Abstract
The genus Flavivirus includes a number of newly recognized viruses that infect and replicate only within mosquitoes. To determine whether insect-specific flaviviruses (ISFs) may infect Culiseta (Cs.) melanura mosquitoes, we screened pools of field-collected mosquitoes for virus infection by RT-PCR targeting conserved regions of the NS5 gene. NS5 nucleotide sequences amplified from Cs. melanura pools were genetically similar to other ISFs and most closely matched Calbertado virus from Culex tarsalis, sharing 68.7% nucleotide and 76.1% amino acid sequence identity. The complete genome of one virus isolate was sequenced to reveal a primary open reading frame (ORF) encoding a viral polyprotein characteristic of the genus Flavivirus. Phylogenetic analysis showed that this virus represents a distinct evolutionary lineage that belongs to the classical ISF group. The virus was detected solely in Cs. melanura pools, occurred in sampled populations from Connecticut, New York, New Hampshire, and Maine, and infected both adult and larval stages of the mosquito. Maximum likelihood estimate infection rates (MLE-IR) were relatively stable in overwintering Cs. melanura larvae collected monthly from November of 2012 through May of 2013 (MLE-IR = 0.7–2.1/100 mosquitoes) and in host-seeking females collected weekly from June through October of 2013 (MLE-IR = 3.8–11.5/100 mosquitoes). Phylogenetic analysis of viral sequences revealed limited genetic variation that lacked obvious geographic structure among strains in the northeastern United States. This new virus is provisionally named Culiseta flavivirus on the basis of its host association with Cs. melanura.
Key Words: : Culiseta melanura, Flavivirus, Insect viruses, Mosquito-only flavivirus, Genomic analysis, Phylogenetic analysis
Introduction
The Flavivirus genus represents a diverse group of viruses that includes a number of important human pathogens, such as dengue virus, Japanese encephalitis virus, tick-borne encephalitis virus, and West Nile virus (WNV). These viruses are maintained in transmission cycles between arthropod vectors and vertebrate hosts, whereas other flaviviruses appear to be limited to infecting insects (Blitvich and Firth 2015). Cell fusing agent was the first insect-specific flavivirus (ISF) discovered 40 years ago from an Aedes (Ae.) aegypti cell line (Stollar and Thomas 1975). Since then, a multitude of ISFs have been isolated and characterized from a wide range of mosquito species worldwide. Some of these include Kamiti River virus (KRV) from Ae. macintoshi in Kenya (Crabtree et al. 2003), Culex flavivirus (CxFV) from Culex (Cx.) pipiens in Japan (Hoshino et al. 2007), Aedes flavivirus (AeFV) from Ae. albopictus and Ae. flavopictus in Japan (Hoshino et al. 2009), Calbertado virus (CLBOV) from Cx. tarsalis and Cx. pipiens in Canada and the United States (Bolling et al. 2011, Tyler et al. 2011), and Palm Creek virus (PCV) from Coquillettidia xanthogaster in Australia (Hobson-Peters et al. 2013). These ISFs form a monophyletic group and are distantly related to the dual-host mosquito-borne and tick-borne flaviviruses and to viruses with no known vector. However, more recently, a second group of ISF-like viruses has been isolated from mosquitoes that replicate solely within arthropod cells but cluster phylogenetically with the dual-host mosquito-borne flaviviruses. This group includes Lammi virus (Huhtamo et al. 2009), Nounane virus (Junglen et al. 2009), Barkedji virus (Kolodziejek et al. 2013), Chaoyang virus (Lee et al. 2013), and Nhumirim virus (Kenney et al. 2014).
