Abstract
Macrophages represent an important component of the tumor microenvironment and play a complex role in cancer progression. These cells are characterized by a high degree of plasticity, and alter their phenotype in response to local environmental cues. While the M1/M2 classification of macrophages has been widely used, the complexity of macrophage phenotypes has not been well studied, particularly in lung cancer. In this study we employed an orthotopic immunocompetent model of lung adenocarcinoma in which murine lung cancer cells are directly implanted into the left lobe of syngeneic mice. Using multi-marker flow cytometry, we defined and recovered several distinct populations of monocytes/macrophages from tumors at different stages of progression. We used RNA-seq transcriptional profiling to define distinct features of each population and determine how they change during tumor progression. We defined an alveolar resident macrophage population that does not change in number and expresses multiple genes related to lipid metabolism and lipid signaling. We also defined a population of tumor-associated macrophages that increase dramatically with tumor, and selectively expresses a panel of chemokine genes. A third population, which resembles tumor-associated monocytes, expresses a large number of genes involved in matrix remodeling. By correlating transcriptional profiles with clinically prognostic genes, we show that specific monocyte/macrophage populations are enriched in genes that predict outcomes in lung adenocarcinoma, implicating these subpopulations as critical determinants of patient survival. Our data underscore the complexity of monocytes/macrophages in the tumor microenvironment, and suggest that distinct populations play specific roles in tumor progression.
Keywords: lung cancer, macrophages, transcriptional profiling, flow cytometry, mouse models
Introduction
Lung cancer remains the leading cause of cancer death in men and women, and most patients die from complications of metastatic disease (1). While tumor initiation is driven by somatic mutations in oncogenic drivers and tumor suppressor genes, progression and metastasis involve critical crosstalk between cancer cells and the tumor microenvironment (TME) (2). Among the diverse cell types of the TME, macrophages have been implicated as important participants and are the most abundant non-tumor cell type in most cancers (3). Studies in several types of cancer, including lung cancer, have demonstrated that macrophage depletion either through pharmacological or genetic approaches results in slower tumor growth, implicating these cells as mediators of tumor progression (4, 5). In lung cancer, several studies have described correlations in macrophage number or phenotype with clinical outcomes (6). Many of these studies have employed the model of M1 and M2 macrophages. This model, which is based on activation of macrophages in vitro, designates a macrophage phenotype as being either “proinflammatory” M1, which is proposed to inhibit cancer progression, or alternatively activated M2, which is pro-angiogenic, immunosuppressive, and promotes cancer progression (7, 8). Although the M1/M2 classification of macrophages serves as a useful starting point, it does not take into account the complexity and plasticity of these cells. Recent studies have demonstrated that distinct populations of macrophages exist in tissues such as the lung, and that these different populations not only express distinct markers but also have different developmental origins (9). For example, the resident alveolar macrophages in the lung are derived from the embryonic yolk sac, whereas the recruited monocytes and macrophages are derived from the bone marrow.
These different populations are likely to play distinct and potentially opposing roles in cancer progression. In fact, tumors are infiltrated by different dynamically changing populations of monocytes/macrophages (8) and identifying key features of each population may help to target them therapeutically.
To study the role of the tumor microenvironment in lung cancer, we have developed an orthotopic immunocompetent mouse model, in which Lewis Lung Carcinoma cells (LLC), which were derived from a spontaneous lung adenocarcinoma in C57BL/6 mice, are directly implanted into the lungs of fully immunocompetent syngeneic C57BL/6 mice. Within 2–3 weeks these cells form a primary tumor that metastasizes to the other lung lobes, mediastinal lymph nodes, liver, and brain. We have previously shown that tumor progression in this model is associated with increases in several monocyte/macrophage populations (10). However, functional differences between these populations have not been identified, and the role of specific monocyte/macrophage subtypes in tumor progression is unknown. In this study, we have used multi-marker flow cytometry to identify and isolate resident populations as well as dynamically expanding populations of monocytes and macrophages. Using RNA-seq we have been able to profile differences in gene expression between these macrophage populations, and determine how these profiles change during tumor progression. Furthermore, using transcriptional profiles, we were able to correlate specific monocyte/macrophage populations to clinical outcomes in lung adenocarcinoma.
Materials and methods
Cells
Lewis lung carcinoma cells stably expressing luciferase (LLC-Luc) were obtained from Caliper Life Sciences and maintained in DMEM (Corning CellGro #10-017-CV) containing 10% fetal bovine serum, penicillin/streptomycin and G418 (500 ng/ml).
Orthotopic mouse model
C57BL/6 mice were obtained from Jackson Laboratory and bred and maintained in the Center for Comparative Medicine at the University of Colorado Denver. Experiments were performed in 10–16 week old males. LLC-Luc cells (1×105 in 25 µl per injection) were suspended in PBS containing 15% Growth Factor Reduced Matrigel (BD Biosciences, cat# 354230) and injected into the parenchyma of the left lung lobe through the rib cage using a 30G needle, as previously described (10). During injection, the lung was directly visualized through a 4–5 mm incision in the skin. After the procedure, the incision was closed using veterinary adhesive. All procedures were performed under protocols approved by the Institutional Animal Care and Use Committee at the University of Colorado Denver.
Preparation of single cell suspensions and flow cytometry
Tumor-bearing mice were sacrificed either 2 or 3 weeks after cancer cell injection, along with uninjected control mice. The lung circulation was perfused with PBS/heparin (20 U/ml, Sigma). Left lung lobes containing tumors were excised and weighed. For control mice, all the lung lobes were harvested for analysis. Tissues from 3–5 mice were pooled, mechanically dissociated with scissors, and incubated for 30 min at 37°C with 3.2 mg/ml Collagenase Type 2 (Worthington, 43C14117B), 0.75 mg/ml Elastase (Worthington, 33S14652), 0.2 mg/ml Soybean Trypsin Inhibitor (Worthington, S9B11099N), and DNAse I 40µg/ml (Sigma). During incubation, samples were placed in a shaking water bath and dispersed by pipetting every 10 minutes. Resulting single-cell suspensions were filtered through 70 µm cell strainers (BD Biosciences) and washed with staining buffer (PBS containing 1% FBS, 2mM EDTA, 10mM HEPES). Samples were subjected to red blood cell lysis, washed with staining buffer, and filtered through 40 µm cell strainers (BD Biosciences). Prior to staining, FcγR was blocked with anti-CD16/CD32 antibody (BD Biosciences) for 10 minutes. Cells were stained for 30–45 min at 4°C with the following antibodies: CD11b-FITC (Clone M1/70, BD Biosciences), SiglecF-PE or AlexaFluor647 (Clone E50-2440, BD Biosciences), Ly6G-PE-Cy7 (Clone 1A8, BD Biosciences), CD64-PE or AlexaFluor647 (Clone X54-5/7.1, BD Biosciences), CD11c-APC-Cy7 (Clone HL3, BD Biosciences), and CD11c-PerCP-Cy5.5 (Clone N418, BioLegend). Flow cytometry analysis and cell sorting were conducted at the University of Colorado Cancer Center Flow Cytometry Core Facility using a Gallios Flow Cytometer (Beckman Coulter) for analysis and XDP-100 or Astrios (Beckman Coulter) cell sorters for sorting. The sorting strategy involved exclusion of debris and cell doublets by light scatter, and dead cells by DAPI (1 µg/ml). Absolute cell counts were obtained using Sphero AccuCount Ultra Rainbow Fluorescent particles 3.8 (Spherotech) per manufacturer’s instructions. Data were analyzed using Kaluza Software (Beckman Coulter).
RNA extraction and RNA-seq
Total RNA was isolated from flow cytometry-sorted cells using QIAshredders and an RNeasy Plus Micro kit (Qiagen). RNA quality and quantity were analyzed using a NanoDrop and Bioanalyzer. RNA-seq library preparation and sequencing were conducted at the Genomics and Microarray Core at the University of Colorado Denver-Anschutz Medical Campus. Libraries were constructed using a NuGen Ovation Human FFPE RNA-Seq Multiplex System kit customized with mouse specific oligos for rRNA removal. Directional mRNA-seq was conducted using the Illumina HiSEQ 2000 system, using the single-read 100 cycles option.
Bioinformatic analysis
RNA-Seq reads were obtained using Illumina Hi-seq analysis Pipeline. Reads quality was checked using FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc). The median number of reads per condition was 24 million. Reads were then processed and aligned to the UCSC Mus musculus reference genome (build mm10) using TopHat v2 (11). TopHat incorporates the Bowtie v2 algorithm to perform the alignment (12). TopHat initially removes a portion of reads based on quality information accompanying each read, then maps reads to the reference genome. The pre-built Mus musculus UCSC mm10 index was downloaded from the TopHat homepage and used as the reference genome. The default parameters for TopHat were used. The aligned read files were processed by Cufflinks v2.0.2 (13). Reads were assembled into transcripts and their abundance was estimated. Cufflinks uses the normalized RNA-Seq fragment counts to measure the relative abundances of transcripts. The unit of measurement is Fragments Per Kilobase of exon per Million fragments mapped (FPKM). Confidence intervals for FPKM estimates were calculated using a Bayesian inference method (13). EdgeR is a Bioconductor software package for examining differential expression of replicated count data (14). Briefly, it used an overdispersed Poisson model to account for both biological and technical variability. Additionally it used Empirical Bayes methods to moderate the degree of overdispersion across transcripts that improving the reliability of inference. Default parameters for EdgeR were used.
