 1.8-fold loss of CD27+CD11b+ cells in the bone marrow, and a threefold loss in the spleen. The bone marrow showed a moderate, 1.4-fold, increase in CD27+CD11b− NK cells, with no significant change in the spleen. This phenotype resembled that observed with SPPL3fl/fl/NKp46-iCre mice, indicating that SPPL3 protease activity is required for normal NK cell maturation.
Figure 9.
SPPL3 protease activity is required in a cell-autonomous manner for normal NK cell maturation. (A) Representative flow panels of the NK cell gate (Lin−CD122+DX5+) in the indicated organs. (B) The absolute number of cells in each maturation stage in NK cells. Pooled data from four independent experiments with n=3–4 mice per genotype. Each data point represents one mouse. *p<0.05, ***p<0.001, ****p<0.0001. Results were analyzed for significance with an unpaired Student’s t-test, statistical significance determined using the Holm-Sidak method, with alpha=5.000%. Each subset was analyzed individually, without assuming a consistent SD).
SPPL3 regulates MGAT5 expression in NK cells
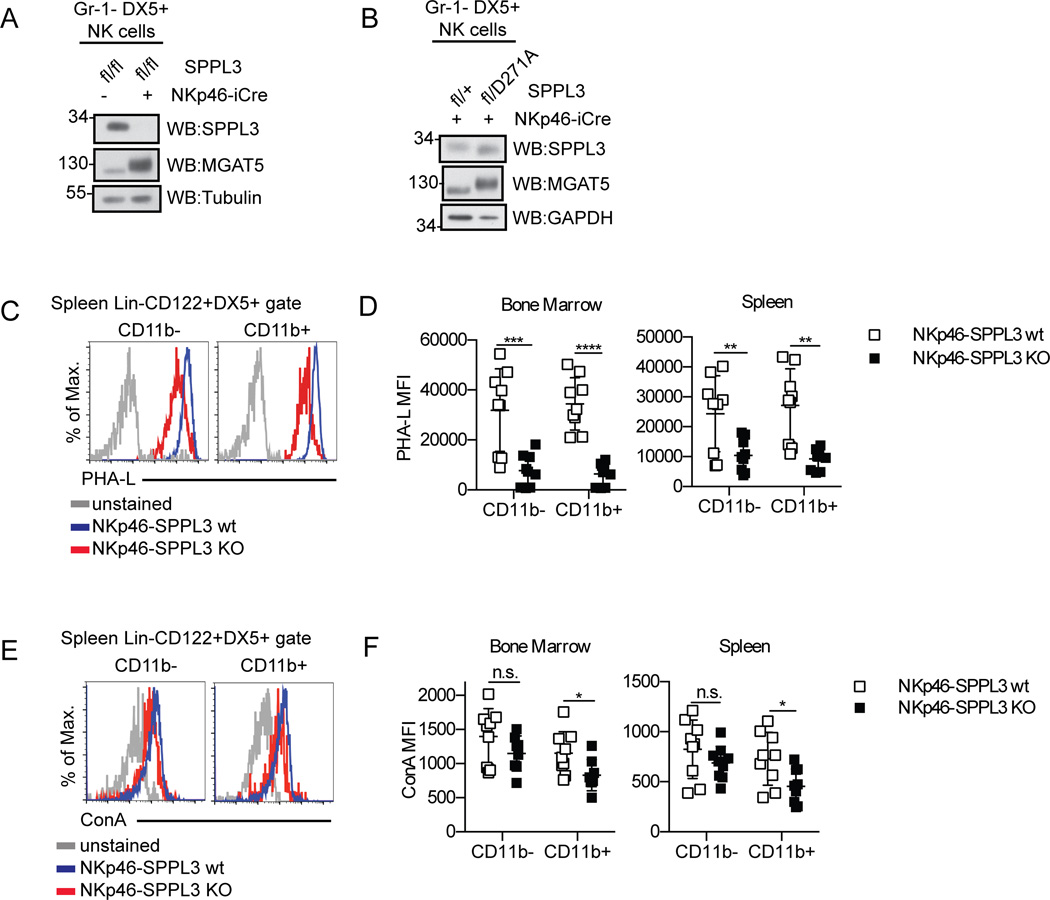
SPPL3 has recently been reported to affect glycosylation in the Golgi through inhibitory cleavage and shedding of glycosyl transferases, such as MGAT5 (21, 22). In SPPL3-deficient MEFs, MGAT5 is retained within the cell and there is an increase in higher-order glycosylation (21). Consistent with previous reports, we found that SPPL3-deficient NK cells isolated from NKp46-SPPL3 KO mice displayed an increase in MGAT5 intracellular expression (Fig. 10 A). We observed an apparent increase in the molecular weight of MGAT5 in SPPL3-deficient and protease-dead SPPL3-expressing NK cells. This is consistent with a previous report which found that in the absence of SPPL3, MGAT5 is retained in the cells as a mature, glycosylated species with slower migration on a gel (21). NK cells from SPPL3fl/D271A/NKp46-iCre mice also showed an increase in MGAT5 expression (Fig. 10 B), confirming a role for the protease activity of SPPL3 in MGAT5 regulation.