CxFV represents the most extensively studied ISF and has been detected in multiple mosquito species from around the world. The virus was found to be highly prevalent in Cx. pipiens and Cx. quinquefasciatus sampled from East Asia, Africa, and the Americas (Hoshino et al. 2007, Morales-Betoulle et al. 2008, Blitvich et al. 2009, Cook et al. 2009, Kim et al. 2009, Farfan-Ale et al. 2010, Bolling et al. 2011, Newman et al. 2011, Machado et al. 2012), and it was infrequently detected in Cx. tarsalis from the United States (Blitvich et al. 2009, Bolling et al. 2011) and Cx. tritaeniorhynchus from Japan (Obara-Nagoya et al. 2013). CxFV appears to perpetuate in mosquito populations by vertical transmission, the passing of the virus from an infected female to her progeny (Saiyasombat et al. 2011, Bolling et al. 2012), and venereal transmission may also play a minor role in viral maintenance (Bolling et al. 2012). Evidence for vertical transmission has also been observed for other ISFs, including KRV (Lutomiah et al. 2007), AeFV (Haddow et al. 2013), and CLBOV (Bolling et al. 2011), on the basis of virus detection in immature mosquitoes.
ISFs may inhibit medically important flaviviruses from establishing infection in mosquitoes through “superinfection exclusion.” This phenomenon occurs when a viral infection establishes itself in a cell, and this infection interferes with the establishment of a secondary infection (Randolph and Hardy 1988, Zou et al. 2009). There are conflicting reports on CxFV-infected mosquitoes' vector competence for WNV. Laboratory studies have suggested that CxFV infection in Cx. pipiens may suppress a secondary infection of WNV up to 7 days postinfection (Bolling et al. 2012). However, prior infection of CxFV was not shown to impact WNV transmission rates in Cx. quinquefasciatus, and simultaneous co-infection actually increased the rate of WNV transmission (Kent et al. 2010). Observations from field studies also fail to support the concept of superinfection exclusion of WNV by CxFV (Newman et al. 2011, Crockett et al. 2012). Nonetheless, other studies in Australia have shown that PCV infection suppresses WNV and Murray Valley encephalitis virus replication in mosquito cell culture (Hobson-Peters et al. 2013). These studies show that although ISFs cannot infect humans, they may impact public health by suppressing or enhancing infection of pathogenic flaviviruses in the mosquito vector.
In this study, we describe a novel ISF infecting Culiseta (Cs.) melanura mosquitoes collected from the northeastern United States. Cs. melanura is a bird-biting mosquito species that serves as the main vector of eastern equine encephalitis virus (EEEV) and a secondary vector of WNV in eastern North America (Scott and Weaver 1989, Andreadis et al. 2004). To determine whether Cs. melanura may be infected by ISFs, we surveyed mosquito populations in the northeastern United States for flavivirus infection using generic and specific PCR primers. The virus was isolated in mosquito cell culture and detected by electron microscopy, and the complete genome was sequenced and compared to other flaviviruses by phylogenetic analysis. We provisionally named it Culiseta flavivirus (CsFV) and describe here the annual cycle of CsFV infection in Cs. melanura females and overwintering mosquito larvae.
Materials and Methods
Mosquito collections
Adult mosquitoes were collected in 91 trapping locations throughout Connecticut as part of the statewide mosquito and arborvirus surveillance program (Andreadis et al. 2004). A dry ice–baited CDC light trap and gravid trap baited with hay-infusion water were set overnight at each site on a weekly rotation from June through October. Additional Cs. melanura were collected in Cheshire, Hillsborough, and Rockingham Counties in New Hampshire and York County, Maine, during June, 2011, and in Oswego County, New York, during May, 2012. Mosquitoes were identified to species on a chill table under a dissecting microscope using diagnostic keys (Andreadis et al. 2005). Female mosquitoes from each trap were pooled by species with a maximum of 50 individuals per pool, and placed into 2.0-mL microcentrifuge tubes containing a copper BB. Pools were stored at −80°C until tested.
