Abstract
Lin28, a well-known RNA-binding protein, regulates diverse cellular properties. All physiological functions of Lin28A characterized so far have been attributed to its repression of let-7 miRNA biogenesis or modulation of mRNA translational efficiency. Here we show that Lin28A directly binds to a consensus DNA sequence in vitro and in mouse embryonic stem cells in vivo. ChIP-seq and RNA-seq reveal enrichment of Lin28A binding around transcription start sites, and a positive correlation between its genomic occupancy and expression of many associated genes. Mechanistically, Lin28A recruits 5-methylcytosine-dioxygenase Tet1 to genomic binding sites to orchestrate 5-methylcytosine and 5-hydroxymethylcytosine dynamics. Either Lin28A or Tet1 knockdown leads to dysregulated DNA methylation and expression of common target genes. These results reveal a surprising role for Lin28A in transcriptional regulation via epigenetic DNA modifications and have implications for understanding mechanisms underlying versatile functions of Lin28A in mammalian systems.
INTRODUCTION
Lin28 influences diverse aspects of cellular activities, including stem cell self-renewal, somatic cell reprogramming, metabolism, organismal growth and regeneration, and carcinogenesis (Shyh-Chang and Daley, 2013). Two paralogs of Lin28 in mammals, Lin28A and Lin28B, exhibit different expression patterns but similar functions. Lin28A is highly expressed in embryos and embryonic stem cells (ESCs), and becomes down-regulated during differentiation. Suppression of let-7 miRNA maturation is among best-studied molecular mechanisms of Lin28A-mediated regulation of gene networks, whereas Lin28A can also influence mRNA translation and splicing (Shyh-Chang and Daley, 2013). One study in C. elegans also suggested binding of Lin28A to the let-7 genomic locus (Van Wynsberghe et al., 2011). Structurally, Lin28A is composed of two classical nucleic acid binding domains: a cold-shock domain (CSD) and a pair of CCHC-type zinc finger motifs (Moss et al., 1997), both of which contribute to the affinity for pre-let7 and repression of let-7 maturation (Loughlin et al., 2012; Nam et al., 2011). Notably, CSD domains in other molecules have been shown to bind DNA (Hudson and Ortlund, 2014), raising the possibility that Lin28A may also exhibit DNA binding activity.
5-methylcytosine (5mC), a key epigenetic modification, regulates gene expression and many cellular processes (Suzuki and Bird, 2008). Ten-eleven translocation (Tet) family proteins can sequentially convert 5mC to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5CaC), leading to DNA demethylation (Wu and Zhang, 2014). The molecular mechanism underlying recruitment of Tet1 to specific genomic loci for region-specific DNA demethylation remains poorly understood. Here we reveal a DNA-binding feature of Lin28A and demonstrate its association with Tet1. Characterization of Lin28A interactions with DNA in mouse ESCs (mESCs) suggests a promoter-binding signature and its role in recruiting Tet1 to co-targeted genomic loci for epigenetic regulation. Our study unravels a DNA-based regulatory role of Lin28A in mESCs and functionally links it to the epigenetic regulator Tet1.
RESULTS
Lin28A binds to specific genomic loci in mouse ESCs and directly binds DNA
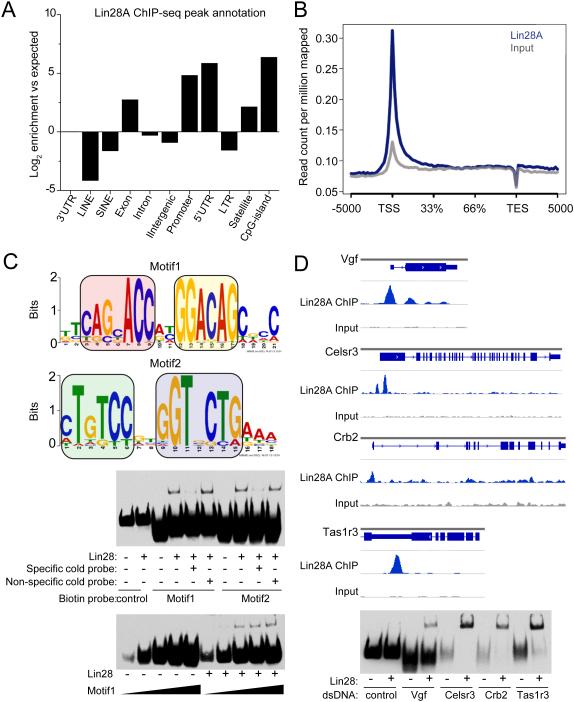
Our previous protein microarray-based screen of the protein-DNA interactome identified Lin28A as one of over 4000 proteins that interact with DNA oligos on chip (Hu et al., 2009). Lin28A, present in both nucleus and cytoplasm of mESCs (Kawahara et al., 2011), is a well-known RNA-binding protein (Shyh-Chang and Daley, 2013). To determine whether Lin28A indeed binds to genomic DNA in mammalian cells, we generated a stable mESC line expressing exogenous FLAG-tagged Lin28A and performed chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq). A total of 7874 high-confidence peaks (with an average false discovery rate FDR = 3.69) were identified (Table S1). Lin28A was enriched at transcription starting sites (TSS)/promoters/5’UTR of annotated genes, followed by gene bodies and CpG islands (Figures 1A-B and S1A; Table S1). Analysis of best-scored ChIP peaks by MEME or Homer predicted a major consensus sequence featuring a bipartite “CAGnACC”-nn-“GGACAG” (Motif 1) and its reverse complementary sequence (Motif 2; Figure 1C and Table S2). Previous structural studies established that Lin28A has two folded domains that recognize two distinct regions of RNA, i.e., GNGA(C/T) and GGAG with variable distance between the two (Loughlin et al., 2012; Nam et al., 2011), similar to our consensus DNA sequence (Figure S1B).