Figure 10.
SPPL3 regulates glycosylation of NK cells. (A, B) Western blot of sorted splenic NK cells (Gr-1−DX5+) after isolation of the indicated genotypes. Representative panels from two independent experiments with n=4–8 mice per genotype. (C, E) Surface staining of PHA-L (C) and ConA (E) on NK cell gate (Lin−CD122+DX5+). Representative flow panels from spleen. (D, F) Median fluorescence intensity of PHA-L (D) and ConA (F) on NK cells from the indicated organs. Pooled data from three experiments with n=2–3 mice per genotype. Each data point represents one mouse. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 Results were analyzed for significance with an unpaired Student’s t-test, statistical significance determined using the Holm-Sidak method, with alpha=5.000%. Each genotype was analyzed individually, without assuming a consistent SD).
These results predicted that enhanced levels of intracellular MGAT5 activity would lead to higher levels of complex glycosylation on proteins expressed on the surface of NK cells. We tested this prediction by staining NKp46-SPPL3 KO and control NK cells with PHA-L, which detects GlcNac-β1,6-mannose glycosylation (31), and Concanavalin A, which detects mannose-containing lectins (32). Unexpectedly, PHA-L surface staining on SPPL3-deficient NK cells from the spleen and bone marrow actually showed lower surface complex glycosylation levels (Fig. 10, C and D). In the bone marrow, PHA-L staining was reduced fourfold on CD11b− NK cells and fivefold on CD11b+ NK cells. In the spleen, PHA-L staining was reduced twofold on CD11b− NK cells and threefold on CD11b+ cells. Concanavalin A surface staining levels on these cells was minimally affected or unaffected (Fig. 10, E and F). The data argue against a simple model in which the phenotype of SPPL3-deficient NK cells is solely explained by enhanced complex glycosylation concomitant with the hyperactivity of MGAT5 and other glycosylation enzymes. Furthermore the reduction in complex glycosylation at the cell surface was consistent across NK cell maturation stages, while the biological effect of SPPL3 deletion was specifically observed in CD11b+ NK cells. Other targets of SPPL3 proteolytic activity are likely responsible for the changes in NK cell biology that result from SPPL3 deletion.
DISCUSSION
Our data establishes the intramembrane aspartyl protease SPPL3 as a key regulator of NK cell maturation. Deletion of SPPL3 in the NK cell lineage leads to a specific loss in the steady state numbers of CD27+CD11b+ and CD27−CD11b+ NK cells. Our analysis suggests that this phenotype results from reduced proliferation of CD27+CD11b− precursors in the bone marrow and reduced survival of CD27+CD11b+ and CD27−CD11b+ NK cells in both the bone marrow and the periphery. Thus, SPPL3 plays specific, cell-autonomous roles that impact each of these stages of NK cell maturation.
SPPL3-deficient NK cells display clear defects in the ability to clear MHC Class I-deficient tumors in vivo and YAC-1 target cells in vitro. Our data demonstrates that at the different stages of NK cell maturation defined by CD27 and CD11b, NK cells possess differential cytotoxic potency on a per-cell basis. Previous studies have arrived at consistent conclusions. From RAG1-deficient animals, CD11blo NK cells show lower YAC-1 lysis than CD11bhi NK cells (9). Also from RAG1-deficient animals, CD27+CD11b+ show higher YAC-1 lysis than CD27−CD11b+ NK cells (7). Our data using NK cells from wild type mice confirms that CD27+CD11b+ NK cells are the most cytotoxic cells in this assay. Additionally, we have shown that CD27+CD11b− and CD27−CD11b+ NK cells have equal cytolytic capabilities, both lower than the double positive NK cells. From this analysis, we estimate that the shift in maturation in SPPL3-deficient mice can account for the reduced cytotoxic potential observed in vitro.