Hierarchical clustering was performed using Cluster 3.0. (15). Gene expression levels were centered on the mean and normalized. Clustering was done using complete linkage and Euclidean distance as the metric. Heat maps were generated using Java TreeView (16). Pathway overrepresentation analysis was conducted using ConsensusPathDB (http://cpdb.molgen.mpg.de/MCPDB) (17, 18). Searches were conducted in KEGG and Reactome databases with cut-off values of p<0.01 and a minimum of 3 overlapping genes. The set of initially selected 2458 differentially expressed genes was used as background. The analyses were additionally confirmed by DAVID (http://david.abcc.ncifcrf.gov/ ) (19, 20), and Gene Ontology (GO) (21, 22).
For comparison with profiles of immune cells published by the Immgen Project Consortium, the following populations from naïve mice were used: lung alveolar macrophages (MF_Lu), lung CD11b+ macrophages (MF_11b+_Lu), blood monocytes classical (Mo_6C+II+_Bl, Mo_6C+II−_Bl), and blood monocytes nonclassical (Mo_6C−II−_Bl, Mo_6C−II+_Bl, Mo_6C−IIint_Bl). The top 100 genes differentially expressed in any pairwise comparison between these populations were downloaded from the website (www.Immgen.org) and combined into a list of 645 genes. The expression of these 645 genes among the populations in this study was examined and subjected to hierarchical clustering. Clusters of genes highly expressed selectively in one cell type were identified. Expression of genes in these clusters was mapped back to the reference Immgen populations using the Immgen website tool “my gene set”.
For the analysis of clinically prognostic genes, the list of ~23,000 genes and their associated survival meta-Z scores for lung adenocarcinoma were downloaded from https://precog.stanford.edu/index.php. The survival scores were used as the metric to rank the genes, and the Gene Set Enrichment Analysis (GSEA, pre-ranked tool) (23, 24) was used to examine enrichment of good or bad prognostic genes in the gene clusters highly expressed in MacA or MacB cells as defined in Fig. 3.
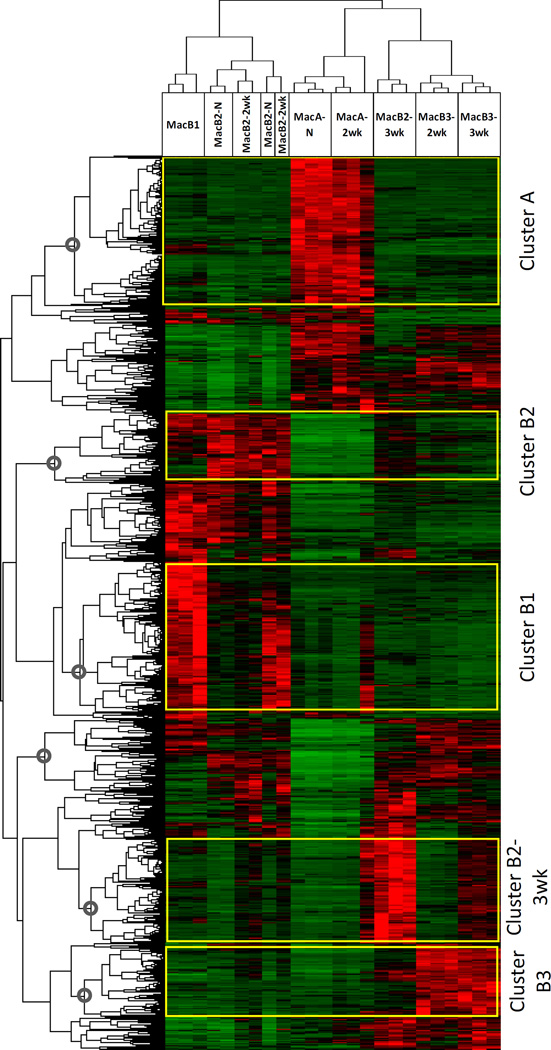
Fig. 3. Global analysis of genes differentially expressed between MacA and MacB populations.
Differentially expressed genes were identified in all pairwise comparisons between the recovered populations from our RNA-seq analysis (2458 genes) Hierarchical clustering was performed on the set of 2458 genes and clusters with high expression in specific populations were identified: Cluster A – genes highly and selectively expressed in MacA-N and MacA-2wk; Cluster B2 –highly expressed in MacB2-N and MacB2-2wk; Cluster B1 –highly expressed in MacB1-N; Cluster B2-3wk –highly expressed in MacB2-3wk; Cluster B3 –highly expressed in MacB3-2wk and MacB3-3wk.
The RNA-seq data reported in this manuscript have been deposited in The NCBI Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/geo/), with the accession number GSE76033.
Quantitative real-time-PCR
Total RNA was isolated from flow cytometry-sorted cells using QIAshredders and an RNeasy Micro kit (Qiagen), and converted into complementary DNA using an iScript cDNA synthesis kit (BioRad). Real time PCR was conducted as previously described (10).
Eicosanoid Measurements
Mice were sacrificed 2.5 weeks after cancer cell injection, along with uninjected control mice, and macrophage populations were recovered by flow cytometry. Immediately after sorting, cells were stimulated with a calcium ionophore, and eicosanoids were extracted and measured by liquid chromatography/tandem mass spectrometry previously described (10). Results were normalized to total protein.
Results
Identification of myeloid cell subsets in the lung during tumor development
Lewis Lung Carcinoma cells (LLC) were injected directly into the left lung lobe of fully immunocompetent syngeneic wild-type C57BL/6 mice. We harvested the whole tumor-bearing left lung lobes 2 and 3 weeks after injection, along with lungs from uninjected control mice. Analysis of weights revealed rapid tumor growth within the lung parenchyma (Fig. 1A). Tissues were processed into single-cell suspensions and analyzed by flow cytometry using myeloid-specific cell-surface markers.
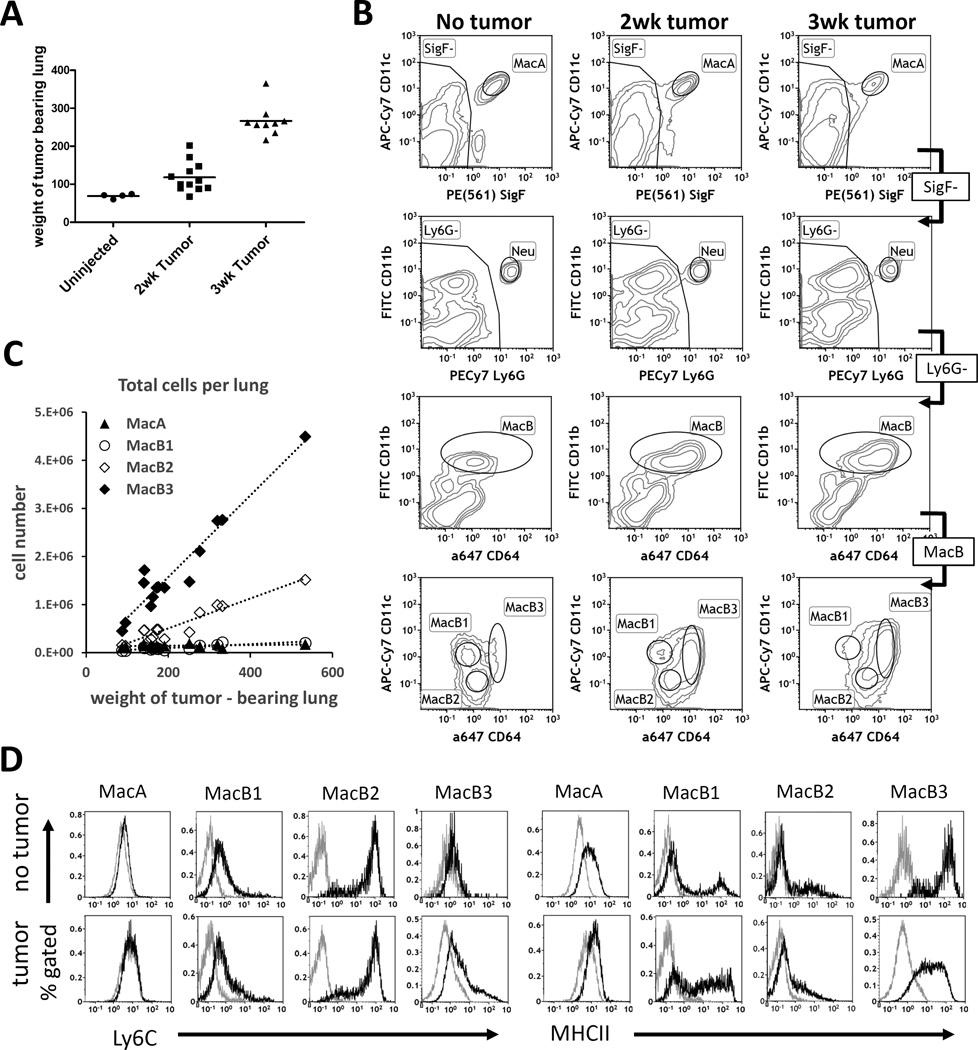
Fig. 1. Identification of myeloid cell populations in lung during tumor growth.