Figure 1. Lin28A binds to specific genomic loci in mESCs and directly to DNA in vitro.
(A) ESCs (E14) stably expressing FLAG-tagged Lin28A were used for ChIP-seq experiments with anti-FLAG antibody. Annotations of Lin28A ChIP-seq peaks to various genomic features are indicated based on their log2 fold enrichment over expected values.
(B) Normalized Lin28A ChIP-seq read densities on UCSC mm9 Refseq gene bodies (n = 20460). Gene bodies were normalized to 0-100% as relative position.
(C) Lin28A ChIP-seq common motifs were predicted using MEME suite from the top 20% of Lin28A peaks (Motif1: E value = 6.3e−156, 42 sites; Motif 2: E value = 9.0e−35, 22 sites). Colored boxes indicate reverse complementary elements of two motifs.
(D-E) ChIP-seq peaks of several representative genomic loci (D) and in vitro verification of Lin28A protein-DNA interaction with gel-shift assay using probes corresponding to these genomic loci (E). Values represent mean ± S.D. (n = 4).
We next used an electrophoretic mobility shift assay (EMSA) to examine direct interaction between purified recombinant Lin28A and DNA motifs (Figure S1C). Lin28A showed similar binding to both the RNA and DNA analog of the pre-let-7 stem-loop, which is the best-characterized Lin28A RNA binding site (Figure S1D). EMSA showed similar Lin28A binding to three out of four promoter sequences identified from ChIP-seq analyses (Figures 1D-E). We further performed ChIP-seq of mESCs using antibodies that recognized endogenous Lin28A (Cho et al., 2012; Wilbert et al., 2012), and found significant overlap between our two datasets, but not with endogenous H3K4me1 or input peaks (Figure S1E). Endogenous Lin28A-Chip mapped reads also displayed significant enrichment on gene TSS/promoters (Figure S1F). Collectively, our in vivo and in vitro analyses identified Lin28A as a DNA-binding protein, in addition to its well-known RNA-binding ability. Unless otherwise specified, all “Lin28A targets” mentioned hereafter refers to DNA associates.
Lin28A is primarily associated with transcribed genes in mESCs
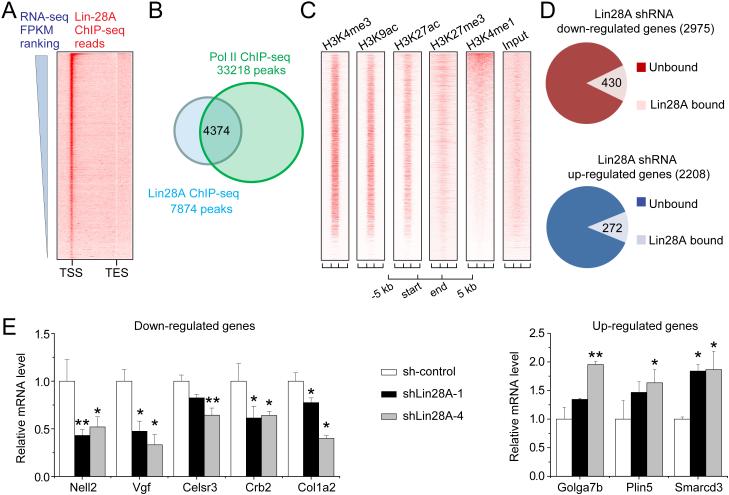
We next asked whether Lin28A occupancy was correlated with gene expression in mESCs. Lin28A binding to TSS displayed a positive correlation with the expression of associated genes (Figure 2A) (Shen et al., 2012). 56% of Lin28A ChIP-seq peaks overlapped with RNA polymerase II (Pol II) peaks (Figure 2B). Furthermore, histone markers that are associated with active transcription, such as H3K4me3, H3K9ac and H3K27ac, were highly enriched at Lin28A binding sites, whereas the transcription repressive marker (H3K27me3) and enhancer marker (H3K4me1) were much more diffusely distributed (Figure 2C).
Figure 2. Lin28A is primarily associated with actively transcribed genes in mESCs.
(A) Lin28A ChIP-seq mapped reads were plotted on gene bodies with their expression levels ranked in a descending order for the heatmap view. mESCs RNA-seq FPKM values (Fragments Per Kilobase of transcript per Million mapped reads) were obtained from a published dataset (Shen et al., 2012).