Virus detection and partial sequencing
In preparation for virus testing, 1.0 mL of phosphate-buffered saline (PBS) containing 0.5% gelatin, 30% rabbit serum, and 1× antibiotic + antimycotic was added to each mosquito pool. Mosquito pools were homogenized using a MM 300 Mixer-Mill (Retsch Laboratory, Irvine, CA) set at 25 cycles per second for 4 min. Samples were centrifuged at 4°C for 7 min at 7000 rpm, and RNA was extracted from a 70-μL aliquot of the supernatant using the Viral RNA Kit (Qiagen, Valencia, CA). RT-PCR was performed using the Titan One Tube RT-PCR System (Roche Diagnostics, Indianapolis, IN) in 25-μL reactions. Primer sequences are given in Table 1. Primer pairs FU2new/cFD3 and ISFfwd/CSFLAVrev were used to detect flavivirus infection initially in Cs. melanura by amplifying portions of the NS5 gene (Kuno et al. 1998). Amplification was as follows: 1 cycle of 50°C for 30 min and 94°C for 2 min, 10 cycles of 94°C for 15 s, 50°C for 30 s, and 68°C for 1 min, followed by 25 cycles of 94°C for 15 s, 50°C for 30 s, and 68°C for 1 min + 5 s per cycle, and 1 cycle of 68°C for 7 min. RT-PCR amplification products were purified using the QIAquick PCR Purification Kit (Qiagen, Valencia, CA) and sequenced at the DNA Analysis Facility (Yale University, New Haven, CT).
Table 1.
Primers Used for RT-PCR and Nucleotide Sequencing
| Primer name | Primer sequence (5′→3′) | Target gene, virus | Amplicon length (bp) | Used for |
|---|---|---|---|---|
| FU2new | GCCGATGACGTAGCCGGTTGGGATAC | NS5, Flavivirus genus | 888 | Sequencing |
| cFD3 | AGCATGTCTTCCGTGGTCATCCA | NS5, Flavivirus genus | ||
| ISFfwd | GGATTATCTGGTACATGTGG | NS5, ISFs | 518 | Sequencing |
| CSFLAVrev | TAAGCCAGGCGTCAATCTCT | NS5, CsFV | ||
| CSFLAVfwd | CCGTACCACAGGAAACTCGT | NS5, CsFV | 255 | Virus detection |
| CSFLAVrev | TAAGCCAGGCGTCAATCTCT | NS5, CsFV | ||
| CSLfwd | CGAGTACGAGGCTCTAGGGT | NS5, CsFV | 913 | Sequencing, virus |
| CSLrev | GACCAGTTGGCATCCACTCA | NS5, CsFV | confirmation |
ISF, insect-specific flavivirus; CsFV, Culiseta flavivirus.
To detect CsFV in all other mosquito pools, primers CSFLAVfwd and CSFLAVrev were used to target specifically and amplify a 255-nucleotide fragment of the NS5 gene of CsFV. The thermal cycling protocol was as follows: 1 cycle of 50°C for 30 min and 94°C for 2 min, 10 cycles of 94°C for 15 s, 58°C for 30 s, and 68°C for 30 s, followed by 25 cycles of 94°C for 15 s, 58°C for 30 s, and 68°C for 30 s + 5 s per cycle, and one cycle of 68°C for 7 min. Primer set CSLfwd/CSLrev was used to retest samples that had produced ambiguous or faint bands with CSFLAVfwd/CSFLAVrev primers and for sequencing and phylogenetic analyses of selected CsFV strains. These primers amplify a 913 bp portion of the NS5 gene under the following parameters: One cycle of 50°C for 30 min and 94°C for 2 min, 10 cycles of 94°C for 15 s, 60°C for 30 s, and 68°C for 1 min, followed by 25 cycles of 94°C for 15 s, 60°C for 30 s, and 68°C for 1 min + 5 s per cycle, and one cycle of 68°C for 7 min. Infection rates per 100 individuals were calculated as bias-corrected maximum likelihood estimates using the Excel Add-In PooledInfRate, version 4.0 (Biggerstaff 2006).