Lewis lung carcinoma cells were injected directly into the lungs of syngeneic C57BL/6 mice, and tumor-bearing left lobes were harvested 2 or 3 weeks after injection, along with lungs from uninjected mice. For each time point, the injection and harvest was repeated 3 times independently, with 3-5 mice pooled each time. (A). Tumor growth: weights of uninjected and tumor-bearing left lung lobes. Uninjected n=12 mice total (weight measurements not taken for all mice), 2wk tumor n= 12 mice, 3wk tumor n=9 mice. (B) Gating strategy to identify MacA, MacB1, MacB2, and MacB3 cells. Representative plots for No-tumor control, 2-week tumor, and 3-week tumor bearing mice. (C) Total absolute numbers of MacA and MacB populations in tumor – bearing left lung lobes quantified in separate experiments. (D) Characterization of Ly6C and MHCII expression in MacA and MacB populations.
In addition to the CD11b and CD11c markers, which are traditionally used for identifying macrophages in the lung (25, 26), our strategy employed SiglecF (SigF) and CD64 markers (9). SiglecF is a marker specific for murine lung resident alveolar macrophages that is not expressed by interstitial or inflammatory macrophages. Although SiglecF also stains eosinophils, their lack of CD11c expression allows us to exclude eosinophils from the monocyte/ macrophage pool (9, 27–29). CD64 is a macrophage-specific marker, and in contrast to F4/80, its expression is low or negative on monocytes, dendritic cells and granulocytes (9).
Using a modification of our previously published gating strategy (10) shown in Fig. 1B, we identified multiple subsets of myeloid cells. In both naïve and tumor-bearing lungs, we identified MacA cells, which bear markers of alveolar macrophages (SiglecF+/CD11c+). Further, we excluded eosinophils (SigF+/CD11c−, Eos gate) and neutrophils (Ly6G+/CD11b+, Neu gate). The remaining CD11b+ cells (MacB gate), were separated into 3 distinct populations based on CD11c and CD64 expression: MacB1 cells (CD11b+/CD64low/CD11c+), MacB2 cells (CD11b+/CD64intermediate/CD11c-), and MacB3 cells (CD11b+/CD64Hi/CD11c+). As shown in the flow cytometry plots, the MacB compartment in naïve lungs was dominated by MacB1 and MacB2 cells, with MacB3 present at very low numbers. Experiments quantifying absolute cell numbers of these populations in the lung, revealed that the total numbers of MacA and MacB1 cells in the left lobe did not change with tumor growth, whereas the MacB2 and MacB3 populations rapidly expanded with increasing tumor size, with the most rapid expansion seen in MacB3 cells (Fig. 1C).
We further characterized MacA and MacB cells using Ly6C and MHCII, which are markers frequently used to phenotype murine myeloid populations by flow cytometry (Fig. 1D). MacA cells, in the presence or absence of tumor, were negative for Ly6C and stained weakly for MHCII, as expected for alveolar macrophages. MacB1 cells expressed low levels of Ly6C in the presence or absence of tumor. In naïve lung, this gate contained both MHCII-negative and MHCII-positive populations, indicating that the MacB1 gate was composed of at least two distinct cell types, with the MHCII-positive cells likely representing CD11b+ dendritic cells (9, 30). In the presence of tumor, the MHCII expression on the cells in the MacB1 gate was heterogeneous, ranging from negative to high. MacB2 cells, both with and without tumor, expressed high levels of Ly6C but had little to no expression of MHCII, which is typical for monocytes (31). MacB3 cells, which in naïve mice were present in very low numbers, were negative for Ly6C and expressed high levels of MHCII, which is consistent with interstitial/infiltrating lung macrophages (31). In the presence of tumor, the MacB3 gate was composed of cells with low expression of Ly6C and intermediate expression of MHCII.
Recovery of cells and transcriptional profiling
To gain further insight into the identity and functional properties of myeloid cells in the lung tumor microenvironment, we performed transcriptome profiling using RNA-seq. We used flow-cytometry based sorting to recover cells from tumor-bearing lungs 2 weeks and 3 weeks after cancer cell injection, as well as from uninjected control mice. RNA was extracted from 3 pools of mice that were independently injected and harvested, with 3–5 mice contributing to each pool. We obtained sufficient cell numbers and RNA quantity to sequence the following populations isolated from 3 independent experiments: from naive mice: MacA cells (MacA-N), MacB1 cells (MacB1-N), MacB2 cells (MacB2-N); from 2 week tumor bearing mice: MacA cells (MacA-2wk), MacB2 cells (MacB2-2wk), MacB3 cells (MacB3-3wk); from 3-week tumor bearing mice: MacB2 (MacB2-3wk), MacB3 cells (MacB3-3wk). We performed RNA-seq profiling on these samples, compared their transcriptional profile to published databases, and analyzed differential gene expression between these populations.
Comparison to Immgen profiles
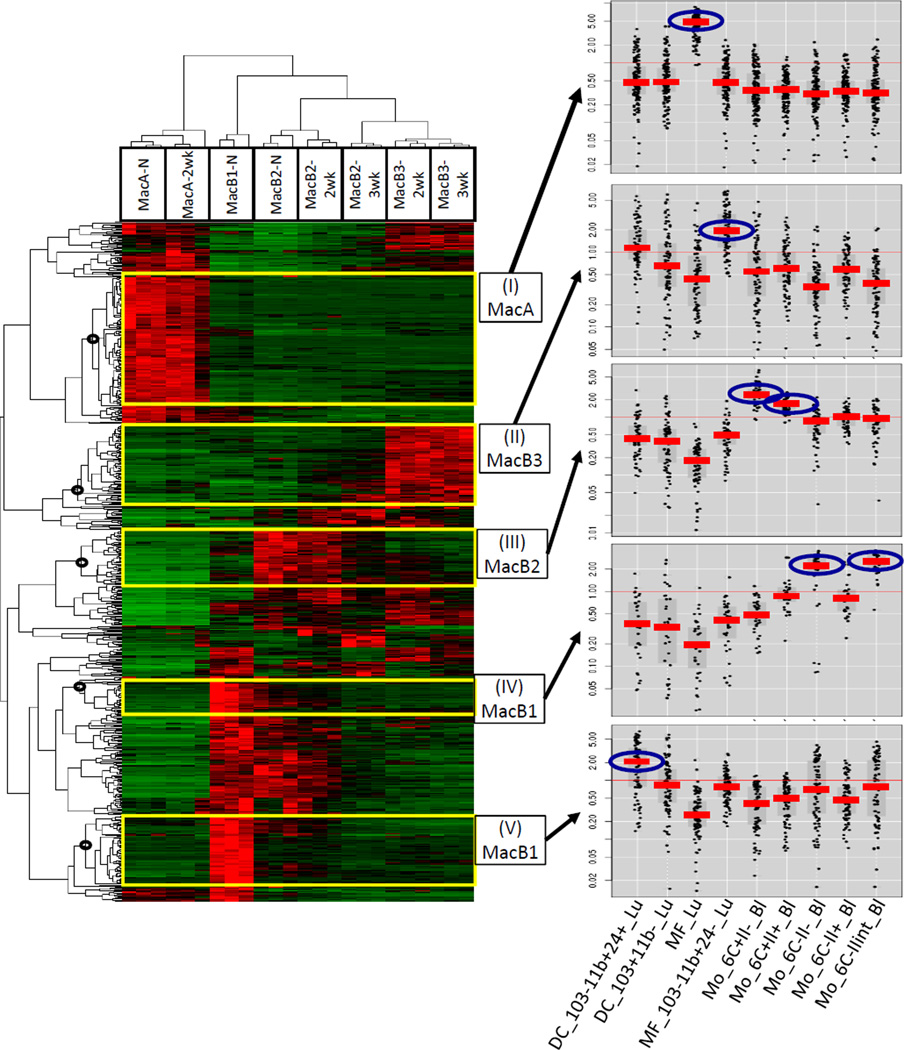
To verify the identity of MacA and MacB populations, we compared their transcriptional profiles to immune cells from naïve mice analyzed by the Immgen Project Consortium (32). As a reference we focused on 9 populations of myeloid cells identified by the Immgen Project: lung macrophages (alveolar macrophages and CD11b+ lung macrophages), lung dendritic cells (CD11b+ and CD11b-) and blood monocytes (Ly6C+/MHCII−, Ly6C+/MHCII+, Ly6C−/MHCII−, Ly6C−/MHCII+, Ly6C−/MHCIIint). We selected 645 genes that were differentially expressed between these 9 reference populations and analyzed their expression in MacA and MacB cells. Using hierarchical clustering, we delineated 5 gene clusters that were highly expressed in a specific MacA or MacB population (Fig. 2). To examine whether these clusters mapped back to specific Immgen cell types, we plotted their expression in the 9 Immgen populations using “My Gene Set” tool (www.ImmGen.org). This analysis indicated that genes specific to MacA-N & MacA-2wk (cluster I) are highly expressed in Immgen alveolar lung macrophages. For MacB1 cells, we defined 2 separate clusters (IV & V). Genes in cluster V map to CD11b+ lung dendritic cells, whereas, genes in cluster IV map to Ly6C−/MHCII− and Ly6C−/MHCIIint monocytes. This indicates that MacB1-N is a mixed population composed of lung CD11b+ dendritic cells and Ly6C− monocytes. MacB2 cells (MacB2-N, MacB2-2wk and MacB2-3wk) express genes in cluster III, and these genes correlate with Ly6C+/MHCII+ and Ly6C+/MHCII− blood monocytes. Lastly, MacB3-2wk and MacB3-3wk express high levels of genes in cluster II, and this cluster maps to CD11b+ lung macrophages. Thus, the genes that are highly expressed in MacA or MacB populations allowed us to map them to specific immune cells based on published transcriptional profiles in naïve mouse. Furthermore, this analysis is in concordance with our immunophenotyping and Ly6C and MHCII expression by flow cytometry.
