Abstract
Remyelination in the central nervous system (CNS) is critical in the treatment of many neural pathological conditions. Unfortunately, the ability to direct and enhance oligodendrocyte (OL) differentiation and maturation remains limited. It is known that microenvironmental signals, such as substrate topography and biochemical signaling, regulate cell fate commitment. Therefore, in this study, we developed a nanofiber-mediated microRNA (miR) delivery method to control oligodendroglial precursor cell (OPC) differentiation through a combination of fiber topography and gene silencing. Using poly(ɛ-caprolactone) nanofibers, efficient knockdown of OL differentiation inhibitory regulators were achieved by either nanofiber alone (20–40%, p < 0.05) or the synergistic integration with miR-219 and miR-338 (up to 60%, p < 0.05). As compared to two-dimensional culture, nanofiber topography enhanced OPC differentiation by inducing 2-fold increase in RIP+ cells (p < 0.01) while the presence of miRs further enhanced the result to 3-fold (p < 0.001). In addition, nanofiber-mediated delivery of miR-219 and miR-338 promoted OL maturation by increasing the number of MBP+ cells significantly (p < 0.01). Taken together, the results demonstrate the efficacy of nanofibers in providing topographical cues and microRNA reverse transfection to direct OPC differentiation. Such scaffolds may find useful applications in directing oligodendrocyte differentiation and myelination for treatment of CNS pathological conditions that require remyelination.
Keywords: Oligodendrocyte precursor cell, Remyelination, Topography, Reverse transfection, Nanofiber, MicroRNA, Gene silencing, Sustained release
1. Introduction
Within the central nervous system (CNS), oligodendrocytes (OL) are the major cell type that contributes to axon myelination. The multilamellar myelin sheaths produced by OL wrap around axons of multiple neurons to support and maintain optimal neuronal signal transmission. Hence disruption of OL structure and myelination, which is commonly seen in traumatic injuries like spinal cord injuries (SCI), will cause failure of neurological function due to hindered or damaged neuronal signal transduction.
In vivo, OL may be derived from oligodendroglial precursor cells (OPCs) that exist endogenously. Much evidence suggests that OPCs residing in the white matter of the spinal cord represent the largest potential source of remyelinating OL [1,2]. However, although the proliferation rate of the OPCs increases significantly post SCI, spontaneous remyelination remains limited, particularly in humans [1,3]. Early in vitro culture experiments have demonstrated that in the absence of signals to maintain undifferentiated OPC, they would rapidly differentiate into OL. Hence, much of the regulation in CNS myelination might be at the level of inhibiting this default pathway. Therefore, strategies that relief some of these inhibitory factors might promote OL differentiation and myelination [4].
MicroRNAs (miRs) are a class of small non-coding RNAs (sncRNA) that contain 21–23 nucleotide base pairs. They play critical roles in various biological processes, including stem/precursor cell differentiation, by silencing the expression of their target mRNAs. To date, many studies have indicated the involvement of miRs during different stages of OPC development [4–6]. In particular, miR-219 and miR-338 have been identified as OL-specific miRs. Specifically, the overexpression of miR-219 and miR-338 promoted OPC differentiation while the knockdown of their expressions inhibited OPC maturation [7]. These miRs function by inhibiting the expression of negative regulators of OPC differentiation, such as PDGFR-α, Sox6, Hes5, FoxJ3 and ZFP238 [7,8]. As such, miR-219 and miR-338 may be potential biochemical cues for controlled delivery to direct OPC differentiation and maturation.
Fiber scaffolds represent a unique class of materials for tissue engineering and regeneration medicine. They mimic the architecture of the natural extracellular matrix (ECM) and provide necessary topographical signals to modulate cell fate [9,10]. Specifically, fiber topography enhanced human Schwann cell maturation and may be beneficial in promoting myelination of the peripheral nervous system (PNS) [11]. Applied to gene silencing, fiber morphology provides a means to alter gene uptake and knockdown efficiencies in cells [12,28]. By incorporating sncRNA directly within fiber constructs, these substrates provide sustained gene silencing in vitro and in vivo [13–16].
In this study, we explore the efficacy of a nanofiber-mediated miR delivery system in enhancing OPC differentiation and maturation. Specifically, miR-219 and miR-338 were incorporated onto poly(ɛ-caprolactone) (PCL) nanofibers by using mussel-inspired bioadhesive 3,4-dihydroxy-L-phenylalanin (DOPA) coating. Due to its latent reactivity which can be exploited for further conjugation of bio-functional molecules, such bio-adhesive coating allowed the easy and efficient incorporation of sncRNA for transfecting neural stem/precursor cells [14]. We hypothesize that the incorporation of miR-219 and miR-338 onto fiber constructs will provide combinatorial topographical and RNA interference (RNAi) signals to silence inhibitory factors that prevent OPC differentiation and maturation. Such an approach will provide control over the OPC fate.