While NKp46-iCre-mediated deletion of SPPL3 allowed us to conclude that SPPL3 exerts a cell-autonomous role in NK cell maturation, SPPL3 deletion in the hematopoietic system with Vav1-iCre did result in a more pronounced buildup of CD27+CD11b− precursors in both the bone marrow and spleen, suggesting the possibility of a cell-nonautonomous role for SPPL3 in other hematopoietic cells that influence the transition from CD11b− NK cells to CD11b+ cells. Alternatively, SPPL3 may also be required cell-autonomously at an early stage in NK cell development, at a step prior to NKp46 expression, that later influences the CD11b− to CD11b+ transition.
Both protease-independent and protease-dependent functions for SPPL3 have recently been described (20–22). The phenotypes observed after expression of the protease-dead SPPL3 D271A allele in NK cells reveal that NK cell maturation relies on the proteolytic activity of SPPL3, and suggest that the protease-independent function of SPPL3 in facilitating store-operated calcium entry is not required in this context. Recently, SPPL3 has been shown to cleave and inhibit several enzymes that mediate complex glycosylation in the Golgi, including MGAT5. We were able to show that in NK cells, MGAT5 levels are indeed controlled by SPPL3 proteolytic activity. However, the increase in MGAT5 does not lead to the predicted increase in complex glycosylation on the surface of NK cells. Rather, a reduced overall level of complex glycosylation is observed, as revealed by PHA-L staining. This pattern is observed on both CD11b− and CD11b+ cells, while the biological effect of SPPL3 deletion is most pronounced on CD11b+ cells, suggesting that the changes in glycosylation do not simply account for the observed phenotypes in maturation and effector function. It is extremely likely that the lack of cleavage of other SPPL3 substrates is responsible for the phenotypes observed in mice lacking SPPL3 in NK cells. One possibility is that a particular substrate must be cleaved by SPPL3 at the CD27+CD11b− stage to maximize the proliferation of these cells while another substrate must be cleaved to promote the survival of the CD11b+ stages. Alternatively, SPPL3 may be required to clear one or more substrates by cleavage, the buildup of which might be toxic to NK cells at specific CD11b− and CD11b+ stages of maturation.
The molecular underpinnings of the terminal maturation program of NK cells are only beginning to emerge. In fact, to our knowledge, the only cell-autonomous pathway associated with a reduction of CD11b+ NK cells so far has been the IL-15-mTOR pathway. While SPPL3-deficient NK cells have a similar phenotype to mTOR-deficient NK cells in terminal maturation as well as proliferation in CD11b− bone marrow cells, our results suggest that SPPL3 functions in a distinct pathway. Activation of mTOR in response to IL-15 is intact in SPPL3-deficient immature NK cells. Additionally, mTOR-deficient NK cells do not show the same changes in cell surface receptor expression that SPPL3-deficient NK cells do. Defining valid substrates for SPPL3 in this process holds high promise for adding to our understanding of the molecular determinants of NK cell maturation and optimal effector function.
The nonconditional deletion of SPPL3 results in postnatal lethality of undetermined cause. Death is likely not caused by autoimmunity or inflammation because conditional deletion of SPPL3 in the hematopoetic- or NK-cell lineages results in healthy pups. The same phenotype observed in our SPPL3 D271A knock-in mice indicates that this role for SPPL3 is also protease-dependent and very likely not related to the regulation of store-operated calcium entry. SPPL3D271A/+ mice are viable and overtly healthy, indicating that the SPPL3 D271A protein is not dominant negative. With the use of appropriate Cre transgenics, the conditional and knock-in mice we have generated should make possible the identification of the cell type in which SPPL3 is required for postnatal viability.
In conclusion, our genetic analysis firmly places SPPL3 in the NK cell maturation pathway and establishes a novel entry point to investigate the molecular determinants that control NK cell biology. The role of SPPL3 in NK cells highlights the expanding roles of intramembrane aspartyl proteases in regulating immune system development and function.
Supplementary Material
Acknowledgments
The authors would like to thank Joseph Sun for the RMA and RMA/s cell lines, Giorgio Trinchieri for the NKp46-iCre mice, Chip Hawkins for performing zygote injections, Xiaoling Zhang for performing the cell sorting, and deMauri Mackie, John Bettridge, and Meiling May for a critical reading of the manuscript.
Footnotes
This work was supported by funds from the Johns Hopkins University Institute for Cell Engineering. CEH was supported by Ruth L. Kirschstein National Research Service Award F31AG043238-01. JLP is a Leukemia and Lymphoma Society Scholar.