Fig. 2. Comparison of transcriptional profiles of MacA and MacB populations to immune cells from naïve mouse published by the Immgen Project Consortium.
MacA and MacB populations were recovered from 3 independently injected and harvested pools of tumor bearing and control mice by flow-cytometry-based sorting. Sufficient cell numbers were obtained for: naive mice: MacA (MacA-N), MacB1 (MacB1-N), MacB2 (MacB2-N); 2 week tumor bearing mice: MacA (MacA-2wk), MacB2 (MacB2-2wk), MacB3 (MacB3-3wk); 3-week tumor bearing mice: MacB2 (MacB2-3wk), MacB3 (MacB3-3wk). RNA was extracted from these populations, transcriptional profiles were assessed by RNA-seq, We constructed a list of 645 genes that were differentially expressed among the following cell types profiled by the Immgen Project: lung alveolar macrophages, lung CD11b+ macrophages, blood monocytes classical (Ly6C+MHCII+, Ly6C+/MHCII−), blood monocytes nonclassical (Ly6C−/MHCII+, Ly6C−/MHCII−, Ly6C−/MHCII int), lung dendritic cells (CD11b+ and CD11b-). The expression of these 645 genes among the MacA and MacB cells was examined by hierarchical clustering and is shown as a heatmap. Clusters of genes highly expressed selectively in one cell type were outlined based on the nodes in the dendrogram (black circles): (I) MacA – cluster expressed in MacA-N and −2wk; (II) MacB3 - cluster expressed in MacB3-2wk and −3wk; (III) MacB2 – cluster expressed in MacB2-N, −2wk, and −3wk; (IV) and (V) 2 separate clusters expressed in MacB1-N. The genes in the clusters were mapped back to the reference Immgen populations using the Immgen website tool “my gene set” (panels on the right). The Immgen population(s) with highest expression is circled in blue. DC_103-11b+24+_Lu: CD11b+ lung dendritic cell; DC_103+11b-_Lu: CD11b- lung dendritic cell; MF_Lu: lung alveolar macrophage; MF_103-11b+24-_Lu: lung CD11b+ macrophage; Mo_6C+II−_Bl: blood monocyte, classical Ly6C+/MHCII−; Mo_6C+II+_Bl: blood monocyte classical Ly6C+/MHCII+; Mo_6C−/II−_Bl : blood monocyte nonclassical Ly6C−/MHCII−; Mo_6C−/II+_Bl : blood monocyte nonclassical Ly6C−/MHCII+; Mo_6C−/IIint_Bl : blood monocyte nonclassical Ly6C−/MHCII intermediate.
Global analysis of differentially expressed genes
The analysis of RNA-seq data allowed us to identify differentially expressed genes between the cell populations and between time-points for respective cell populations. In brief, in a pairwise analysis a differentially expressed gene was expected to have expression level FPKM > 5 in at least one condition, and have >2.5 fold change in log2 expression with FDR adjusted p-value <0.05 between the two conditions. In all pairwise comparisons between cell types and /or time points, we identified 2458 genes that were differentially expressed. (Supplemental Table 1). The large number of differentially expressed genes suggests that although all the analyzed cell populations belong to the mononuclear phagocyte lineage, significant biological differences exist between them.
To identify relationships between genes and among MacA and MacB populations, we performed unbiased hierarchical clustering of all 2458 differentially expressed genes and visualized the data using a dendrogram and a heatmap (Fig. 3). The top dendrogram indicates that the individual replicates of the cell populations cluster together. Interestingly, the replicates of MacB2-N and MacB2-2wk were closely clustered. This may reflect the fact that in 2-week samples, the harvested tissue was composed of smaller tumors with a larger contribution from the uninvolved lung. Otherwise, genes could be grouped based on their expression into 5 broad clusters: up- (or down) regulation of gene expression in MacA-N & MacA-2wk (Cluster A); MacB1-N (Cluster B1); MacB2-N and MacB2-2wk (Cluster B2); MacB2-3wk (Cluster B2-3wk); and MacB3-2wk & MacB3-3wk (Cluster B3).
We conducted pathway overrepresentation analysis of the gene clusters using web-based bioinformatic tools at ConsensusPathDB (http://consensuspathdb.org/) (17, 18), searching in the KEGG and Reactome databases. The analyses were additionally confirmed by DAVID (http://david.abcc.ncifcrf.gov/ ) (19, 20), and Gene Ontology (GO) (21, 22) analyses independently. Below, we discuss characteristic pathway signatures of individual clusters in greater detail.
MacA cells
We first focused on genes highly expressed by MacA-N and MacA-2wk (See Supplemental Table 1). Although MacA cells do not increase in number during tumor growth in our model, they represent pre-existing resident alveolar macrophages that are unique to the lung and may play a critical role in early stages of lung cancer or in the setting of slowly developing tumors. In Cluster A, pathway overrepresentation analysis highly ranked multiple gene sets related to lipid metabolism (Fig. 4A). To follow up on the pathway overrepresentation results, we selected consensus genes from the top-ranking five lipid metabolism pathways, and we examined their expression across the populations in the study. The heatmap in Fig. 4B shows 28 lipid metabolism genes that are differentially expressed. Eighteen of these genes formed a cluster that was highly expressed in MacA cells but was expressed at low levels in the other populations. Thus, both the overrepresentation analysis and the analysis of expression of lipid pathway genes across MacA and MacB populations revealed that lipid metabolism is a specific gene expression signature of MacA cells.
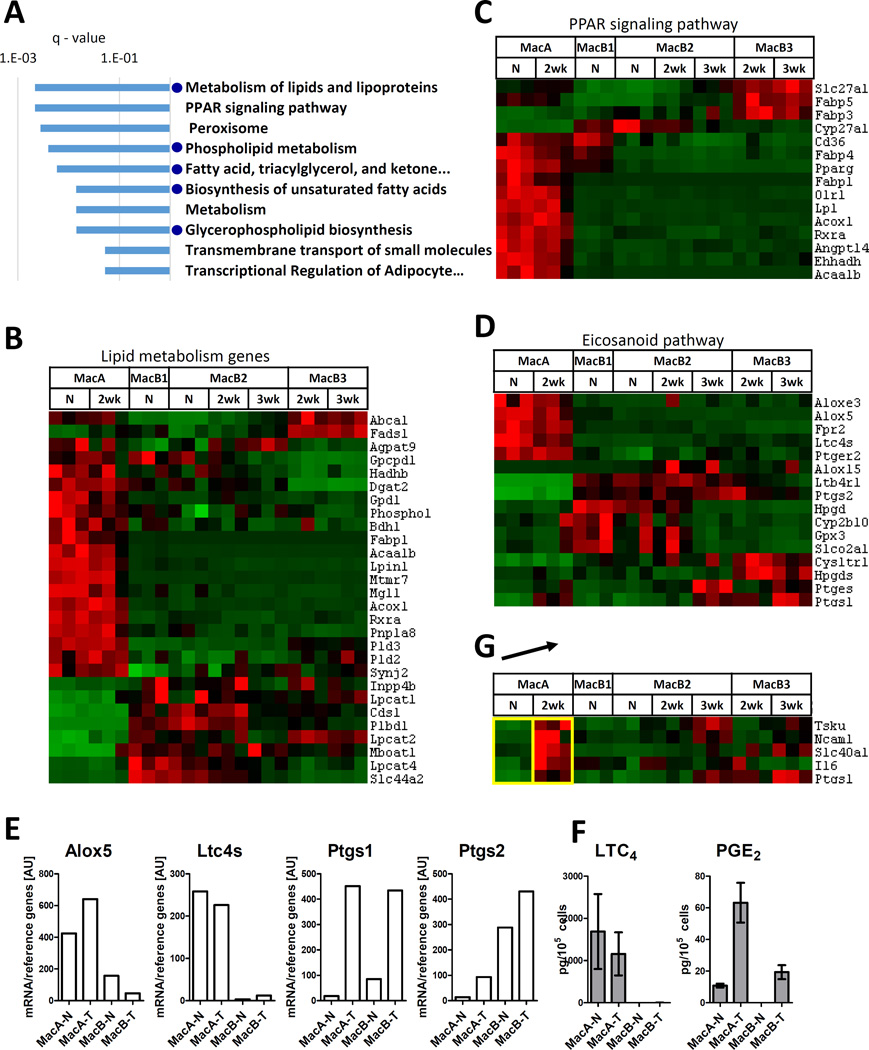
Fig. 4. Analysis of MacA cells.
(A) Top 10 KEGG/Rectome database pathways overrepresented in gene Cluster A (genes highly expressed in MacA-N and MacA-2wk cells). Blue dots indicate pathways related to lipid metabolism. (B) Levels of differentially expressed genes related to lipid metabolism. Gene list was based on consensus between the top ranking lipid metabolism pathways. (C) Levels of differentially expressed genes related to PPAR signaling. Gene list was based on the KEGG set “PPAR signaling pathway”. (D) Levels of differentially expressed genes in the eicosanoid pathway. The key leukotriene pathway enzymes (Alox5, Ltc4s) and prostaglandin pathway enzymes (Ptgs1, Ptgs2) are indicated. (E) Levels of mRNA of Alox5, Ltc4s, Ptgs1, Ptgs2. The cells were recovered form a separate group of mice, in which the MacB subpopulations were pooled, and mRNA was extracted and quantified by RT-qPCR. Levels were normalized to reference genes (geometric average of beta-actin, Gapdh, 18s and UbqC). (F) Production of LTC4 and PGE2 by MacA and MacB cells recovered by flow cytometry and stimulated in vitro with calcium ionophore (G). Genes upregulated in MacA-2wk compared to MacA-N. The arrow indicates the 2 populations being compared. Shown are only genes that met the differential expression criteria in this comparison, and whose average expression in MacA was higher than in the MacB populations.