Virus isolation and propagation
Ae. albopictus (C6/36) cells were grown in 25-cm2 flasks containing minimum essential medium (MEM), 5% fetal bovine serum (FBS), 1× nonessential amino acids, and antibiotics + antimycotics, and held at 28°C, 5% CO2. African green monkey (Vero E6) cells and baby hamster kidney (BHK-21) cells were maintained in 25-cm2 flasks containing MEM, 5% FBS, and antibiotics + antimycotics at 37°C, 5% CO2. CsFV was initially isolated in C6/36 cell culture by inoculating 100 μL of mosquito pool homogenate onto confluent cells and by splitting cell cultures for three passages at a 1:5 dilution every 7 days. CsFV isolate 502-13, passage 3, was used to assess its ability to propagate in mosquito (C6/36) and mammalian (Vero E6 and BHK-21) cells by inoculating 100 μL of virus into each cell line in triplicate. Cell cultures were maintained for 7 days, and then 100 μL of the culture medium was used for each subsequent passage for a total of three passages.
Electron microscopy
Five-day-old C6/36 cell cultures previously inoculated with CsFV and cultures that were not inoculated (negative controls) were centrifuged, infused with a 5% agar solution, and fixed overnight at 4°C in a 2.5% (vol/vol) glutaraldehyde +2% paraformaldehyde solution containing 0.1% (wt/vol) CaCl2 and 1% (wt/vol) sucrose in 100 mM sodium cacodylate buffer (pH 7.4). Samples were postfixed in aqueous 1% (wt/vol) OsO4, dehydrated through an ascending ethanol and acetone series, and embedded in LX-112/Araldite (Ladd Research Industries, Williston, VT). Thin sections were poststained with 5% (wt/vol) uranyl acetate in 50% (vol/vol) methanol followed by Reynold's lead citrate and examined in a Zeiss EM 10C electron microscope at an accelerating voltage of 100 kV.
Genomic sequencing
Total RNA (15 μL) from virus-infected C6/36 cell culture supernatants (virus isolate 502-13) were prepared for whole genome sequencing, as previously described (Grubaugh et al. 2015). In brief, the samples were treated with DNase, amplified using the Ovation RNA-Seq System V2 (NuGEN, San Carlos, CA), sheared using the Covaris S2 Focused-ultrasonicator (Covaris, Woburn, MA), and prepared using the Ovation Ultralow Library Kit (NuGEN, San Carlos, CA). Agencourt RNAclean XP beads (Beckman Coulter, Brea, CA) were used for all purification steps. Finished libraries were analyzed for correct size distribution using the 2100 Bioanalyzer (Agilent, Santa Clara, CA) and were quantified using the SYBR Green I–based qPCR Library Quantification Kit (KAPA Biosystems, Wilmington, MA). Paired-end reads of 100 nucleotides were generated using the Illumina HiSeq 2500 platform at Beckman Coulter Genomics (Danvers, MA). De-multiplexed paired-end reads were assembled de novo using Trinity (Grabherr et al. 2011), and the putative viral contigs were aligned and edited manually using Geneious version 7.0.6 (Kearse et al. 2012). The reference-guided aligner MOSAIK (Lee et al. 2014) was used to verify the accuracy of the assembled viral genome.
Phylogenetic analysis
Sequence alignments were generated in MEGA 6.0 based on nucleotide or translated amino acid sequences. Full-length polyprotein amino acid sequences were aligned using the ClustalW algorithm. Ambiguous positions in the alignment were removed using Gblocks (Castresana 2000), and 1449 amino acid positions were retained in the final alignment. A second alignment was generated from an 863-nucleotide portion of the NS5 gene from 22 CsFV strains. The optimal substitution model for each dataset was identified after performing maximum likelihood fits of 24 different models in MEGA. Phylogenetic trees were reconstructed by maximum likelihood analysis in MEGA and support for individual nodes was obtained by performing 1000 bootstrap replicates.