2. Materials and methods
2.1. Materials
Polycaprolactone (PCL, Mw: 80,000), 3, 4-dihydroxy-L-phenylalanine (DOPA), Dnase I, 2, 2, 2-trifluoroethanol (TFE, ≥99.0%), 10% formalin, Triton X-100 and fluoromount were purchased from Sigma-Aldrich. LIVE/DEAD® cell viability kit, Alexa-Fluor 488 goat anti-mouse, Alexa-Fluor 568 goat anti-rabbit antibodies, scramble miR, miR-219, miR-338-3p and miR-338-5p mimics were purchased from Life Technologies. TransIT-TKO was purchased from MirusBio. 1× Tris-EDTA (TE, PH = 7.4) was purchased from 1st Base, Singapore. RNeasy mini kit was purchased from Qiagen. M-MLV reverse transcriptase was purchased from Promega. IQ SYBR Green Supermix was purchased from Bio-Rad. All primers were purchased from AITbiotech, Singapore. Rabbit anti-NG2 chondroitin sulfate proteoglycan (AB5320), rabbit anti-oligodendrocyte transcription factor 2 (Olig2) (AB9610), mouse anti-receptor interacting protein (RIP) (MAB1580) and mouse anti-myelin basic protein (MBP) (MAB387) antibodies were purchased from Merck. Small ribonucleic acid and 488-labeled double stranded oligonucleotides (ODN), both of similar size as miR (i.e. 21–23 bp), were purchased from AITbiotech. All other reagents were purchased from Invitrogen.
2.2. Fabrication and characterization of nanofibers
For electrospinning of nanofibers, PCL was dissolved in a mixture (v/v = 4:1) of TFE and TE to obtain a 12 wt.% solution. The homogenized mixture was loaded into a syringe and injected at a fixed rate of 1.0 ml/h by a syringe pump (New Era pump Systems Inc., USA). Positive 10 kV (Gamma High Voltage, USA) were applied to the solution and rotating collector respectively. The distance between the syringe and the collector was 12 cm. The nanofibers were collected on a negative 4 kv charged aluminum foil, which was wrapped around a rotating wheel (2400 rpm). The morphology of dried nanofiber was evaluated by scanning electron microscopy (SEM) (JOEL, JSM-6390LA, Japan) under an accelerating voltage of 5 kV after sputter-coating with platinum. The average fiber diameters were calculated by measuring 100 fibers using Image J (NIH, USA).
2.3. Preparation of miR-loaded PCL nanofibers
PCL nanofibers were cut to fit 24-well plate (2 cm2 area) and pre-wet with deionized water overnight. Thereafter all nanofibers were immersed into 0.5 mg/ml DOPA dissolved in poly-DOPA coating buffer (10 mM bicine and 50 mM NaCl, pH = 8.5) and agitated at 120 rpm on an orbital shaker for 4 h. The nanofibers were then rinsed with deionized water to remove residual monomer and lyophilized overnight. Thereafter, 4 μg of miR mimics was complexed with 6 μl of TransIT-TKO and incubated at room temperature for 15 min. The complexes were then dropped onto UV-sterilized nanofibers for a complete absorption at 37 °C for 30 min. Next, all miR-absorbed nanofibers were coated with laminin at 10 μg/cm2 for 2 h. All PCL nanofibers were divided into 3 groups: TKO with scrambled negative miR (denoted as scrambled miR), TKO with miR-219 (denoted as miR-219), and TKO with equal mass of miR-219, miR-338-3p and miR-338-5p (denoted as miR-219/miR-338). Conventional poly-D, L-ornithine (p-DL-o) coated coverslips were used as the two-dimensional (2D) culture control, according to the reported protocol [17].
2.4. Primary OPC isolation, culture, viability and reverse transfection
The isolation of primary OPCs from rats was approved by the Institutional Animal Care and Use Committee (IACUC) at Nanyang Technological University, Singapore. According to the reported protocol [17], the cortices from P1–2 neonatal rats were digested with 0.01% trypsin and 0.01 mg/ml Dnase I at 37 °C for 15 min. After being homogenized mechanically, the suspension was passed through a 70 μm cell strainer (BD Biosciences, USA) and seeded in T75 flasks at a density of 10 million cells. After a 7-day culture, the OPCs were purified by shaking on an orbital shaker for 18–20 h at 37 °C. Following that, the cell suspension was passed through a 20 μm sterile screening pouch (Sefar American, USA), and the OPCs were then seeded onto poly-DOPA-coated PCL nanofibers at a density of 50,000 cells per scaffold. Two-dimensional p-DL-o coated coverslips were used as the control, on which the OPCs were treated with a bolus transfection by 50 nM miRs at 1:1 (v/v) of miR:TKO. The purity of isolated OPCs was ~95.84 ± 1.65%, verified by NG2 immunofluorescent staining (Supplementary Fig. 2). At indicated time point post-transfection, three independent samples for each experimental group were collected for live-dead assay using LIVE/DEAD® cell viability kit. The total numbers of live and dead OPCs were quantified using ImageJ software. All other nanofibers and coverslips were collected for real-time PCR or immunofluorescent staining.
2.5. In vitro characterization of small ribonucleic acid loaded nanofibers
Poly-DOPA coated nanofibers that were loaded with RNA/TKO complexes were incubated at 37 °C for 30 min followed by washing with 1 ml of PBS. After treating the supernatant with 1 μg/ml of heparin solution, the concentration of the de-complexed RNA in the supernatant was determined by RiboGreen® assay. Thereafter, the fluorescence intensity was measured using a microplate reader (Tecan®, Infinite 200, Austria) and the mass of RNA was determined. The mass of RNA in the supernatant was termed mwashed while the mass of initially loaded RNA was denoted as mloaded. The initial RNA loading efficiency was then calculated as (mloaded − mwashed)/mloaded × 100%. Following that, the nanofibers were incubated in 3 ml of PBS at 37 °C for up to 30 days. At indicated time points, 1 ml of supernatant was collected and replaced with 1 ml of fresh PBS. The concentration of RNA in the supernatant was measured as indicated above. To determine the drug distribution, 488-labeled RNA was loaded onto the nanofibers in the same manner as indicated above, followed by visualization under a confocal microscope (Zeiss LSM710).