(B) Lin28A ChIP-seq peaks substantially overlapped with RNA polymerase II ChIP-seq peaks (Handoko et al., 2011).
(C) Lin28A-binding sites were enriched with active histone marks, but were diffused with histone modifications for repressive or enhancer markers. Regions between start and end define boundaries of Lin28A-binding sites. ChIP-seq mapped reads of various histone modifications either from ENCODE or generated in-house were plotted in heat-map views. The Lin28A binding sites were ranked by peak intensity.
(D) Replicated samples from stable mESC lines expressing Lin28A shRNA or its scrambled control were subjected to RNA-seq. 2975 genes were substantially down-regulated in Lin28A KD mESCs compared to the scrambled control (log2 FPKM ratio < −0.5; upper panel). Among those down-regulated genes upon Lin28A depletion, 430 genes were directly bound by Lin28A based on Lin28A ChIP-seq analysis (p = 0.02; Chi Squared Test). 2208 genes were up-regulated (log2FPKM ratio > 0.5) upon Lin28A KD in mESCs, with 272 genes directly bound by Lin28A (lower panel; p = 0.00004; Chi Squared Test).
(E) Examples of genes either down-regulated or up-regulated in mESCs with Lin28A KD. GAPDH was used as an internal control, and relative mRNA values were calculated based on comparison with the scrambled control. Two different Lin28A shRNAs were used. Values represent mean ± S.D. (n = 4; *p < 0.05; **p < 0.01; Student’s t test). See also Figure S2.
To assess the functional role of Lin28A in regulating gene expression directly, we generated a stable mESC line with Lin28A knockdown (KD) via lentivirus-mediated shRNA expression and performed RNA-seq (Figures S2A-B; Table S1). Expression of pluripotency genes Oct4, Nanog, Sox2 and Klf4 was not significantly altered by Lin28A KD (Figure S2C). Among 2975 genes that were at least 20.5 fold down-regulated by Lin28A KD when compared to the scrambled shRNA control (sh-control), 430 genes were bound by Lin28A on promoters or gene bodies (p = 0.02; Chi Squared Test; Figure 2D). In addition, 2208 genes were at least 20.5 fold up-regulated in Lin28A KD mESCs, 272 of which were Lin28A-bound (p = 0.00004; Chi Squared Test; Figure 2D). The influence of Lin28A KD on mRNA levels of representative target genes was confirmed by quantitative PCR (Figure 2E). To distinguish Lin28A-dependent transcriptional regulation from post-transcriptional regulation, we took advantage of a recently released analytical tool, iRNA-seq, which performs a genome-wide assessment of acute transcriptional activity based on analysis of intron coverage from total RNA-seq data (Madsen et al., 2015). We found 55-64% overlap with standard RNA-seq analysis (Figure S2D), indicating a prime role of Lin28A in acute transcription regulation.
Lin28A forms a complex with Tet1 to recruit Tet1 to their genomic co-targets
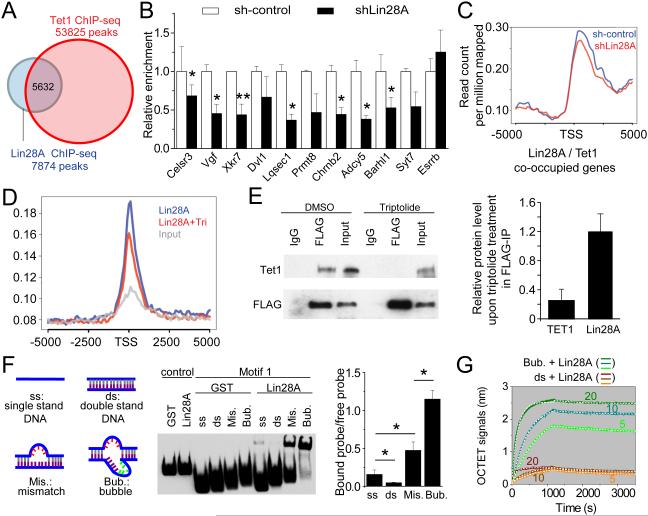
To understand how Lin28A regulates gene expression, we compared genome-wide Lin28A distribution with published databases. Interestingly, more than half (49.1% to 71.5%) of Lin28A binding sites were shared by Tet1 in mESCs (Williams et al., 2011; Wu et al., 2011; Xu et al., 2011) (Figures 3A and S3A). TSS and gene body regions co-occupied by Lin28A and Tet1 showed similar enrichment of histone modifications associated with active transcription as those bound by Lin28A (Figure S3B). Furthermore, Tet1 binding motifs predicted by MEME of ChIP-seq analysis (Wu et al., 2011) also shared similarity to the 5’ portion “CAGCACC” within the identified Lin28A motif (Figure S3C and 1C; Table S2).
Figure 3. Lin28A is required to recruit Tet1 to their common genomic target sites.