Another pathway that ranked highly in Cluster A was “Peroxisome proliferator receptor (PPAR) signaling” (KEGG). A heatmap visualizing all differentially expressed genes belonging to this pathway shows that out of 16 “PPAR signaling” genes, 12 were highly expressed in MacA cells (Fig. 4C).
We have previously shown that MacA cells in tumor-bearing lungs produce leukotrienes, which are a sub-family of lipid inflammatory eicosanoids derived from arachidonic acid (10). Thus, in Fig. 4D we examined the expression of known genes in the eicosanoid pathway (KEGG “Arachidonic acid metabolism”) (33, 34), of which 16 were differentially expressed across the populations in this study. The key leukotriene metabolism genes, 5-lipoxygenase (Alox5) and LTC4 synthase (Ltc4s) were highly expressed in MacA cells from both tumor- and non-tumor-bearing lungs. In contrast, the key enzyme in prostaglandin production pathway, cyclooxygenase-2 (COX-2, Ptgs2) was specifically downregulated in MacA cells compared to other populations. Cyclooxygenase-1 (COX-1, Ptgs1) was expressed at low levels in MacA cells from lungs without tumor; however, its expression was upregulated in MacA-2wk. For these key enzymes, we confirmed the mRNA-seq data in a separate group of mice using quantitative RT-PCR (Fig. 4E). To confirm the gene expression data functionally, we recovered MacA and Mac B cells from control and tumor-bearing mice by flow cytometry, stimulated these cells in vitro with a calcium ionophore, and measured eicosanoid production by liquid chromatography/tandem mass spectrometry (Fig. 4F). The production of leukotriene C4 (a product of 5-lipoxygenase and LTC4 synthase) was specific to MacA cells, both in tumor-bearing and non-tumor-bearing lungs. Furthermore, the production of PGE2 (a product of Ptgs1/COX-1 or Ptgs2/COX-2) was increased in MacA cells from tumor-bearing mice vs. control, which is in agreement with increased Ptgs1 expression by RNA-seq. Surprisingly, MacB cells produced low levels of PGE2, but this may be due to the non-physiological stimulation. Among the receptors, MacA expressed high levels of Ptger2, the receptor for prostaglandin E2 and Fpr2, the receptor for lipoxins. In contrast, the receptor for LTB4 (Ltb4r1, BLT1) was specifically downregulated in MacA cells compared to other populations, as was the receptor for cysteinyl leukotrienes (Cysltr1).
Such a combination of enzymes and receptors suggests that among the studied populations, MacA cells would be the major producers of leukotrienes, but would not respond to these products. However, they would be responsive to lipoxins, which are the anti-inflammatory products of the lipoxygenase pathway, and to PGE2. The data also suggest that MacA in naïve lung do not produce prostaglandins, however they gain that capability in the setting of tumor through the upregulation of COX-1. These data are consistent with increased levels of prostaglandins seen in tumor-bearing mice, which are abrogated when cPLA2, the rate limiting enzyme in eicosanoid production, is deleted (10, 35).
Comparing MacA-2wk to MacA-N, we identified 89 genes that were upregulated in tumor-bearing compared to control lungs, and one gene that was down-regulated (Supplemental Table 2). However, even though the differential expression was statistically significant, the absolute expression levels of the upregulated genes remained significantly lower in MacA-2wk than in MacB populations (Supplemental Figure 1). It is possible that the increased levels of these genes reflect overall dysregulation of gene expression in MacA cells from tumor-bearing lungs, rather than biologically significant induction. Out of the 89 upregulated genes, only 5 (Ncam1, Slc40a1, Il6, Tsku, Ptgs1) had above-average absolute expression levels in MacA-2wk cells compared to other populations (Fig. 4G). As discussed above, the increased expression of Ptgs1 (COX-1), which was confirmed by qPCR and by increased PGE2 production in vitro (Fig. 4E and 4F), indicates that MacA cells from tumor-bearing lungs, may gain the potential to produce prostaglandins. IL-6 has been implicated in many types of cancer, and increased levels are associated with poorer overall survival in lung cancer (36). We have previously demonstrated increased levels of IL-6 in our model, and shown that cancer cells can induce expression in bone marrow-derived macrophages in vitro (35). Interestingly, IL-6 appears to be selectively expressed by MacA in the setting of tumor.
MacB3 cells
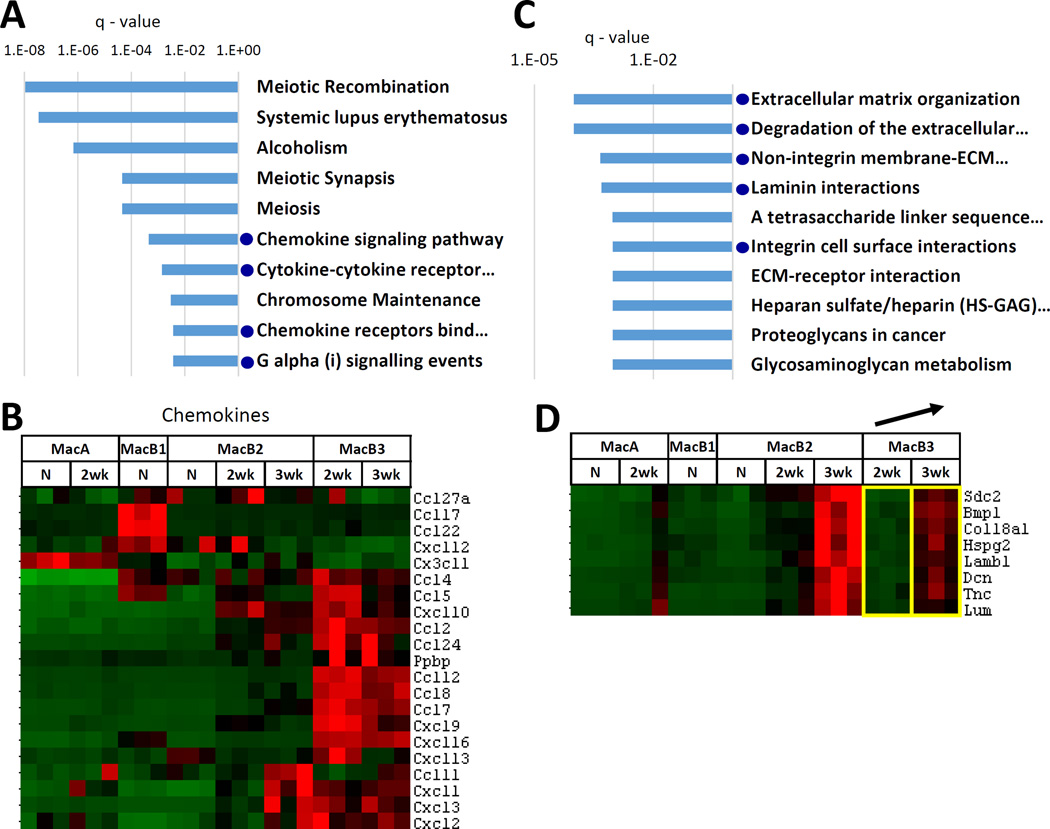
We next focused on MacB3 cells, which increase rapidly with tumor growth and constitute the major component of the lung tumor microenvironment, particularly at the 3 wk time point. As indicated by pathway overrepresentation analysis, Cluster B3 (genes highly expressed both in MacB3-2wk and MacB3-3wk) was enriched in pathways related to chemokine and cytokine signaling (Fig. 5A, Supplemental Table 1). To confirm this result independently of KEGG and Reactome databases, we chose a published panel of chemokine genes (37) and examined these in MacA and MacB populations. As shown in the heatmap in Fig. 5B, out of 21 differentially expressed chemokine genes, 16 clustered as highly expressed in MacB3s. Of those 16, six were also highly expressed in MacB2-3wk (Fig. 5B). Thus, the analysis of gene cluster B3 suggests that MacB3 cells play a critical role in communication between the diverse cells of the TME through the secretion of multiple chemokines. The other highly ranking pathways (“Meiotic recombination”, “Systemic lupus erythematosus”, “Alcoholism”, “Chromosome maintenance”) all contained 14 histone genes that overlapped with Cluster B3, without any other genes pointing to a specific function.
Fig. 5. Analysis of MacB3 cells.
(A) Top 10 KEGG/Reactome pathways overrepresented in gene Cluster B3 (genes highly expressed in MacB3-2wk and MacB3-3wk cells). Blue dots indicate pathways related to chemokine signaling. (B) Levels of differentially expressed chemokine genes in MacA and MacB populations. (C) Analysis of genes upregulated in MacB3 cells with tumor progression – comparing MacB3-3wk to MacB3-2wk. Top 10 KEGG/Reactome pathways overrepresented among the 35 upregulated genes. Blue dots indicate extracellular matrix related pathways. (D) Specific genes that were upregulated in MacB3-3wk compared to MacB3-2wk and that belonged to the extracellular matrix – related pathways. The yellow boxes indicate the 2 populations being compared.