2.6. Real-time PCR
At day 2 post-transfection, OPCs were lysed by a TRIzol® reagent and RNA was extracted using a RNeasy mini kit. Thereafter 100 ng of RNA was used for reverse transcription using M-MLV transcriptase according to the manufacturer’s protocol. Real-time PCR was carried out using iQ SYBR Green Supermix in a StepOnePlus™ system (Applied Biosystems, USA) with the following program: 10 min at 95 °C, 15 s at 95 °C followed by 1 min at 60 °C for 40 cycles. The sequences of the primers are shown in Table 1 and β-actin was used as the housekeeping gene. All our primers showed similar amplification efficiency under the parameters used. Hence the ΔΔCt method was used for fold change analysis. All the results were normalized by the Ct value of OPCs that were treated with scrambled miR in two-dimensional p-DL-o group.
Table 1.
Sequences of primers for real-time PCR.
| β-Actin | Forward | 5′-ACGGTCAGGTCATCACTATCG-3′ |
| Reverse | 5′TGCCACAGGATTCCATACCCAG-3′ | |
| pdgfr-α | Forward | 5′-CGTCTGGTCTTATGGCGTTCTG-3′ |
| Reverse | 5′-TCTCTTTTCGGGTTCACTGTTCC-3′ | |
| foxj3 | Forward | 5′-TCAGTTCTTCACACAGACGGGC-3′ |
| Reverse | 5′-TATGAGGATAACCAGGGGGTGG-3′ | |
| zfp238 | Forward | 5′-TGAAGACGAAGGCGAAGATGAC-3′ |
| Reverse | 5′-AGGGGCTGGCTACTGTTTTTCC-3′ | |
| sox6 | Forward | 5′-TGGTATGAAGATGGACGGCG-3′ |
| Reverse | 5′-TGTTGTTGTTGTTGGGGAAA-3′ |
2.7. Immunofluorescent staining
At indicated time points, the OPCs on nanofibers and coverslips were fixed with 10% formalin, permeabilized with 0.1% Triton-X 100 in PBS for 20 min and blocked with 1.5% goat serum at room temperature for 1 h. The samples were then incubated with primary antibody overnight at 4 °C followed by a secondary antibody for 1 h at room temperature. The primary antibodies used were: rabbit anti-NG2 (1:500), mouse anti-MBP (1:200), mouse anti-RIP (1:500) and rabbit anti-Olig2 (1:1000). The secondary antibodies used were: Alexa-Fluor 488 goat anti-mouse (1:700) and Alexa-Fluor 568 goat anti-rabbit (1:1000). The nuclei were counterstained with DAPI. The samples were mounted and imaged under a confocal microscope (Zeiss LSM710). At least 700 cells were counted in each sample using ImageJ. The proportion of RIP+ or MBP+ cells was then expressed as a percentage of Olig2+ oligodendrocyte lineage cells, which indicates all stages of oligodendrocytes.
2.8. Statistical analysis
All the values were represented as mean ± S.E.M. After verifying, the one-way ANOVA and the Tukey post-hoc tests were used when the samples had equal variance. Otherwise the Kruskal–Wallis and Mann–Whitney U-tests were used for comparison between more than two groups. For a paired comparison, Student’s t-test was used.
3. Results
3.1. Characterization of PCL nanofibers
Supplementary Fig. 3 shows the structure of the poly(DOPA) coated electrospun PCL nanofiber scaffolds. The average diameter of the poly(DOPA) coated nanofiber was 305 ± 49 nm. As compared to the non-coated nanofiber scaffolds (average fiber diameter of 292 ± 37 nm, Fig. 1A), the poly(DOPA) coating was uniform and did not mask or alter any of the underlying nanofiber topography. The average thickness of the nanofiber scaffolds was 151.62 ± 8.75 μm.
Fig. 1.
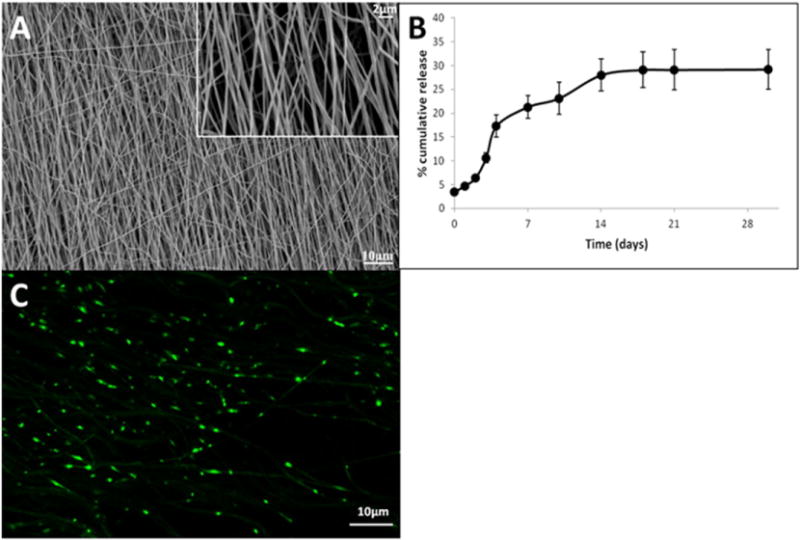
Characteristics of PCL nanofibers. (A) Scanning electron micrograph of scaffolds; (B) RNA release profile (mean ± S.E.M.); (C) RNA distribution on PCL nanofiber scaffolds.