(A) Genome-wide Lin28A-binding regions substantially overlapped with Tet1-binding regions identified by ChIP-seq (Wu et al., 2011).
(B-C) Knocking down Lin28A decreased Tet1 binding on their co-target genes. An mESC line with HA-Tet1 knocked into the endogenous Rosa locus was used for the ChIP experiment (B). Lin28A was knocked down by siRNA transfection. Relative enrichment on amplicons was calculated by comparing to the scrambled control. Esrrb is a negative control amplicon without Lin28A binding. Values represent mean ± S.D. (n = 3; *p < 0.05; **p < 0.01; Student’s t test). Also shown is the endogenous Tet1 Chip-seq result on genome average of Tet1 binding to TSS for all co-target genes with or without Lin28A KD (C).
(D-E) Triptolide treatment reduced genome-wide Lin28A binding on Lin28A-bound gene promoters in mESCs (D) and disrupted interaction of Lin28A and Tet1 (E). mESCs were treated with DMSO (control) or 1 μM triptolide for 5 hours before ChIP-seq (D) or IP-western analyses of Tet1 or Lin28A (E). Mapped ChIP-seq reads on 5 kb upstream and downstream of Lin28A-bound TSS are shown in (D). Using ImageJ, we quantified the IP-western blots (E) and calculated the relative amount of immunoprecipitated Tet1/Lin28A with input cell lysate, then compared this ratio between control and triptolide-treated samples. Values represent mean ± S.D. (n = 5; *p < 0.05; Student’s t test).
(F) Lin28A bindings to various forms of Motif1. Binding affinities of Lin28A with single-strand (ss) DNA, double-strand (ds) DNA, double strand with 5-bp mismatch (Mis.), and a “bubble” structure (Bub.) were examined by gel-shift assay. Values represent mean ± S.D. (n = 3; *p < 0.05; **p < 0.01; Student’s t test).
(G) Real-time measurement of KD for binding of Lin28A to bubble-DNA and dsDNA by Octet® QKe system. The concentrations for recombinant Lin28A protein were 5, 10 and 20 μg/ml for different curves. The concentration for bubble-DNA or dsDNA was 500 nM. These data were used to extract association and dissociation curves for 3 concentrations of Lin28A protein and fit the curves by 1:1 model to obtain KD.
See also Figure S3.
Next, we explored the mechanism underlying co-occupancy of Lin28A and Tet1 on target genes. We used a loss-of-function approach to determine whether depletion of one protein would affect the chromatin-binding property of the other in mESCs that expressed either HA-tagged Tet1 knocked into the Rosa locus (Figure S3D) or exogenous FLAG-tagged Lin28A. For a group of 10 genomic loci that showed Lin28A occupancy in ChIP-seq, Lin28A KD decreased Tet1 enrichment on common targets by 30-60% (Figure 3B). ChIP-seq of Lin28A KD mESCs using Tet1 antibodies also showed a similar pattern of decreased Tet1 binding to common targets (Figure 3C). In contrast, Tet1 KD did not affect Lin28A binding to chromatin (Figure S3E). Together, our analyses revealed significant co-occupancy across the genome between Lin28A and Tet1 in mESCs, and a requirement for Lin28A to recruit Tet1 to common genomic targets, but not vice versa.
To further elucidate the mechanism of Lin28A-mediated recruitment of Tet1 to genomic loci, we tested whether the two proteins reside in the same complex in HEK293T cells co-expressing exogenous Lin28A and Tet1, as well as in mESCs with or without exogenously expressed Lin28A. Co-immunoprecipitation analysis demonstrated the biochemical association between Lin28A and Tet1 in HEK293T cells and mESCs (Figures S3F-H). The catalytic domain of Tet1 (Tet1-CD) was sufficient to form a complex with Lin28A, but neither its catalytic activity nor the presence of DNA molecules was required for formation of the complex (Figures S3I-J).
We then sought to investigate the regulatory mechanism for recruiting Lin28A to its genomic binding loci. Given the positive correlation between Lin28A genomic occupancy at TSS/promoters and gene expression, as well as its co-occupancy with Pol II (Figures 2A-B), we asked whether active transcription is required. We blocked transcription initiation, specifically at the step of unwinding double strand DNA, by treatment with RNA Polymerase II inhibitor Triptolide (Bensaude, 2011). Triptolide decreased Lin28A binding to chromatin (Figure 3D), and attenuated Lin28A-Tet1 complex formation in mESCs (Figure 3E), suggesting that active transcription is required for Lin28A to bind chromatin and associate with Tet1.
Based on the bipartite character of the known Lin28A-RNA structure (Loughlin et al., 2012; Nam et al., 2011) and similarity between DNA and RNA consensus sequences (Figure S1B), we predicted that a “bubble” structure would strengthen the direct Lin28A-DNA interaction. We created a “bubble” structure by inserting 5-bp random sequences on top of the mismatched DNA oligo (Figures 3F and S3K). Indeed, EMSA showed that Lin28A displayed the strongest binding to the DNA with “bubble” structures containing both mismatches and insertions, with weaker binding to mismatch-only DNA, single-strand DNA, and double-strand DNA oligo of motif 1 (Figure 3F). To quantitatively assess Lin28A affinity to different forms of DNA in vitro, we performed real-time KD measurements and found a high affinity of KD of 72 ± 25 nM for Lin28A-“bubble” DNA binding and 186 ± 100 nM for Lin28A-dsDNA binding (Figure 3G). Together, our results suggest a hierarchical model in which Lin28A recognizes transcription bubbles, then recruits Tet1 onsite.