Results
Virus isolation and characterization
To determine whether Cs. melanura mosquitoes may be infected by ISFs, we initially screened seven pools consisting of 249 mosquitoes collected in Connecticut using PCR primers (FU2new and cFD3) targeting a highly conserved region of the Flavivirus NS5 gene. Three Cs. melanura pools yielded bands of the expected size (888 bp). The amplification product from one pool was sequenced to design new primers (ISFfwd and CSFLAVrev) to amplify an overlapping region of the NS5 gene of 518 bp. A partial NS5 nucleotide sequence (960 bp) was compared to sequences on GenBank and most closely matched CLBOV from Cx. tarsalis, sharing 68.7% nucleotide and 76.1% amino acid sequence identity. This level of sequence divergence is consistent with distinct viral species for the Flavivirus genus (Kuno et al. 1998). The virus was provisionally named Culiseta flavivirus (CsFV) on the basis of its host association with Cs. melanura.
To isolate and propagate CsFV, positive mosquito pools were inoculated into C6/36 cell cultures and blind-passaged three times. Two virus isolates (50-13 and 502-13) were identified when screened by RT-PCR using virus-specific primers (CSFLAVfwd and CSFLAVrev); however, cytopathic effect was not apparent in virus-positive cell cultures. CsFV isolate 50-13 was then inoculated and serial passaged in mosquito (C6/36) and mammalian (Vero E6 and BHK-21) cell lines. CsFV appeared to replicate only within the mosquito cell line. CsFV RNA was detected by RT-PCR after three passages in C6/36 cells, but was not detected after the second and third passage in Vero and BHK cells.
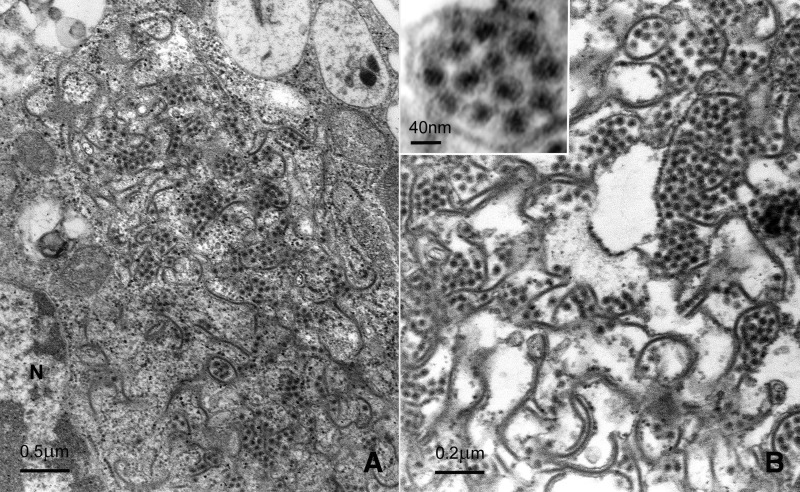
When examined by transmission electron microscopy, infected C6/36 cell cultures revealed spherical virus particles surrounded by a faint outer envelope membrane measuring 40 nm in diameter. These were located within the cisternae of the endoplasmic reticulum (Fig. 1A) and were not observed in noninoculated cell cultures. Naked nucleocapsid precursors were not detected. Dense accumulations of mature virus particles associated with distended cisternae endoplasmic reticulum characterized by intercisternal invaginations of tubular membranes were further seen in disrupted cells (Fig. 1B). These characteristics are consistent with insect cell lines infected with a flavivirus (Grimley 1991).
FIG. 1.
Section of 5-day-old C6/36 cell cultures infected with CsFV. (A) Spherical virus particles associated with cisternae of the endoplasmic reticulum. N, cell nucleus. Magnification, 31,000 ×. (B) Dense accumulations of mature virus particles associated with distended cisternae of endoplasmic reticulum and intercisternal tubular membranes. Magnification 48,000 ×. (Inset) Higher magnification of mature enveloped virions. Magnfication, 155,000 ×.