The loading efficiency of RNA was 98.7 ± 0.15%. As shown in Fig. 1B, after an initial burst release of ~3.5%, a sustained release of RNA was obtained. The in vitro cumulative release reached a plateau after 2 weeks followed by a negligible release thereafter, which indicated that the remaining RNA was retained on the scaffolds. As indicated in Fig. 1C, a fairly uniform distribution of RNA over the nanofiber scaffolds was obtained.
3.2. Viability of OPC on nanofiber scaffolds
The representative LIVE-DEAD assay images of OPC on the nanofibers and 2D controls are shown in Fig. 2A. Quantification of these images indicated no significant differences between day 4 and day 7 on the nanofibers, for the numbers of live cells, dead cells and total cell number. However, on 2D coverslip, significantly higher numbers of live cells and dead cells and total cell number were observed. Although the total number of OPCs was higher on the nanofibers on day 4, OPCs proliferated much faster on 2D thereafter, resulting in a significantly higher number of cells at day 7.
Fig. 2.
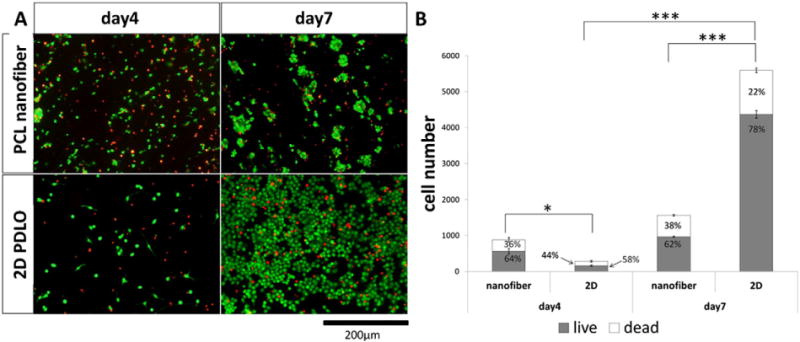
Live-dead assay results. (A) Representative fluorescent microphotographs and (B) quantification of cell number on PCL scaffold and 2D PDLO coverslips (mean ± S.E.M., N = 3). *p < 0.05 and ***p < 0.001 (Student’s t-test).
3.3. Target gene knockdown
Fig. 3 shows the extent of gene silencing by miRs after 2 days of nanofiber-mediated transfection versus conventional bolus transfection on 2D culture. Specifically, FoxJ3, Sox6, ZFP238 and PDGFR-α, the downstream targets of the miRs, were evaluated. In general, as compared to scrambled miR treatment, miR-219 or miR-219/miR-338 transfection resulted in successful knockdown of these targets regardless of substrate topography and transfection methods. The treatment with miR-219/miR-338 cocktail appeared more efficient since gene silencing was significantly enhanced versus miR-219, except for ZFP238, in 2D culture. Nevertheless, miR-219 alone seems more efficient on 2D than on nanofibers. Overall, nanofiber topography reduced markers’ expressions (except PDGFR-α) as compared to 2D cultures (p < 0.05 for scrambled miR on nanofiber vs. 2D). This implied that fiber topography might enhance OL differentiation.
Fig. 3.
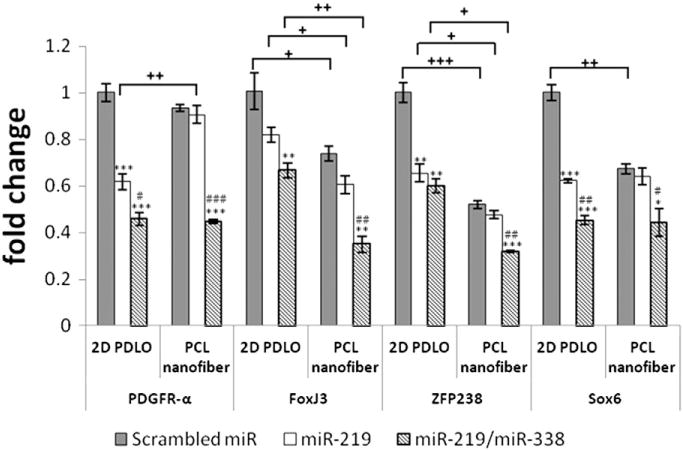
Target gene knockdown on day 2 (mean ± S.E.M., N = 3). *p < 0.05 (ANOVA), **p < 0.01 (ANOVA) and ***p < 0.001 (ANOVA) compared with respective scrambled miR group. #p < 0.05 (ANOVA), ##p < 0.01 (ANOVA) and ###p < 0.001 (ANOVA) compared with respective miR-219 group. +p ≤ 0.05 (Student’s t-test or Mann–Whitney U test), ++p < 0.01 (Student’s t-test) and +++p < 0.001 (Student’s t-test) between two groups.