Lin28A coordinates with Tet1 to regulate cytosine modification dynamics and gene expression
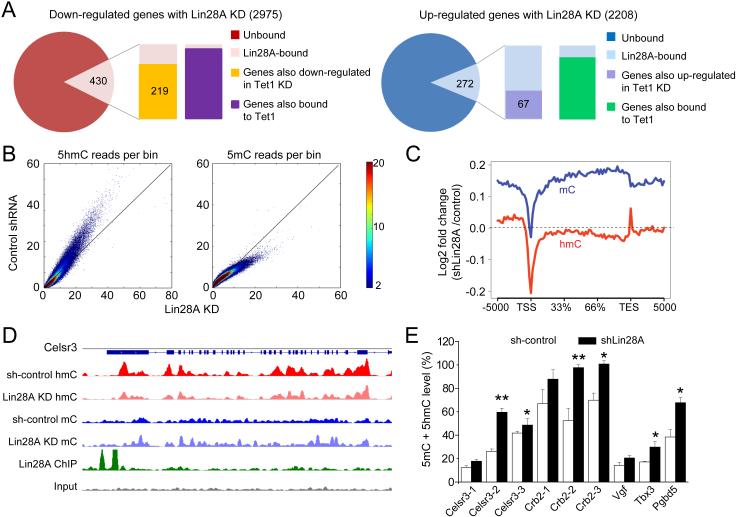
To investigate co-regulation of gene expression by Lin28A and Tet1, we analyzed RNA-seq results from Lin28A KD and Tet1 KD in mESCs. Among 430 genes that were bound by Lin28A and displayed reduced expression with Lin28A KD (Figure 2D), 219 genes (50.9%) showed similar reduced expression with Tet1 KD (Xu et al., 2011) (Figure 4A). In contrast, among 272 genes that were bound by Lin28A and up-regulated by Lin28A KD, only 67 genes (24.6%) were up-regulated by Tet1 KD (Figure 4A). This observation indicates a ~2-fold dominance of Lin28A-mediated coordination of Tet1 for their positively-regulated co-targets. This result is consistent with the previous finding that Tet1 KD in mESCs leads to up-regulation and down-regulation of different genes (Huang et al., 2014; Williams et al., 2011; Wu et al., 2011). Over 95% of up- or down-regulated genes bound by Lin28A were also bound by Tet1 on promoters or intragenic regions (Figure 4A) and co-binding sites exhibited similar compositions of CpG island abundance (Figure S4A).
Figure 4. Lin28A coordinates with Tet1 to regulate DNA modification dynamics on target genes.
(A) Among a total of 2975 down-regulated genes upon Lin28A KD, 430 genes were bound by Lin28A. Among those, 219 genes were also shown to be down-regulated in Tet1 KD mESCs (Xu et al., 2011). In comparison, 272 genes out of 2208 up-regulated genes upon Lin28A KD were bound by Lin28A. Only 67 were also shown to be up-regulated in Tet1 KD mESCs. The majority of Lin28A-bound genes with expression changes were co-bound by Tet1. RNA-seq values were analyzed by Pearson's Chi-squared test with Yates' continuity correction (p < 2.2e−16).
(B) enome-wide 5hmC and 5mC reads in mESCs upon Lin28A KD and their scrambled controls were counted within 10 kb bins for the whole mouse genome. All bins were normalized to sequencing coverage and plotted to compare the overall trend of 5hmC and 5mC abundance between Lin28A KD mESCs versus their scrambled control.
(C) The log2 fold change ratio of 5hmC (red) and 5mC (blue) mapped reads on gene bodies between Lin28A KD and scrambled control were calculated and plotted. The dashed line indicates non-changing points with log2 (fold change) = 0. Plots below the dashed line indicate reduced value in Lin28A KD compared to the scrambled control, and vice versa.
(D) Celsr3 locus as an example of a Lin28A-bound gene that showed reduced expression, and inverse changes of 5hmC and 5mC on the gene body upon Lin28A KD.
(E) Quantification of 5mC levels by methylation-specific HpaII/MspI digestion and T4-βGT mediated distinguishing of 5hmC and 5mC, followed with quantitative PCR. Values represent mean ± S.D. (n = 4; *p < 0.05; **p < 0.01; Student’s t test). See also Figure S4.