Genomic sequence and phylogenetic analysis
The complete genome of CxFV was sequenced and assembled (GenBank acc. no. KT599442). The sequence was based on a total of 6.8 million reads, of which 58.8% aligned to CsFV. The genome consists of 10,864 nucleotides with a primary open reading frame (ORF) from nucleotides 92 to 10,282 that encodes a 3396-amino acid polyprotein. The CsFV coding region was most similar to that of Naikwogo virus, PCV, and CxFV (49.1–49.4% nucleotide identity); however, the complete polyprotein sequence of the aforementioned CLBOV was not included in this analysis because it was not available on GenBank. Common to other ISFs, a “slippery” programmed −1 ribosomal frameshift motif (GGAUUUU) was detected at nucleotide position 3343 near the expected NS1 and NS2A junction (Blitvich and Firth 2015). This generated a second predicted overlapping ORF encoding a novel 247-amino-acid protein, previously termed fifo, from nucleotides 3508 to 4248 (Firth et al. 2010).
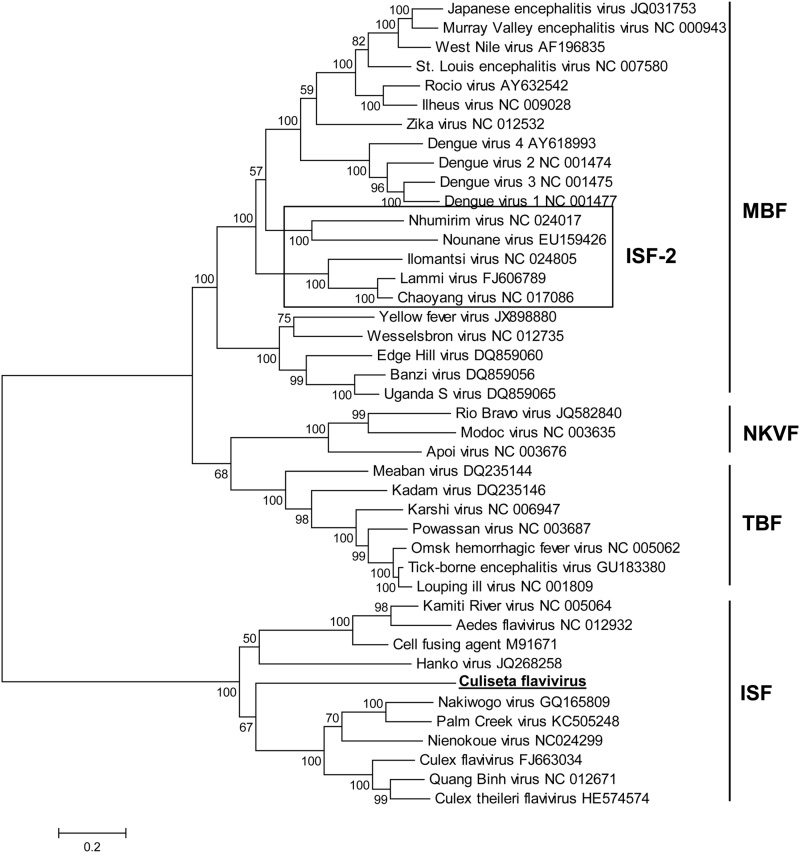
Phylogenetic relationships of 42 flaviviruses were evaluated by maximum likelihood analysis of polyprotein amino acid sequences, and the resulting phylogenetic tree is shown in Figure 2. The overall topology of the tree was similar to that of other analyses, with viruses clustering into groups defined by host range—ISFs, tick-borne flaviviruses, mosquito-borne flaviviruses, and no known vector flaviviruses (Cook et al. 2012). The recently discovered ISF-like viruses (ISF-2) fell within the mosquito-borne flavivirus group, as previously described (Kenney et al. 2014, Blitvich and Firth 2015). CsFV belonged to the classical ISF group but represents a distinct evolutionary lineage that grouped with ISFs associated with Culex, Mansonia, and Coquillettidia spp. mosquitoes (Nakiwogoa virus, PCV, Nienokoue virus, CxFV, Quang Binh virus, and Culex theileri flavivirus).
FIG. 2.
Maximum likelihood tree based on polyprotein amino acid sequences. Labels include virus names and corresponding GenBank accession numbers. Numbers at nodes indicate bootstrap support values. MBF, mosquito-borne flaviviruses; NKVF, no known vector flaviviruses; TBF, tick-borne flaviviruses; ISF, insect-specific flaviviruses.