3.4. Immunostaining of OPC differentiation and maturation markers
a) Effects of nanofiber-mediated miR transfection on OPC differentiation
RIP is an immature OL marker. Here, we evaluated its expression levels as an indicator of the extent of OPC differentiation. As shown in Fig. 4, nanofiber topography enhanced the RIP expressions significantly regardless of miR treatment. When coupled with miRs, the synergistic effect of RNAi and fiber topography improved the differentiation outcomes by greater than 2 fold (Fig. 4B). Both cocktail treatment of miR-219/miR-338 and miR-219 treatment alone appeared beneficial only when introduced by nanofiber-mediated reverse transfection.
Fig. 4.
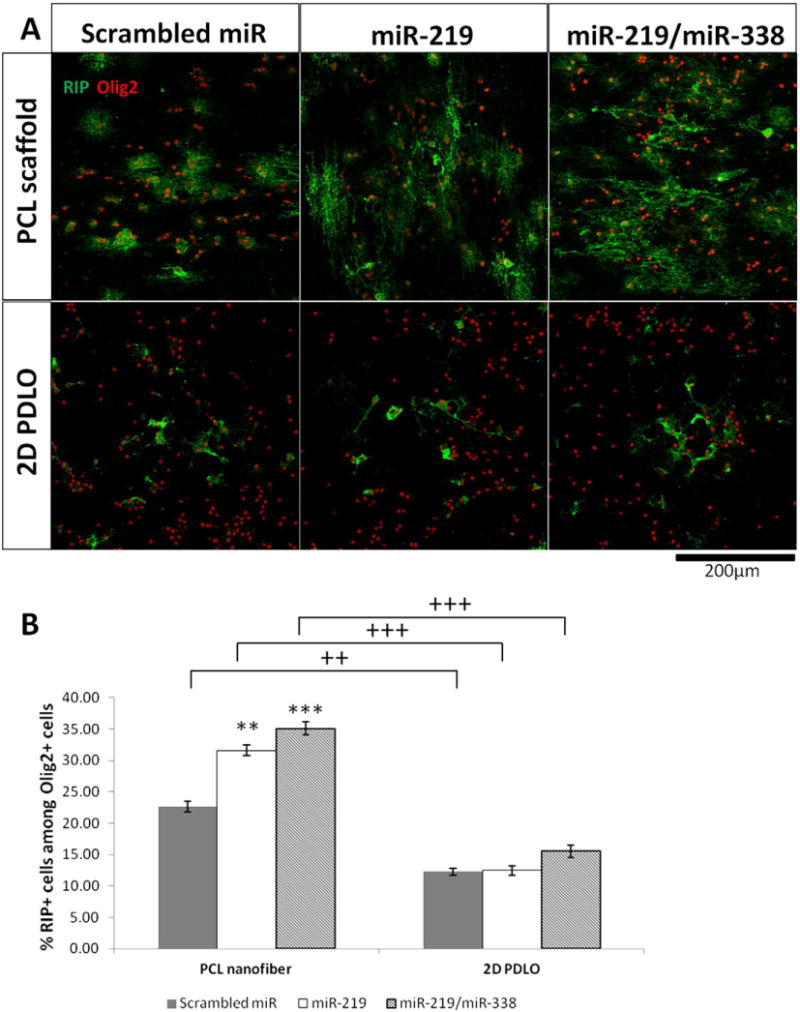
Immunofluorescent staining (day 4) of immature OL marker RIP, indicating enhanced differentiation on PCL scaffold. (A) Representative fluorescent images and (B) Quantification of RIP+ cells among Olig2+ oligodendrocyte lineage cells. **p < 0.01 and ***p < 0.001 versus respective scrambled miR group (ANOVA, mean ± S.E.M., N = 3). ++p < 0.01 and +++p < 0.001 between two groups with same miR treatment (Student’s t-test).
b) Effects of nanofiber-mediated miR transfection on OL maturation
MBP is a marker for mature OL. Here, we evaluated its expression levels as an indicator of the extent of OL maturation. As indicated in Fig. 5, scrambled miR only induced a very basal level of MBP expression on both nanofiber and 2D coverslips. Once combined with the miR-219 or miR-219/miR-338 transfection, the efficiencies of nanofiber-mediated maturation increased significantly by ~2–3 fold on day 4 and 3–5 fold on day 7. Comparatively, the enhancement in MBP+ cells was less significant on 2D bolus transfection controls (1–2 fold on day 7). As shown in Fig. 5B, fiber topography enhanced miR-219/miR-338 cocktail effect more than miR-219 treatment only. Altogether, the nanofiber-mediated reverse transfection facilitated OL maturation at both time points.
Fig. 5.