To further assess the physiological role of the Lin28A-Tet1 association, we analyzed genome-wide 5hmC and 5mC levels in Lin28A KD and sh-control mESCs. We employed previously established methods for selective chemical labeling and enrichment for 5hmC-containing DNA, and methylated DNA immunoprecipitation, coupled with high-throughput sequencing techniques. Lin28A KD resulted in a modest genome-wide decrease of 5hmCs, concurrent with an increase of 5mC levels in mESCs (Figures 4B-C and S4B). Analysis of multiple target genes co-regulated by Lin28A and Tet1 showed consistent patterns of decreased 5hmCs and increased 5mCs at gene bodies (Figures 4D and S4C). An increase in methylation levels upon Lin28A KD was confirmed by T4-βGT-coupled methylation-specific HpaII/MspI digestion analysis on multiple Lin28A-Tet1 co-targets (Figure 4E). Collectively, our data indicated that Tet1 recruitment by Lin28A promotes 5mC-to-5hmC conversion and demethylation at gene bodies.
DISCUSSION
In this study, we identified Lin28A DNA-binding properties both in vitro and in vivo, and its association with Tet1 in regulating gene expression. Our results suggest a hierarchical model in which Lin28A first recognizes DNA consensus sequences within active transcriptional bubbles and recruits Tet1 to regulate gene expression by modulating cytosine modification states. Our study unravels the unique chromatin-association features of Lin28A on a genome-wide scale, and further links Lin28A to epigenetic DNA modifications and associated gene regulation. Our findings have significant implications for understanding molecular mechanisms underlying versatile roles of Lin28A in stem cell pluripotency, differentiation and transformation.
Lin28A protein contains a Cold Shock Domain, one of the most ancient nucleic acid binding domains with the ability to interact with a wide range of single-stranded RNA and DNA (Hudson and Ortlund, 2014). Consistent with this idea, xtrLin28B CSD has been shown to preferentially bind to single-stranded pyrimidine-rich oligonucleotides of both DNA and RNA with up to eight bases in vitro (Mayr et al., 2012). Our in vivo ChIP-seq analyses identified the consensus binding sequences (“CAGnACC”-nn-“GGACAG”) for Lin28A in mESCs, which are highly similar to the previously identified “GGAG(A)” motif found in Lin28A-bound pre-miRNA and mRNA. These results suggest a shared binding mechanism of Lin28A to nucleic acids, which makes it challenging to distinguish the RNA-binding activity from its DNA-binding activity via mutation analysis. Notably, Lin28A is not the only protein that possesses dual binding affinity to DNA and RNA. For example, recent studies have implicated a transcriptional regulatory role for RNA-binding proteins (Mo et al., 2013; Yue et al., 2010) as well as a functional link between DNA methyltransferase Dnmt1 and RNA (Di Ruscio et al., 2013). Together, these results argue for reconsidering the traditional boundary between RNA- and DNA-binding proteins and it will be interesting to examine other classical RNA-binding proteins for their potential DNA-associated function.
Our results also reveal a molecular mechanism targeting Tet1 to specific genomic loci for epigenetic regulation. Although Tet1 possesses an N-terminal CXXC structure typical for DNA binding, this domain does not seem to recognize specific target gene sequences for Tet1 (Frauer et al., 2011). Previously identified Tet1-interacting proteins, such as Mbd3, PcG, Sin3A and OGT, mostly lack a specific DNA-recognition capacity, making them more “functional” partners than “recruiters” of Tet1 (Wu and Zhang, 2014). Lin28A represents a qualitatively different type of Tet1-associated proteins, with an ability to bind active chromatin through consensus sequences and guide Tet1 in targeting specific genomic loci to regulate gene expression. After recruiting Tet1 onsite, Lin28A could help Tet1 maintain a hypomethylated state of active gene promoters or, alternatively, Tet1 may further proceed along the gene body for 5mC to 5hmC conversion while Lin28A remains at the promoter to function primarily as a recruiter. Since no direct protein-protein interaction between Lin28A and Tet1 was detected in the GST pulldown assay (data not shown), intermediate molecules might combine with these two to form the protein complex. In addition, it is possible that some Tet1 could form a complex with Lin28 before being recruited to target transcription loci. Analogous to our findings for Tet1, IDAX recognizes unmethylated CpG and interacts with the Tet2 catalytic domain to recruit Tet2 onsite (Ko et al., 2013). It is likely that there are multiple mechanisms to recruit Tet1 to gene targets and more factors to be identified.
In summary, our study links the well-known RNA-binding protein Lin28A to epigenetic DNA modifications and establishes a molecular mechanism targeting Tet1 to specific genomic loci for epigenetic regulation. Our findings provide a mechanistic context to understand the role of Lin28A in the regulation of diverse biological processes, and a direction for future investigation.
EXPERIMENTAL PROCEDURES
Cell Cultures
The E14 mESC line was cultured on MEF feeder cells (Ma et al., 2008). mESCs were transfected by electroporation using Nucleofector® Kit from Lonza (VPH-1001). Lentiviruses were generated as described (Duan et al., 2007). mESCs were infected with the lentiviral supernatant and selected by 3 μg/ml puromycin for 3 days.