In naïve lung, the MacB3 cells are present in very low numbers; thus we were unable to recover sufficient numbers of cells for RNA-seq analysis. However, these cells increased rapidly with tumor growth, between 2 weeks and 3 weeks. We identified 35 genes that were upregulated and 4 that were downregulated in MacB3 cells at 3 weeks compared to 2 weeks (Supplemental Table 2). Overrepresentation analysis indicated enrichment of pathways related to extracellular matrix (Fig. 5C). Interestingly, while upregulation of the ECM-related genes in MacB3 at 3 weeks was statistically significant, these genes had highest expression in MacB2 cells at 3 weeks (Fig. 5D). This suggests that ECM production is upregulated in the tumor microenvironment at 3 weeks, with MacB2 cells being the major producers, but MacB3 cells also contributing.
MacB2 and MacB1 cells
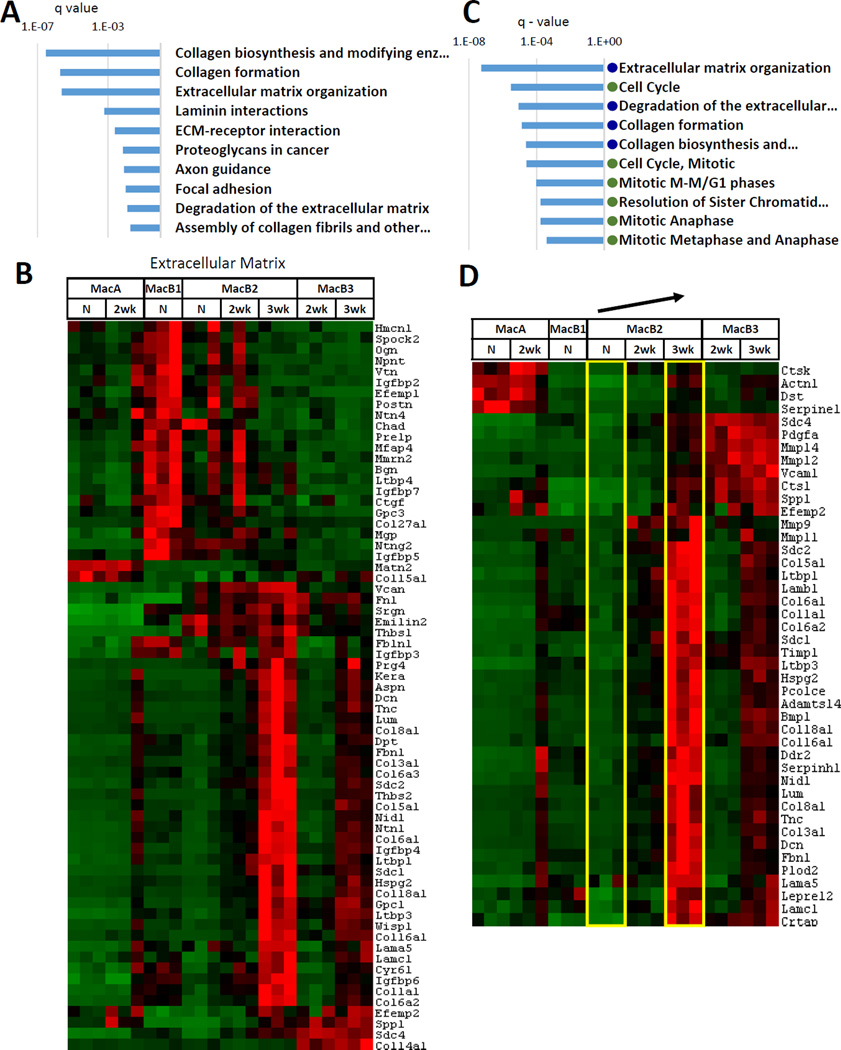
MacB2 cells also increase substantially during tumor growth. In our initial clustering of 2458 differentially expressed genes (Fig. 3), genes that were highly expressed in MacB2 at 3 weeks formed a cluster separate from genes highly expressed in MacB2 from uninjected lung or from 2-week tumor. In this cluster (B2-3wk), pathways that were highly ranked by overexpression analysis were related to extracellular matrix (ECM), particularly collagen (Fig. 6A, Supplemental Table 1).
Fig. 6. Analysis of MacB2-3wk.
(A) Top 10 KEGG/Reactome pathways overrepresented in gene Cluster B2-3wk (genes highly expressed in MacB2-3wk cells). (B) Levels of differentially expressed extracellular matrix genes. (C) Analysis of genes upregulated in MacB2 cells with tumor progression. Comparison of MacB2-3wk to MacB2-N. Top 10 KEGG/Reactome pathways overrepresented among the 472 upregulated genes. Blue dot indicates extracellular matrix related pathways. Green dot indicates pathways related to cell cycle. (D) Specific genes that were upregulated in MacB2-3wk compared to MacB2-N and that belonged to the extracellular matrix – related pathways. The arrow indicates the 2 populations being compared.
To follow up on this result, we examined a published collection of ECM proteins (38, 39) of which 61 were differentially expressed in this study (Fig. 6B). Interestingly, this analysis revealed two distinct gene clusters, one highly expressed in MacB2-3wk (34 genes) and one highly expressed in MacB1-N cells (21 genes). The MacB2-3wk cluster contained several genes previously associated with tumor progression, such as collagens, fibronectin, tenascin C, and versican. (40, 41). As discussed above, these genes were also upregulated in MacB3 cells at 3 weeks (Fig. 5D).
We examined changes in gene expression in MacB2 as a function of tumor progression. In comparison to MacB2-N, 87 genes were upregulated and 3 were downregulated in MacB2-2wk cells. However, most of these genes were not selectively expressed in MacB2 cells. Comparison of MacB2-N to MacB2-3wk found that 472 genes were upregulated and 97 were downregulated in MacB2-3wk cells (Supplemental Table 2). Overrepresentation analysis indicated that the set of upregulated genes was enriched in pathways related to extracellular matrix and pathways related to cell cycle regulation (Fig. 6C, D). Since expression of ECM proteins was a distinctive feature of MacB2-3wk, this result is consistent with upregulation of these genes during tumor progression. The downregulated genes contained genes related to biological oxidation, and enriched pathways included “Phase I – Functionalization of compounds”, “Drug metabolism – cytochrome P450”, and “Biological oxidations”.
Analysis of genes highly expressed in MacB1 (Cluster B1) or in MacB2 from uninjected lung and from 2-week tumor (Cluster B2) did not provide robust and repetitive results, such as those observed for MacA, MacB2-3wk and MacB3 cells.
Relationship to M1/M2 Macrophages
Since studies focusing on clinical outcomes frequently use the M1/M2 designation of macrophages, we examined the relationship of our populations with commonly used M1 and M2 markers. This analysis has shown that ArgI (M2 marker) was expressed at highest levels in MacB3 (at both 2 and 3 weeks), whereas Nos2 (M1 marker) was expressed in both MacB2 (2 and 3 weeks) as well as MacB3 (2 and 3 weeks), with the highest levels in MacB2-3wk. Analysis of a broader set of M1/M2 markers (42) indicated that none of our populations preferentially expresses either M1 or M2 markers (Supplemental Figure 2). This analysis demonstrates the complexity of macrophage phenotypes in vivo and poor correlation with the M1/M2 designation, which was based on in vitro activation of bone-marrow derived macrophages (42).
Correlation with genes prognostic in human lung adenocarcinoma
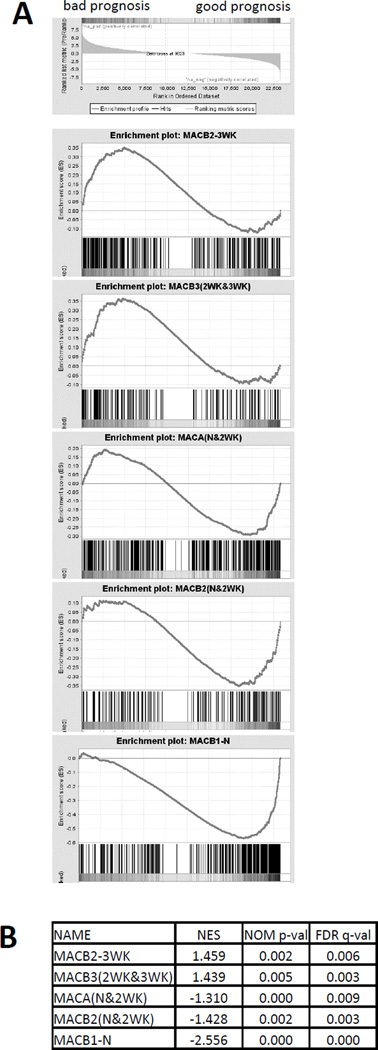
Finally, a recent study has used a bioinformatics approach to establish a system to examine associations between gene expression in human tumors and patient survival across multiple types of cancer (PRECOG) https://precog.stanford.edu/ (43). We used the data from the PRECOG database to test whether MacA or MacB populations are enriched in genes correlated with good or bad clinical outcomes in lung adenocarcinoma. We ranked ~23,000 human genes according to their survival scores, and we used Gene Set Enrichment Analysis (GSEA, pre-ranked tool) (23, 24) to examine whether the clusters associated with MacA or MacB populations (defined in Fig. 3) are enriched in genes correlated with good or bad prognosis. The GSEA plots and normalized enrichment scores (Fig. 7A, B) indicate that MacB2-3wk and MacB3 (2wk and 3wk) are enriched in genes correlated with bad clinical outcomes. In contrast, MacB1-N, MacA and the early stage MacB2 (N and 2wk) were enriched in genes predicting good clinical outcomes, with MacB1-N having the highest enrichment score. These data suggest that prognostic genes identified in the analysis of whole tumors are highly expressed in specific macrophage populations, suggesting that enrichment of these populations in a tumor may predict clinical outcome.