SUPPLEMENTAL MATERIAL
Two supplemental figures demonstrating the gating strategy for NK cells, as well as additional phenotyping of NKp46-iCre mice can be found online.
REFERENCES
- 1.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat. Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 2.Lodolce JP, Boone DL, Chai S, Swain RE, Dassopoulos T, Trettin S, Ma A. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity. 1998;9:669–676. doi: 10.1016/s1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]
- 3.Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, Matsuki N, Charrier K, Sedger L, Willis CR, Brasel K, Morrissey PJ, Stocking K, Schuh JC, Joyce S, Peschon JJ. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J. Exp. Med. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, Lindsten T, Reiner SL. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huntington ND, Vosshenrich CAJ, Di Santo JP. Developmental pathways that generate natural-killer-cell diversity in mice and humans. Nat. Rev. Immunol. 2007;7:703–714. doi: 10.1038/nri2154. [DOI] [PubMed] [Google Scholar]
- 6.Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113:5488–5496. doi: 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- 7.Hayakawa Y, Smyth MJ. CD27 Dissects Mature NK Cells into Two Subsets with Distinct Responsiveness and Migratory Capacity. J. Immunol. 2006;176:1517–1524. doi: 10.4049/jimmunol.176.3.1517. [DOI] [PubMed] [Google Scholar]
- 8.Narni-Mancinelli E, Chaix J, Fenis A, Kerdiles YM, Yessaad N, Reynders A, Gregoire C, Luche H, Ugolini S, Tomasello E, Walzer T, Vivier E. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad. Sci. U. S. A. 2011;108:18324–18329. doi: 10.1073/pnas.1112064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S, Iizuka K, Kang H-SP, Dokun A, French AR, Greco S, Yokoyama WM. In vivo developmental stages in murine natural killer cell maturation. Nat. Immunol. 2002;3:523–528. doi: 10.1038/ni796. [DOI] [PubMed] [Google Scholar]
- 10.Friedmann E, Lemberg MK, Weihofen A, Dev KK, Dengler U, Rovelli G, Martoglio B. Consensus analysis of signal peptide peptidase and homologous human aspartic proteases reveals opposite topology of catalytic domains compared with presenilins. J. Biol. Chem. 2004;279:50790–50798. doi: 10.1074/jbc.M407898200. [DOI] [PubMed] [Google Scholar]
- 11.Voss M, Schröder B, Fluhrer R. Mechanism, specificity, and physiology of signal peptide peptidase (SPP) and SPP-like proteases. Biochim. Biophys. Acta. 2013;1828:2828–2839. doi: 10.1016/j.bbamem.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 12.Lemberg MK, Bland FA, Weihofen A, Braud VM, Martoglio B. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J. Immunol. Baltim. Md 1950. 2001;167:6441–6446. doi: 10.4049/jimmunol.167.11.6441. [DOI] [PubMed] [Google Scholar]
- 13.Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 14.Hage FEl, Stroobant V, Vergnon I, Baurain J-F, Echchakir H, Lazar V, Chouaib S, Coulie PG, Mami-Chouaib F. Preprocalcitonin signal peptide generates a cytotoxic T lymphocyte-defined tumor epitope processed by a proteasome-independent pathway. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10119–10124. doi: 10.1073/pnas.0802753105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beisner DR, Langerak P, Parker AE, Dahlberg C, Otero FJ, Sutton SE, Poirot L, Barnes W, Young MA, Niessen S, Wiltshire T, Bodendorf U, Martoglio B, Cravatt B, Cooke MP. The intramembrane protease Sppl2a is required for B cell and DC development and survival via cleavage of the invariant chain. J. Exp. Med. 2013;210:23–30. doi: 10.1084/jem.20121072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergmann H, Yabas M, Short A, Miosge L, Barthel N, Teh CE, Roots CM, Bull KR, Jeelall Y, Horikawa K, Whittle B, Balakishnan B, Sjollema G, Bertram EM, Mackay F, Rimmer AJ, Cornall RJ, Field MA, Andrews TD, Goodnow CC, Enders A. B cell survival, surface BCR and BAFFR expression, CD74 metabolism, and CD8- dendritic cells require the intramembrane endopeptidase SPPL2A. J. Exp. Med. 2013;210:31–40. doi: 10.1084/jem.20121076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneppenheim J, Dressel R, Hüttl