RNA Extraction, Quantitative-RT-PCR and Genomic DNA Extraction
Total RNA was extracted from mESCs by RNeasy (Qiagen), and 1 μg total RNA was primed with Oligo(dT)20 primers (Invitrogen) for cDNA synthesis, followed by Q-PCR analysis (Guo et al., 2011). Total genomic DNA of mESCs was extracted by Qiagen DNeasy Blood and Tissue Kit (Guo et al., 2011).
Protein Co-immunoprecipitation and Chromatin Immunoprecipitation (ChIP)
Co-immunoprecipitation was carried out with antibodies to FLAG (M2, Sigma), V5 (Invitrogen), Lin28A (Abcam) or Tet1 (Millipore) as described (Kim et al., 2009). Five million mESCs were used for Chip following procedures as described (Ma et al., 2009) with antibodies to FLAG (M2, Sigma), HA (Sigma), Lin28A (Abcam) or Tet1 (Millipore), or normal mouse or Rabbit IgG (Millipore) as a control.
Electrophoretic Mobility Shift Assay (EMSA)
Each binding reaction was carried out with 100 fmol of biotinylated dsDNA probe and 1 pmol of purified protein (Guo et al., 2014). Human Lin28A protein was purified as GST-His6 fusion proteins from yeast (Zhu et al., 2001). Control DNA is a 60 bp biotin end-labeled EBNA duplex in the Pierce kit. Sequences of 5’-biotinylated DNA probes are: Motif1 ssDNA (sense), 5’-CTTCTGGCGCTGTCCATGGTGCTGAACCCAG-3’, antisense 5’-CTGGGTTCAGCACCATGGACAGCGCCAGAAG-3’; Motif2 ssDNA (sense), 5’-GAA ACTGTCCGTGGTGCTGAAACACAGCAGCA-3’, antisense 5’-TGCTGCTGTGTTTCAGCACCACGGACAGTTTC-3’. dsDNA was made by annealing sense strand and antisense strand ssDNA. Mismatch duplex of motif1 was constructed by annealing sense strand with mutant antisense strand (5’-CTGGGTTCAGCACCATTTTTTGCGCCAGAAG-3’); bubble duplex of motif1 was made by annealing mutant sense strand (5’-CTTCTGGCGCTGTCCATCACAAGGTGCTGAACCCAG-3’) with mutant antisense strand (5’-CTGGGTTCAGCACCATTTTTTGCGCCAGAAG-3’).
KD Measurement by Octet® QKe system
We followed the manufacturer’s protocol of Octet® QKe systems (FortéBIO) for real-time, label-free analysis of affinity, kinetics and concentration from biomolecular interactions in 96 microplates. Data analysis software of the OCTET QK system was used to extract association and dissociation curves for Lin28A protein and fit curves by 1:1 model to obtain the KD for each condition.
5hmC-specific Enrichment and Methylated DNA Immunoprecipitation (MeDIP)
Five μg of genomic DNA were used for selective chemical labeling and enrichment for 5hmC-containing DNA (Song et al., 2011). MeDIP experiments were performed according to the manufacturer’s protocol (Active Motif).
Library Preparation and High-throughput Sequencing
Enriched DNA from ChIP, 5hmC-capture and MeDIP was subjected to library construction using the NEBNext ChIP-Seq Library Prep Reagent Set for Illumina according the manufacturer’s protocol. 50-cycle single-end sequencings were performed using Illumina HISeq 2000. RNA-seq libraries were generated from duplicated samples per condition using the Illumina TruSeq RNA Sample Preparation Kit v2 following manufacturer’s protocol. The RNA-seq libraries were sequenced as 50-cycle pair-end runs using Illumina HISeq 2000.
Bioinformatics
Bioinformatics analyses for ChIP-seq, 5hmC-seq and MeDIP-seq followed methods described (Szulwach et al., 2011; Yao et al., 2014). Annotation and motif analysis were performed by Hypergeometric Optimization of Motif EnRichment (HOMER) v4.7 (http://homer.salk.edu/homer/) (Heinz et al., 2010) or MEME suite 4.10.1 (http://meme-suite.org/tools/meme) (Bailey et al., 2009). Acute transcription was further evaluated using iRNA-seq package (Madsen et al., 2015).
Quantification of 5mC/5hmC by Glucosylation and HpaII/MspI Digestion
Two μg of gDNA were treated with Epimark 5hmC and 5mC Analysis Kit (New England Biolabs) according to the manufacturer’s protocol.
Supplementary Material
AKNOWLEDGEMENTS
We thank Mingjie Zhang, Kimberly Christian and members of Song, Ming and Jin laboratories for discussion, Shaohui Hu for suggestions on EMSA experiments, and L. Liu and Y. Cai for technical support. This work was supported by grants from NIH (NS047344 and MH087874) and MSCRF to H.S.; from NIH (NS079625 and HD073162) to P.J; from MSCRF and NIH (NS048271 and MH105128) and MSCRF to G.-l.M.; by postdoctoral fellowships from MSCRF to Y.Z., N.S.K.; by a postdoctoral fellowship from the National Ataxia Foundation to B.Y.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBER
The accession number for Lin28 ChIP-seq, RNA-seq, 5mC and 5hmC-seq data reported in this study is GEO: GSE74254.