Fig. 7. Correlation of MacA and MacB transcriptional profiles to prognostic genes in human lung adenocarcinoma.
A) Gene Set Enrichment Analysis (GSEA) plots - ~23,000 human genes were ranked according to their survival scores in lung adenocarcinoma as listed in the PRECOG database, and the GSEA pre-ranked analysis tool was used to examine enrichment in good or bad prognosis genes in the gene clusters highly expressed in MacA or MacB cells as defined in Fig. 3. (B) GSEA enrichment scores: NES – normalized enrichment score, NOM p-val – nominal p-value, FDR q-val – false discovery rate
Discussion
Macrophages have been implicated as critical components of the tumor microenvironment, controlling tumor initiation, progression and metastasis. In this study we have taken an unbiased approach to begin to define the complexity of macrophage phenotypes in the setting of lung cancer progression. We have employed an immunocompetent orthotopic model of cancer progression in which interactions between macrophages, cancer cells, and adaptive immune cells are present. Our study has defined at least 4 separate monocyte/macrophage populations and used transcriptional profiling to begin to define their function. Our results reveal that each population has a distinct gene expression signature, suggesting that it plays a specific role in cancer progression.
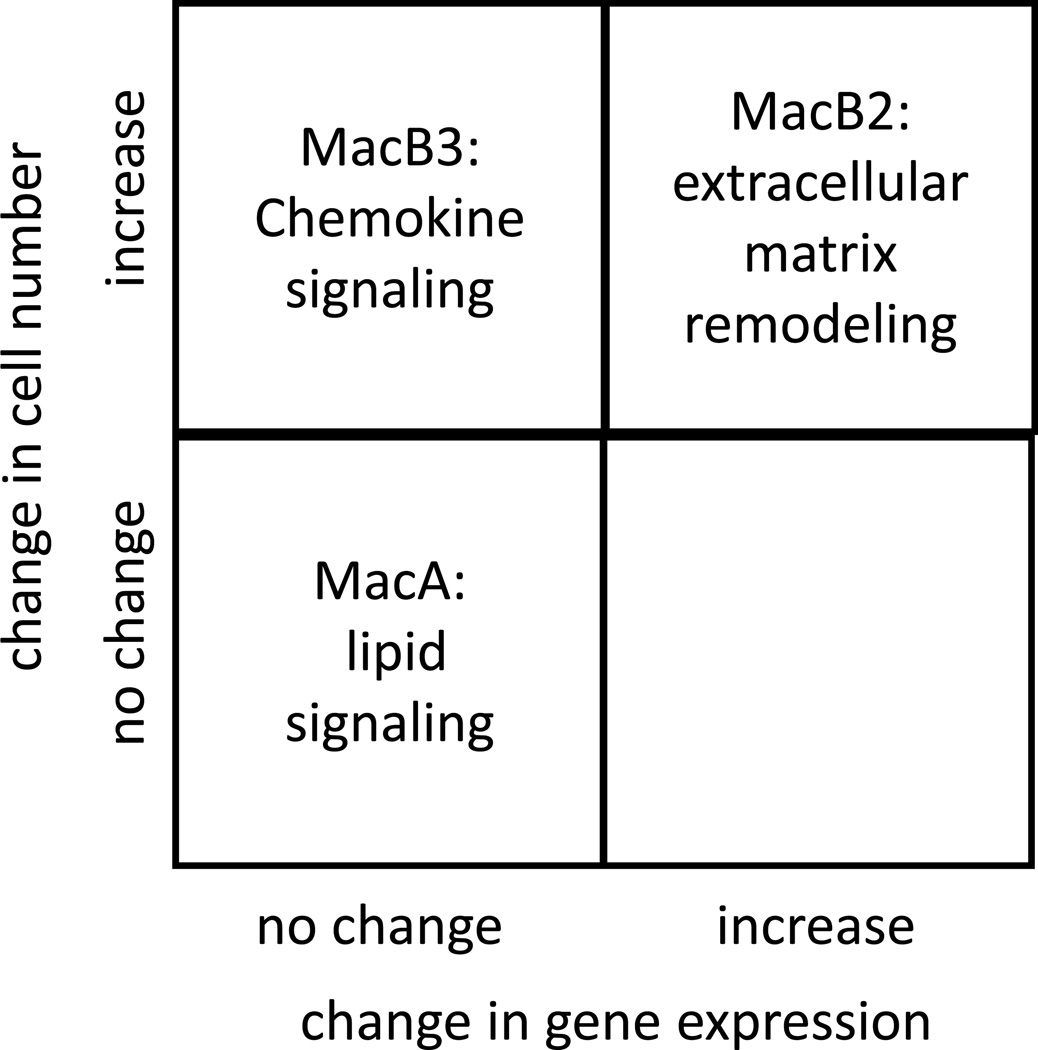
Our data underscore that the tumor microenvironment is shaped both by increases in specific cell populations, as well as by gene expression changes in each of these populations (Fig. 8). The population designated MacA represents resident alveolar macrophages. This is confirmed by correlating these cells with the Immgen signature. This population does not change significantly in number during tumor progression. However, there are modest but potentially important changes in gene expression associated with the presence of the tumor. The gene expression signature implies that this MacA population may be the major mediator of leukotriene production, and the induction of COX-1 suggests that these cells also contribute to the increase in prostaglandin production observed in lung cancer progression (10). While we confirmed the gene expression data by measuring LTC4 and PGE2 production in MacA cells, one limitation of this experiment is that the cells were analyzed outside of their physiological environment. Another gene selectively expressed in the MacA from tumor-bearing lungs is IL-6, which plays a complex role in cancer progression.
Fig. 8. Overview of the changes in cell number and gene expression of key monocyte and macrophage populations identified in the lung tumor microenvironment.
Cells that we have designated as MacB, especially MacB2 and MacB3, represent populations that increase during tumor progression in our model. Our analysis suggests that these cells are involved in specific functions regulating tumor progression. In the absence of tumor, these cells are present in very low numbers in the lung, suggesting that they are recruited from the circulation in response to factors produced by cancer cells. MacB3 cells, which bear markers of infiltrating macrophages, expressed high levels of multiple cytokines at both time points examined. This would suggest that these cells may orchestrate the recruitment of other stromal cells during tumor progression. This may induce a “feed-forward” loop, in which early recruitment of MacB3 cells leads to additional waves of recruitment of other stromal cells. Interestingly, at the later time point, we observed increases in genes encoding extracellular matrix and enzymes that remodel the ECM. Similar ECM-related pathways were also detected in MacB2 cells, which represent tumor-associated monocytes, and which show similar increases during tumor progression. Thus these distinct populations of macrophages can alter the tumor microenvironment both by recruiting additional cells and by remodeling the physical properties of the TME.
Correlating our data with analysis of human tumors, it appears that genes associated with better clinical outcomes are expressed at earlier time points, whereas genes associated with poor outcomes are expressed at later time points. Although studies have suggested that these changes are a result of a phenotypic switch in macrophage populations interacting with cancer cells, our data indicate a more complex situation. Distinct populations of macrophages expand during cancer progression. Thus a change in a specific marker will reflect alterations in existing populations due to phenotypic modulation, as well as increases in populations that express these markers constitutively. For example, studies have demonstrated increased production of TGF-β1 by macrophages during cancer progression. From our analysis, the level of TGF-β1 production is not altered in any of our populations (not shown), but marked increases in the numbers of MacB2 and MacB3 cells during progression will result in increased numbers of TGF-β1producing cells. However, increases in several ECM proteins will be a result of increased numbers of ECM-producing macrophages, as well as upregulation of production in these cells during tumor progression.
The populations we have defined do not easily conform to the “M1/M2” paradigm. While some M1/M2 markers are enriched in certain populations, using a more extensive list of markers shows that these genes are expressed across multiple populations. In addition, a model in which macrophages recruited to the site of the tumor as M1/anti-tumorigenic macrophages undergo phenotypic changes to an M2/pro-tumorigenic phenotype is not supported by our data. Based on the PRECOG analysis, most of the populations we have defined retain an expression signature associated with either good or bad clinical outcomes throughout tumor progression. An exception is the MacB2 population, which appears to be associated with a good outcomes at early time points, but undergoes changes in gene expression leading to an association with a bad outcome at later time points.
To our knowledge, this is one of the first studies to molecularly define the diversity of macrophages in lung cancer in an immunocompetent setting. The major conclusions of our analysis is that distinct macrophage populations express specific gene signatures, which predicts that they play specific and distinct roles in cancer progression. However, the actual function of these cells will need to be validated in experiments that selectively deplete a specific macrophage subpopulation in the tumor microenvironment. Our studies indicate that recruited populations of macrophages generally aid tumor progression, suggesting that selectively blocking their recruitment may represent a therapeutic strategy to inhibit lung cancer progression. A limitation of this study is the use of only one lung cancer model. However, we would argue that our orthotopic injection model is an appropriate model to answer questions concerning progression of advanced lung cancer and the role of specific cells in the lung tumor microenvironment. Other models would less accurately reflect the tumor microenvironment of the lung (e.g. subcutaneous flank injections) or would result in a well-defined primary tumor (e.g. tail vein injections). Another limitation of these studies is the use of a single lung cancer cell line. Targeted therapies in lung adenocarcinoma are based on identification of specific oncogenic drivers that drive proliferation of cancer cells in a cell-autonomous fashion. However, there is little known as to how specific drivers affect the tumor microenvironment. It will be of interest to determine if the changes in number and gene expression profiles of the macrophage populations we have defined are dependent on the oncogenic drivers of the cancer cells. For our orthotopic model, this will require a panel of murine lung cancer cells that express the drivers that have been identified in human lung cancer. Thus, our report provides a framework for studies of macrophage populations in other models of lung cancer, including spontaneous models in which specific oncogenic drivers are activated.