S, Lüllmann-Rauch R, Engelke M, Dittmann K, Wienands J, Eskelinen E-L, Hermans-Borgmeyer I, Fluhrer R, Saftig P, Schröder B. The intramembrane protease SPPL2a promotes B cell development and controls endosomal traffic by cleavage of the invariant chain. J. Exp. Med. 2013;210:41–58. doi: 10.1084/jem.20121069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lückerath K, Kirkin V, Melzer IM, Thalheimer FB, Siele D, Milani W, Adler T, Aguilar-Pimentel A, Horsch M, Michel G, Beckers J, Busch DH, Ollert M, Gailus-Durner V, Fuchs H, Hrabe de Angelis M, Staal FJT, Rajalingam K, Hueber A-O, Strobl LJ, Zimber-Strobl U, Zörnig M. Immune modulation by Fas ligand reverse signaling: lymphocyte proliferation is attenuated by the intracellular Fas ligand domain. Blood. 2011;117:519–529. doi: 10.1182/blood-2010-07-292722. [DOI] [PubMed] [Google Scholar]
- 19.Friedmann E, Hauben E, Maylandt K, Schleeger S, Vreugde S, Lichtenthaler SF, Kuhn P-H, Stauffer D, Rovelli G, Martoglio B. SPPL2a and SPPL2b promote intramembrane proteolysis of TNFalpha in activated dendritic cells to trigger IL-12 production. Nat. Cell Biol. 2006;8:843–848. doi: 10.1038/ncb1440. [DOI] [PubMed] [Google Scholar]
- 20.Makowski SL, Wang Z, Pomerantz JL. A protease-independent function for SPPL3 in NFAT activation. Mol. Cell. Biol. 2015;35:451–467. doi: 10.1128/MCB.01124-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voss M, Künzel U, Higel F, Kuhn P-H, Colombo A, Fukumori A, Haug-Kröper M, Klier B, Grammer G, Seidl A, Schröder B, Obst R, Steiner H, Lichtenthaler SF, Haass C, Fluhrer R. Shedding of glycan-modifying enzymes by signal peptide peptidase-like 3 (SPPL3) regulates cellular N-glycosylation. EMBO J. 2014;33:2890–2905. doi: 10.15252/embj.201488375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn P-H, Voss M, Haug-Kröper M, Schröder B, Schepers U, Bräse S, Haass C, Lichtenthaler SF, Fluhrer R. Secretome analysis identifies novel signal Peptide peptidase-like 3 (Sppl3) substrates and reveals a role of Sppl3 in multiple Golgi glycosylation pathways. Mol. Cell. Proteomics MCP. 2015;14:1584–1598. doi: 10.1074/mcp.M115.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 2002;1(119 Suppl):S97–S101. doi: 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 24.de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soderquest K, Powell N, Luci C, van Rooijen N, Hidalgo A, Geissmann F, Walzer T, Lord GM, Martín-Fontecha A. Monocytes control natural killer cell differentiation to effector phenotypes. Blood. 2011;117:4511–4518. doi: 10.1182/blood-2010-10-312264. [DOI] [PubMed] [Google Scholar]
- 27.Jaeger BN, Donadieu J, Cognet C, Bernat C, Ordoñez-Rueda D, Barlogis V, Mahlaoui N, Fenis A, Narni-Mancinelli E, Beaupain B, Bellanné-Chantelot C, Bajénoff M, Malissen B, Malissen M, Vivier E, Ugolini S. Neutrophil depletion impairs natural killer cell maturation, function, and homeostasis. J. Exp. Med. 2012;209:565–580. doi: 10.1084/jem.20111908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marçais A, Cherfils-Vicini J, Viant C, Degouve S, Viel S, Fenis A, Rabilloud J, Mayol K, Tavares A, Bienvenu J, Gangloff Y-G, Gilson E, Vivier E, Walzer T. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat. Immunol. 2014;15:749–757. doi: 10.1038/ni.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voss M, Fukumori A, Kuhn P-H, Künzel U, Klier B, Grammer G, Haug-Kröper M, Kremmer E, Lichtenthaler SF, Steiner H, Schröder B, Haass C, Fluhrer R. Foamy virus envelope protein is a substrate for signal peptide peptidase-like 3 (SPPL3) J. Biol. Chem. 2012;287:43401–43409. doi: 10.1074/jbc.M112.371369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cummings RD, Kornfeld S. Characterization of the structural determinants required for the high affinity interaction of asparagine-linked oligosaccharides with immobilized Phaseolus vulgaris leukoagglutinating and erythroagglutinating lectins. J. Biol. Chem. 1982;257:11230–11234. [PubMed] [Google Scholar]
- 32.Cummings RD, Etzler ME. Antibodies and Lectins in Glycan Analysis. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. 2nd. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.