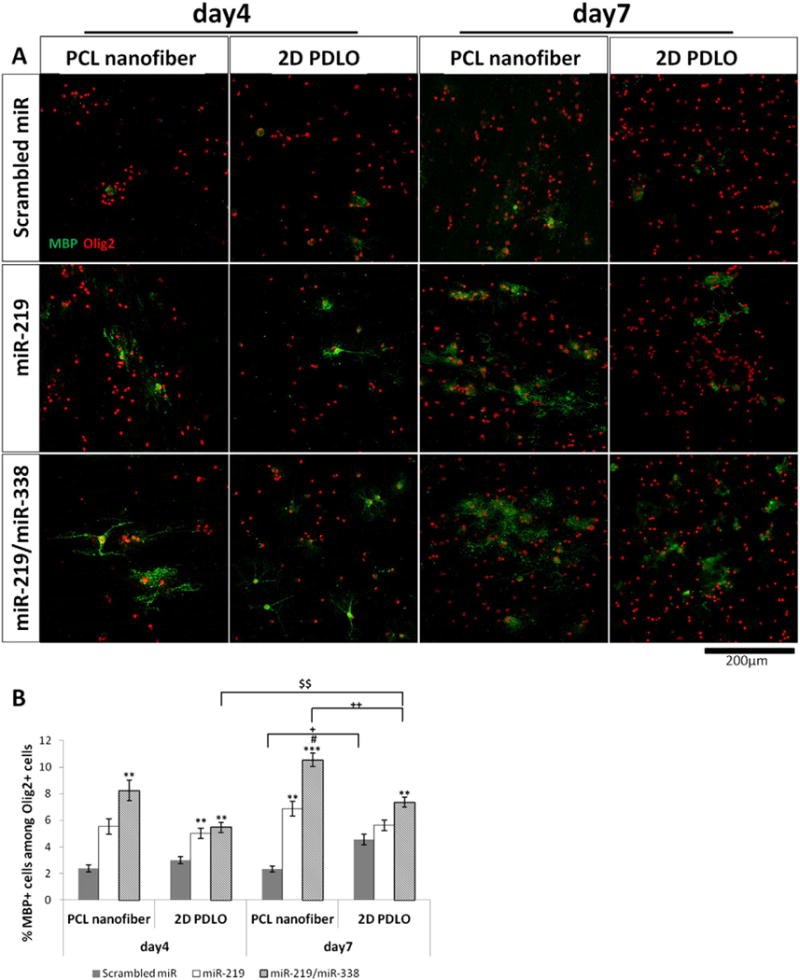
Immunofluorescent staining of mature OL marker MBP, indicating enhanced differentiation on PCL nanofiber (day 4 and day 7). (A) Representative fluorescent images, and (B) quantification of MBP + cells among Olig2 + oligodendrocyte lineage cells. **p < 0.01 and ***p < 0.001 versus respective scrambled miR group; #p < 0.05 versus respective miR-219 group (ANOVA, mean ± S.E.M., N = 3). +p < 0.05 and ++p < 0.01 between two groups (Student’s t-test). $$p < 0.01 (Student’s t-test).
4. Discussion
The OL plays significant roles in maintaining the functionality of the CNS. Hence, methods that provide the ability to understand and direct OPC–OL differentiation and maturation are attractive. Within the CNS, the OL interacts closely with axons, which provide guidance for OL migration, adhesion and myelination. Topographically, nanofibers mimic the architecture of axons and may help recapitulate the OL–axon interactions. As such, nanofibers have served as biomimicking platforms for understanding neuronal–glial interaction and OL myelination [18,19]. In this study, we expanded the applications of nanofiber substrates by incorporating miRs to provide synergistic topographical and biochemical signaling to control the OPC fate.
SncRNA, such as siRNA and miRs, provide biochemical signals to control cell phenotype [27]. Once incorporated within the nanofiber scaffolds, enhanced gene transfection efficiency and prolonged gene-silencing duration (N14 days vs. 5–7 days by bolus delivery in vitro) could be achieved [15]. Such constructs could provide long term gene silencing in vitro [13–15] and in vivo [15]. Therefore, in this study, we expand this promising platform for sustained miR delivery as a strategy for enhancing OL differentiation and maturation.
Although miR and siRNA are chemically similar, a more pronounced release of miRs was observed in this study versus our previous work [14] (~30% vs. ~10% cumulative release over 30 days). Here, poly-DOPA coating was adopted as compared to the more commonly encountered polydopamine coating. Comparatively, under alkaline pH (8.5), the unique carboxylic groups in poly-DOPA enabled the coating solution to be more evenly dispersed [20]. Polydopamine, however, frequently resulted in undesirable aggregate formation [21]. In addition, a different transfection reagent, TKO (vs. lipofectamine RNAimax), was used for successful OPC transfection. Correspondingly, a difference in nucleic acid release profile was obtained.
So far, the exact mechanism of polymerization and binding of DOPA is still unclear. Under neutral or slightly basic conditions, the negative charge of poly(DOPA) may interact with the positively charged TKO/miR complex via electro-statistic interactions. O-quinone groups on 3,4-dihydroxyl-L-phenylalanin (DOPA), can also react with amino (via Schiff base) or thiol groups (via Michael addition) that may exist in TKO/miR complexes [14]. Regardless of mechanism, efficient gene knockdown was still achieved, which further induced efficient OPC differentiation and OL maturation. TKO/miR complexes that were released from the scaffolds may be taken up by cells as soluble complexes, while a portion may have been taken up directly by cells, as surface absorbed complexes, after cells adhered onto the surface of the scaffolds. Further detailed analyses by live cell imaging with fluorescently tagged complexes should be carried out to understand the exact cellular uptake mechanisms.
As compared to 2D culture, fiber topography decreased OPC proliferation significantly. In addition, the expressions of FoxJ3, ZFP238 and Sox6 were also significantly reduced in OPCs that were cultured on fiber constructs (Fig. 3, scrambled miR on 2D PDLO vs. PCL nanofiber, p < 0.05). These results suggest that fiber topography alone could enhance OPC differentiation. Coupled with the fact that fiber topography enhanced human Schwann cell maturation [11], the results suggest the potential of fiber scaffolds for remyelination applications.