Prevalence of CsFV in field-collected mosquitoes
To evaluate the host range of CsFV in field-collected mosquitoes, we tested 3620 mosquitoes representing 20 species that were collected in Connecticut from 2012 to 2013 (Table 2). CsFV was detected in 24 pools of Cs. melanura (n = 584, 33 pools) by RT-PCR, and the maximum likelihood estimate-infection rate (MLE-IR) was 9.2/100 mosquitoes. All other mosquito species were negative by RT-PCR. CsFV was then detected in additional pools of Cs. melanura collected from Maine, New Hampshire, and New York (Table 3). The MLE-IR was similar among states and ranged from 8.7 to 9.6/100 mosquitoes. CsFV was detected in male and female mosquitoes from New York and both sexes yielded similar MLE-IRs (9.4–9.5/100 mosquitoes). Culiseta virus was detected solely in Cs. melanura mosquitoes and was found in all of the northeastern states surveyed.
Table 2.
Detection of Culiseta Flavivirus in Female mosquitoes Collected in Connecticut 2012–2013
| Mosquito species | No. mosquitoes | No. mosquito pools | No. pools positive | MLE-IR/100 mosquitoes (95% CI) |
|---|---|---|---|---|
| Aedes cinereus | 182 | 7 | 0 | |
| Aedes vexans | 421 | 9 | 0 | |
| Anopheles punctipennis | 153 | 5 | 0 | |
| Anopheles walker | 115 | 4 | 0 | |
| Coquillettidia perturbans | 239 | 8 | 0 | |
| Culex pipiens | 366 | 20 | 0 | |
| Culex salinarius | 272 | 6 | 0 | |
| Culex restuans | 124 | 6 | 0 | |
| Culiseta melanura | 584 | 33 | 24 | 9.2 (6.0–14.5) |
| Culiseta minnesotae | 5 | 5 | 0 | |
| Culiseta morsitans | 23 | 16 | 0 | |
| Ochlerotatus canadensis | 260 | 9 | 0 | |
| Ochlerotatus cantator | 16 | 1 | 0 | |
| Ochlerotatus japonicus | 15 | 2 | 0 | |
| Ochlerotatus sollicitans | 26 | 1 | 0 | |
| Ochlerotatus taeniorhynchus | 135 | 3 | 0 | |
| Ochlerotatus triseriatus | 128 | 4 | 0 | |
| Ochlerotatus trivittatus | 122 | 4 | 0 | |
| Psorophora ferox | 188 | 6 | 0 | |
| Uranotaenia sapphirina | 246 | 9 | 0 | |
| Total | 3620 | 158 | 24 |
Table 3.
Detection of Culiseta Flavivirus in Cs. melanura Collected from Maine, New Hampshire, and New York During 2011–2012
| State | Sex | No. mosquitoes | No. mosquito pools | No. pools positive | MLE-IR/100 mosquitoes (95% CI) |
|---|---|---|---|---|---|
| Maine | F | 164 | 12 | 8 | 9.6 (4.5–22.2) |
| New Hampshire | F | 251 | 26 | 12 | 8.7 (4.7–15.9) |
| New York | F | 90 | 9 | 6 | 9.5 (4.1–20.8) |
| New York | M | 102 | 11 | 7 | 9.4 (4.4–18.8) |
MLE-IR, maximum likelihood estimate-infection rate; CI, confidence interval.