Finally, it is important to extend these studies to human lung tumors. In this regard, it is encouraging that we were able to correlate transcriptional profiles of specific macrophage populations with either bad or good prognostic genes derived from bioinformatic studies in human lung adenocarcinoma. Thus, our studies form the basis to develop a panel of markers to characterize macrophage populations in human tumors. Earlier studies have shown that the location of macrophages in lung tumors may determine their role in cancer progression. Increased numbers of macrophages within the tumor itself was associated with a favorable prognosis, whereas increased numbers of macrophages in the stroma surrounding the tumor was associated with an unfavorable prognosis (44). The development of a panel of markers specific for MacB3 and MacB2 cells will be required to define their localization, and may be of value in predicting clinical outcomes in human disease.
Supplementary Material
Acknowledgments
We would like to thank the members of the University of Colorado Cancer Center Flow Cytometry Core for their assistance with flow cytometry and the Genomics and Microarray Core for their assistance with RNA-seq.
Financial support: NIH/NCI (R01 CA162226, R01 CA164780, R01 CA108610 to R.A. Nemenoff), Colorado Lung SPORE (P50 CA058187), Ruth L. Kirschtein NRSA Award T32CA17468 (T Sippel) and Department of Veterans Affairs (CDA 1IK2BX001282-01A1 to H. Li). The Flow Cytometry and the Genomics and Microarray Shared Resources receive support from the NIH/NCI (University of Colorado Cancer Center Support Grant P30 CA046934). SD acknowledges funding from Boettcher Foundation and United Against Lung Cancer Foundation.
Abbreviations
- TME
tumor microenvironment
- LLC
Lewis Lung Carcinoma
- COX
cyclooxygenase
- ECM
extracellular matrix
- GSEA
gene set enrichment analysis
- FPKM
Fragments Per Kilobase of exon per Million fragments mapped
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Kenny PA, Lee GY, Bissell MJ. Targeting the tumor microenvironment. Front Biosci. 2007;12:3468–3474. doi: 10.2741/2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaynagetdinov R, Sherrill TP, Polosukhin VV, Han W, Ausborn JA, McLoed AG, McMahon FB, Gleaves LA, Degryse AL, Stathopoulos GT, Yull FE, Blackwell TS. A critical role for macrophages in promotion of urethane-induced lung carcinogenesis. J Immunol. 2011;187:5703–5711. doi: 10.4049/jimmunol.1100558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma SK, Chintala NK, Vadrevu SK, Patel J, Karbowniczek M, Markiewski MM. Pulmonary alveolar macrophages contribute to the premetastatic niche by suppressing antitumor T cell responses in the lungs. J Immunol. 2015;194:5529–5538. doi: 10.4049/jimmunol.1403215. [DOI] [PubMed] [Google Scholar]
- 6.Remark R, Becker C, Gomez JE, Damotte D, Dieu-Nosjean MC, Sautes-Fridman C, Fridman WH, Powell CA, Altorki NK, Merad M, Gnjatic S. The non-small cell lung cancer immune contexture. A major determinant of tumor characteristics and patient outcome. Am J Respir Crit Care Med. 2015;191:377–390. doi: 10.1164/rccm.201409-1671PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allavena P, Sica A, Solinas G, Porta C, Mantovani A. The inflammatory micro-environment in tumor progression: the role of tumor-associated macrophages. Critical reviews in oncology/hematology. 2008;66:1–9. doi: 10.1016/j.critrevonc.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 9.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma'ayan A, Chua WJ, Hansen TH, Turley SJ, Merad M, Randolph GJ. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature immunology. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poczobutt JM, Gijon M, Amin J, Hanson D, Li H, Walker D, Weiser-Evans M, Lu X, Murphy RC, Nemenoff RA. Eicosanoid profiling in an orthotopic model of lung cancer progression by mass spectrometry demonstrates selective production of leukotrienes by inflammatory cells of the microenvironment. PLoS One. 2013;8:e79633. doi: 10.1371/journal.pone.0079633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 16.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 17.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–D717. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB--a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder--open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan M, Li WC, Vlahos R, Maxwell MJ, Anderson GP, Hibbs ML. Distinct macrophage subpopulations characterize acute infection and chronic inflammatory lung disease. J Immunol. 2012;189:946–955. doi: 10.4049/jimmunol.1200660. [DOI] [PubMed] [Google Scholar]
- 26.Janssen WJ, Barthel L, Muldrow A, Oberley-Deegan RE, Kearns MT, Jakubzick C, Henson PM. Fas determines differential fates of resident and recruited macrophages during resolution of acute lung injury. Am J Respir Crit Care Med. 2011;184:547–560. doi: 10.1164/rccm.201011-1891OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens WW, Kim TS, Pujanauski LM, Hao X, Braciale TJ. Detection and quantitation of eosinophils in the murine respiratory tract by flow cytometry. J Immunol Methods. 2007;327:63–74. doi: 10.1016/j.jim.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sung SS, Fu SM, Rose CE, Jr, Gaskin F, Ju ST, Beaty SR. A major lung CD103 (alphaE)-beta7 integrin-positive epithelial dendritic cell population expressing Langerin and tight junction proteins. J Immunol. 2006;176:2161–2172. doi: 10.4049/jimmunol.176.4.2161. [DOI] [PubMed] [Google Scholar]
- 29.Zhang JQ, Biedermann B, Nitschke L, Crocker PR. The murine inhibitory receptor mSiglec-E is expressed broadly on cells of the innate immune system whereas mSiglec-F is restricted to eosinophils. European journal of immunology. 2004;34:1175–1184. doi: 10.1002/eji.200324723. [DOI] [PubMed] [Google Scholar]
- 30.Desch AN, Randolph GJ, Murphy K, Gautier EL, Kedl RM, Lahoud MH, Caminschi I, Shortman K, Henson PM, Jakubzick CV. CD103+ pulmonary dendritic cells preferentially acquire and present apoptotic cell-associated antigen. J Exp Med. 2011;208:1789–1797. doi: 10.1084/jem.20110538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becher B, Schlitzer A, Chen J, Mair F, Sumatoh HR, Teng KW, Low D, Ruedl C, Riccardi-Castagnoli P, Poidinger M, Greter M, Ginhoux F, Newell EW. High-dimensional analysis of the murine myeloid cell system. Nature immunology. 2014;15:1181–1189. doi: 10.1038/ni.3006. [DOI] [PubMed] [Google Scholar]
- 32.Heng TS, Painter MW C Immunological Genome Project. The Immunological Genome Project: networks of gene expression in immune cells. Nature immunology. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 33.Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation. Nature reviews. Immunology. 2015;15:511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiser-Evans MC, Wang XQ, Amin J, Van Putten V, Choudhary R, Winn RA, Scheinman R, Simpson P, Geraci MW, Nemenoff RA. Depletion of cytosolic phospholipase A2 in bone marrow-derived macrophages protects against lung cancer progression and metastasis. Cancer Res. 2009;69:1733–1738. doi: 10.1158/0008-5472.CAN-08-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao C, Yu Z, Guo W, Liu Q, Wu Y, Li Y, Bai L. Prognostic value of circulating inflammatory factors in non-small cell lung cancer: a systematic review and meta-analysis. Cancer Biomark. 2014;14:469–481. doi: 10.3233/CBM-140423. [DOI] [PubMed] [Google Scholar]
- 37.Balkwill FR. The chemokine system and cancer. J Pathol. 2012;226:148–157. doi: 10.1002/path.3029. [DOI] [PubMed] [Google Scholar]
- 38.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 39.Ricard-Blum S. The collagen family. Cold Spring Harb Perspect Biol. 2011;3:a004978. doi: 10.1101/cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theocharis S, Klijanienko J, Giaginis C, Rodriguez J, Jouffroy T, Girod A, Point D, Tsourouflis G, Satre-Garau X. Peroxisome proliferator-activated receptor-gamma in mobile tongue squamous cell carcinoma: associations with clinicopathological parameters and patients survival. J Cancer Res Clin Oncol. 2010 doi: 10.1007/s00432-010-0882-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, Locati M, Mantovani A, Martinez FO, Mege JL, Mosser DM, Natoli G, Saeij JP, Schultze JL, Shirey KA, Sica A, Suttles J, Udalova I, van Ginderachter JA, Vogel SN, Wynn TA. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, Diehn M, West RB, Plevritis SK, Alizadeh AA. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welsh TJ, Green RH, Richardson D, Waller DA, O'Byrne KJ, Bradding P. Macrophage and Mast-Cell Invasion of Tumor Cell Islets Confers a Marked Survival Advantage in Non-Small-Cell Lung Cancer. J Clin Oncol. 2005;23:8959–8967. doi: 10.1200/JCO.2005.01.4910. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.