During OPC differentiation into OL, 43 potential functional miRs have been identified [22]. Most notably, miR-219 and miR-338 increased during OL differentiation. They target transcription factors that maintain undifferentiated and proliferative states of OPC [7,8]. Consistently, our results showed that nanofiber-mediated miR-219 and miR-338 reverse transfection efficiently silenced the inhibitory factors of OL differentiation by 50–60% after 48 h of transfection (Fig. 3), which is more efficient than other works also delivering miR (~25%) [23].
As compared to 2D bolus transfection, nanofiber scaffolds induced lower extents of relative gene silencing except cocktail treatment on FoxJ3 (Fig. 3 and Supplementary Fig. 1, scrambled miR vs. miR-219 and miR-219/miR-338). One possible reason may be the lower amounts of miRs that were released from the scaffolds vs. bolus delivery (~135 ng from nanofibers at day 0, Fig. 2B vs. ~333 ng for 50 nM of bolus delivery). Another potential reason may be the fact that fiber topography alone decreased the expression of the inhibitory factors. Hence, with a lower basal expression level of inhibitory factors of OPC differentiation, subsequent gene silencing with miR-219 and miR-219/miR-338 cocktail elicited lower extents of relative gene knockdown. Despite the lower gene silencing efficiencies, the subsequent extent of OPC differentiation and OL maturation were enhanced on miR-incorporated nanofiber substrates vs. 2D cultures. The results, therefore, demonstrated the advantage of using nanofiber-mediated miR delivery approach to direct cell fate commitment in OPC.
RIP+ cells represent an early differentiation stage of OPCs. On nanofiber with scrambled miR treatment, 22% of RIP+ cell was obtained while on 2D culture with the same treatment, only 12% was achieved. The significantly higher proportion of RIP+ cells on nanofiber vs. that obtained by bolus transfection in conventional 2D culture, both in our works and that by Zhao et al. [7], suggested that fiber topography played an important role in OPC differentiation. Comparatively, miR treatment appeared more potent in promoting OL maturation than OPC differentiation. Specifically, as compared with scrambled miR treatment, miR-219 or miR-219/miR-338 cocktail treatments induced 3–5 folds increase in MBP expression (Fig. 5), while only less than 2-fold enhancement in RIP expression was observed (Fig. 4). One possible reason may be the fact that OPC differentiation (as indicated by RIP+ cells) is more sensitive to substrate topography, hence masking any additional effects by miR treatment. In general, miR-219/miR-338 cocktail treatment appeared more beneficial in directing OPC differentiation and maturation, especially when coupled with nanofiber topography.
Fiber alignment and diameter alter gene uptake and gene silencing efficiencies in cells [12]. In addition, fiber diameter also affected OL myelination [18]. Therefore, future works focusing on fiber diameter and/or orientation effect on nanofiber-mediated OPC differentiation and maturation could provide useful insights to direct the behavior and cell fate commitment of OPC.
As a proof of concept, PCL was used in this study. Further translational applications can extend to the choice of different materials for specific applications. In terms of direct scaffold implantation, both our and others’ studies have already demonstrated that electrospun scaffolds can be directly implanted into the spinal cord for SCI treatment [24,25]. As for applications where cell harvesting is required, e.g. multiple sclerosis, alternative choice of materials like thermo-sensitive chitosan may be explored, where cells seeded on thermo-sensitive electrospun fibers could be obtained simply by dissolving the scaffolds at low temperatures, e.g. 4 °C [26].
5. Conclusion
In this study, we introduced a nanofiber-mediated microRNA delivery system to direct OL differentiation and maturation by gene silencing. As compared to conventional bolus transfection on 2D cultures, our nanofiber-mediated reverse transfection platform more efficiently promoted cell fate commitment in OPC. Such constructs may find useful applications in priming OPC for transplantation and remyelination in a variety of neurological pathologies, such as SCI and multiple sclerosis.
Supplementary Material
Acknowledgments
This work was supported by the National Medical Research Council (NMRC) CBRG Grant (NMRC/CBRG/0002/2012).
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jconrel.2015.03.005.