SUPPLEMENTARY INFORMATION
Supplementary information for this article includes supplementary experimental procedures, four figures and two tables.
AUTHORS CONTRIBUTION
Y.Z., B.Y., J.S., P.J. and H.S. conceived the project, L.L. conducted 5hmC-capture and MeDIP experiments, and constructed sequencing libraries. B.Y. performed bioinformatic analyses. Y.Z. made FLAG-Lin28A-expressing and Lin28A KD mESC lines, and performed EMSA experiments. B.Y and Y.Z. performed ChIP experiments. N.S.K provided HA-Tet1 mESCs. Y.Z. and N.S.K performed IP-Westernblot experiments. Q.S. provided Lin28A recombinant protein. S.L. and Q.S. performed KD measurement. X.C. conducted Lin28-DNA structure modeling and provided recombinant Tet1-CD protein. H.W. performed statistic analyses. S.H., H.Z. J.Q. and J.W. assisted with EMSA and protein microarray experiments. Y.Z., B.Y., G-l.M., P.J. and H.S. wrote the manuscript. All authors discussed results and commented on the manuscript.
REFERENCES
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic acids research. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J, Ha M, Kim YK, Kim VN. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012;151:765–777. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, Liu XB, Yang CH, Jordan JD, Ma DK, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauer C, Rottach A, Meilinger D, Bultmann S, Fellinger K, Hasenoder S, Wang M, Qin W, Soding J, Spada F, et al. Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. PLoS One. 2011;6:e16627. doi: 10.1371/journal.pone.0016627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, Zhong C, Hu S, Le T, Fan G, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2014;17:215–222. doi: 10.1038/nn.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nature genetics. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, Woodard C, Wang H, Jeong JS, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J, Zepeda-Martinez JA, Pape UJ, Jacobsen SE, Peters B, et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2014;111:1361–1366. doi: 10.1073/pnas.1322921111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson WH, Ortlund EA. The structure, function and evolution of proteins that bind DNA and RNA. Nature reviews Molecular cell biology. 2014;15:749–760. doi: 10.1038/nrm3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara H, Okada Y, Imai T, Iwanami A, Mischel PS, Okano H. Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated let-7 family microRNA biogenesis in early neural differentiation. J Biol Chem. 2011;286:16121–16130. doi: 10.1074/jbc.M110.199166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Duan X, Liu CY, Jang MH, Guo JU, Pow-anpongkul N, Kang E, Song H, Ming GL. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63:761–773. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, Segal MF, Li H, Koh KP, Lahdesmaki H, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin FE, Gebert LF, Towbin H, Brunschweiger A, Hall J, Allain FH. Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat Struct Mol Biol. 2012;19:84–89. doi: 10.1038/nsmb.2202. [DOI] [PubMed] [Google Scholar]
- Ma DK, Chiang CH, Ponnusamy K, Ming GL, Song H. G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem cells. 2008;26:2131–2141. doi: 10.1634/stemcells.2008-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen JG, Schmidt SF, Larsen BD, Loft A, Nielsen R, Mandrup S. iRNA-seq: computational method for genome-wide assessment of acute transcriptional regulation from total RNA-seq data. Nucleic Acids Res. 2015;43:e40. doi: 10.1093/nar/gku1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr F, Schutz A, Doge N, Heinemann U. The Lin28 cold-shock domain remodels pre-let-7 microRNA. Nucleic Acids Res. 2012;40:7492–7506. doi: 10.1093/nar/gks355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo S, Ji X, Fu XD. Unique role of SRSF2 in transcription activation and diverse functions of the SR and hnRNP proteins in gene expression regulation. Transcription. 2013;4 doi: 10.4161/trns.26932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12:395–406. doi: 10.1016/j.stem.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, Pasquinelli AE. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol. 2011;18:302–308. doi: 10.1038/nsmb.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbert ML, Huelga SC, Kapeli K, Stark TJ, Liang TY, Chen SX, Yan BY, Nathanson JL, Hutt KR, Lovci MT, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell. 2012;48:195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, Barbera AJ, Zheng L, Zhang H, Huang S, et al. Genome-wide Regulation of 5hmC, 5mC, and Gene Expression by Tet1 Hydroxylase in Mouse Embryonic Stem Cells. Mol Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B, Lin L, Street RC, Zalewski ZA, Galloway JN, Wu H, Nelson DL, Jin P. Genome-wide alteration of 5-hydroxymethylcytosine in a mouse model of fragile X-associated tremor/ataxia syndrome. Human molecular genetics. 2014;23:1095–1107. doi: 10.1093/hmg/ddt504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue X, Schwartz JC, Chu Y, Younger ST, Gagnon KT, Elbashir S, Janowski BA, Corey DR. Transcriptional regulation by small RNAs at sequences downstream from 3' gene termini. Nat Chem Biol. 2010;6:621–629. doi: 10.1038/nchembio.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.