References
- 1.McDonald JW, Belegu V. Demyelination and remyelination after spinal cord injury. J Neurotrauma. 2006;23:345–359. doi: 10.1089/neu.2006.23.345. [DOI] [PubMed] [Google Scholar]
- 2.Emery B. Regulation of oligodendrocyte differentiation and myelination. Science. 2010;330:779–782. doi: 10.1126/science.1190927. [DOI] [PubMed] [Google Scholar]
- 3.Cao Q, He Q, Wang Y, Cheng X, Howard RM, Zhang Y, DeVries WH, Shields CB, Magnuson DS, Xu XM, Kim DH, Whittemore SR. Transplantation of ciliary neurotrophic factor-expressing adult oligodendrocyte precursor cells promotes remyelination and functional recovery after spinal cord injury. J Neurosci. 2010;30:2989–3001. doi: 10.1523/JNEUROSCI.3174-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emery B. Transcriptional and post-transcriptional control of CNS myelination. Curr Opin Neurobiol. 2010;20:601–607. doi: 10.1016/j.conb.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Bhalala OG, Srikanth M, Kessler JA. The emerging roles of microRNAs in CNS injuries, Nature reviews. Neurology. 2013;9:328–339. doi: 10.1038/nrneurol.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He X, Yu Y, Awatramani R, Lu QR. Unwrapping myelination by microRNAs. Neuroscientist. 2012;18:45–55. doi: 10.1177/1073858410392382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, Wang F, Appel B, Lu QR. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65:612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, Barres BA. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Downing TL, Soto J, Morez C, Houssin T, Fritz A, Yuan F, Chu J, Patel S, Schaffer DV, Li S. Biophysical regulation of epigenetic state and cell reprogramming. Nat Mater. 2013;12:1154–1162. doi: 10.1038/nmat3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chew SY, Low WC. Scaffold-based approach to direct stem cell neural and cardiovascular differentiation: an analysis of physical and biochemical effects. J Biomed Mater Res A. 2011;97:355–374. doi: 10.1002/jbm.a.33064. [DOI] [PubMed] [Google Scholar]
- 11.Chew SY, Mi R, Hoke A, Leong KW. The effect of the alignment of electrospun fibrous scaffolds on Schwann cell maturation. Biomaterials. 2008;29:653–661. doi: 10.1016/j.biomaterials.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yau WW, Long H, Gauthier NC, Chan JK, Chew SY. The effects of nanofiber diameter and orientation on siRNA uptake and gene silencing. Biomaterials. 2014;37C:94–106. doi: 10.1016/j.biomaterials.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Cao H, Liu T, Chew SY. The application of nanofibrous scaffolds in neural tissue engineering. Adv Drug Deliv Rev. 2009;61:1055–1064. doi: 10.1016/j.addr.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Low WC, Rujitanaroj PO, Lee DK, Messersmith PB, Stanton LW, Goh E, Chew SY. Nanofibrous scaffold-mediated REST knockdown to enhance neuronal differentiation of stem cells. Biomaterials. 2013;34:3581–3590. doi: 10.1016/j.biomaterials.2013.01.093. [DOI] [PubMed] [Google Scholar]
- 15.Rujitanaroj PO, Jao B, Yang J, Wang F, Anderson JM, Wang J, Chew SY. Controlling fibrous capsule formation through long-term down-regulation of collagen type I (COL1A1) expression by nanofiber-mediated siRNA gene silencing. Acta Biomater. 2013;9:4513–4524. doi: 10.1016/j.actbio.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rujitanaroj PO, Wang YC, Wang J, Chew SY. Nanofiber-mediated controlled release of siRNA complexes for long term gene-silencing applications. Biomaterials. 2011;32:5915–5923. doi: 10.1016/j.biomaterials.2011.04.065. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Balasubramaniyan V, Peng J, Hurlock EC, Tallquist M, Li J, Lu QR. Isolation and culture of rat and mouse oligodendrocyte precursor cells. Nat Protoc. 2007;2:1044–1051. doi: 10.1038/nprot.2007.149. [DOI] [PubMed] [Google Scholar]
- 18.Lee S, Leach MK, Redmond SA, Chong SY, Mellon SH, Tuck SJ, Feng ZQ, Corey JM, Chan JR. A culture system to study oligodendrocyte myelination processes using engineered nanofibers. Nat Methods. 2012;9:917–922. doi: 10.1038/nmeth.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, Chong SY, Tuck SJ, Corey JM, Chan JR. A rapid and reproducible assay for modeling myelination by oligodendrocytes using engineered nanofibers. Nat Protoc. 2013;8:771–782. doi: 10.1038/nprot.2013.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei H, Ren J, Han B, Xu L, Han L, Jia L. Stability of polydopamine and poly(DOPA) melanin-like films on the surface of polymer membranes under strongly acidic and alkaline conditions. Colloids Surf B: Biointerfaces. 2013;110:22–28. doi: 10.1016/j.colsurfb.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Shin YM, Lee YB, Shin H. Time-dependent mussel-inspired functionalization of poly(L-lactide-co-varepsilon-caprolactone) substrates for tunable cell behaviors. Colloids Surf B: Biointerfaces. 2011;87:79–87. doi: 10.1016/j.colsurfb.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28:11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen MK, Jeon O, Krebs MD, Schapira D, Alsberg E. Sustained localized presentation of RNA interfering molecules from in situ forming hydrogels to guide stem cell osteogenic differentiation. Biomaterials. 2014;35:6278–6286. doi: 10.1016/j.biomaterials.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu T, Houle JD, Xu J, Chan BP, Chew SY. Nanofibrous collagen nerve conduits for spinal cord repair. Tissue Eng A. 2012;18:1057–1066. doi: 10.1089/ten.tea.2011.0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurtado A, Cregg JM, Wang HB, Wendell DF, Oudega M, Gilbert RJ, McDonald JW. Robust CNS regeneration after complete spinal cord transection using aligned poly-L-lactic acid microfibers. Biomaterials. 2011;32:6068–6079. doi: 10.1016/j.biomaterials.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang JM, Leong KW. Myogenic induction of aligned mesenchymal stem cell sheets by culture on thermally responsive electrospun nanofibers. Adv Mater. 2007;19:2775–2779. doi: 10.1002/adma.200602159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yau WW, Rujitanaroj PO, Lam L, Chew SY. Directing stem cell fate by controlled RNA interference. Biomaterials. 33:2608–2628. doi: 10.1016/j.biomaterials.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 28.Cao H, Marcy G, Goh EL, Wang F, Wang J, Chew SY. The effects of nanofiber topography on astrocyte behavior and gene silencing efficiency. Macromol Biosci. 2014;12:666–674. doi: 10.1002/mabi.201100436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