Seasonal and spatial dynamics of CsFV in Connecticut
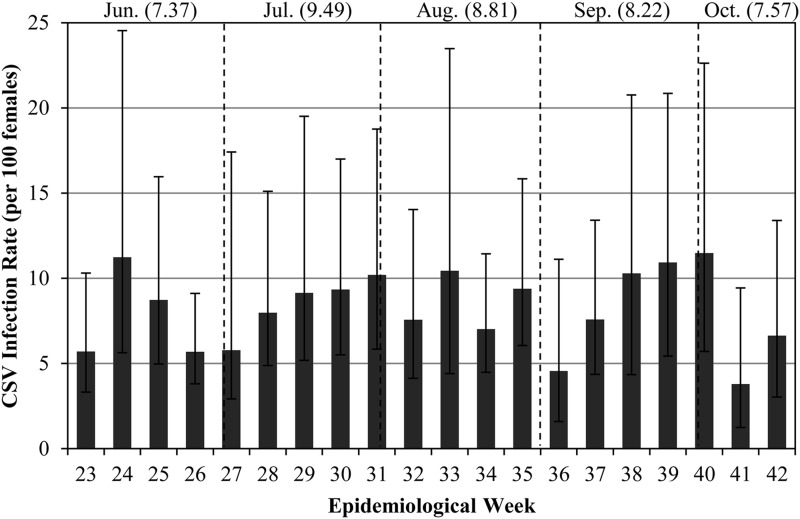
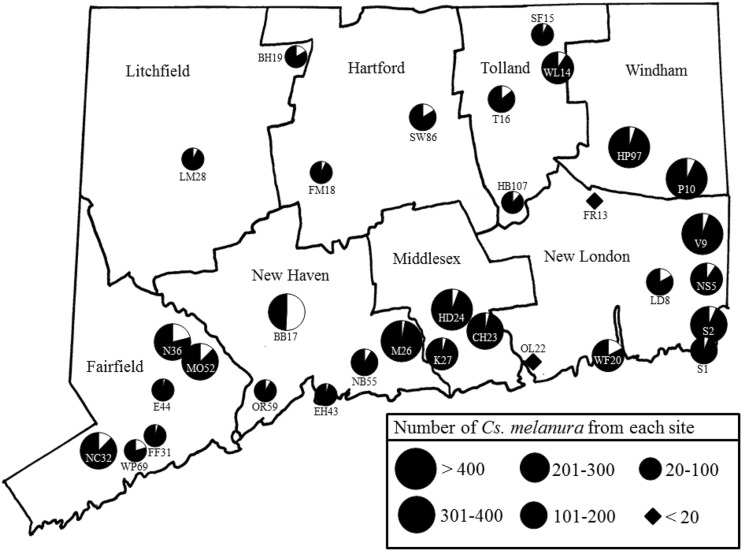
To characterize the seasonal and spatial dynamics of CsFV in Connecticut, additional Cs. melanura were collected and tested from 32 sites from June 3 through October 17, 2013. Each site was trapped on a 7- to 10-day rotation as part of the statewide mosquito surveillance program. A total of 7178 Cs. melanura females were processed as 474 mosquito pools for virus testing by RT-PCR, and 274 of these pools tested positive for CsFV. The MLE-IR for all of the tested Cs. melanura adults was 8.5 (per 100 mosquitoes). For the temporal and spatial analyses, two of 32 sites, OL22 and FR13, were omitted due to the low number of Cs. melanura trapped at each site (<20 mosquitoes). CsFV was detected during all 20 weeks of the study (Fig. 3), and although MLE-IRs fluctuated from week to week, they remained fairly stable, ranging from 3.8 to 11.5. In addition, the MLE-IR was calculated for each site and plotted on a map of Connecticut (Fig. 4). CsFV was present at all 30 sites, and the site infection rates ranged from 3.4 to 50.9.
FIG. 3.
Weekly infection rates of Culiseta flavivirus (CsFV) in Cs. melanura females collected at 30 sites in Connecticut between June and October of 2013. Error bars show 95% confidence intervals. Numbers in parentheses indicate infection rates for each month.
FIG. 4.
County map of Connecticut showing geographic location of Cs. melanura infected with Culiseta flavivirus (CsFV) during 2013. Pie charts show the estimated proportion of infected Cs. melanura (indicated by white sector) per 100 individuals at each site. Pie chart size indicates the total number of Cs. melanura trapped at each site during the study period. Infection rates are not shown for sites (OL22 and FR13) that produced fewer than 20 Cs. melanura.
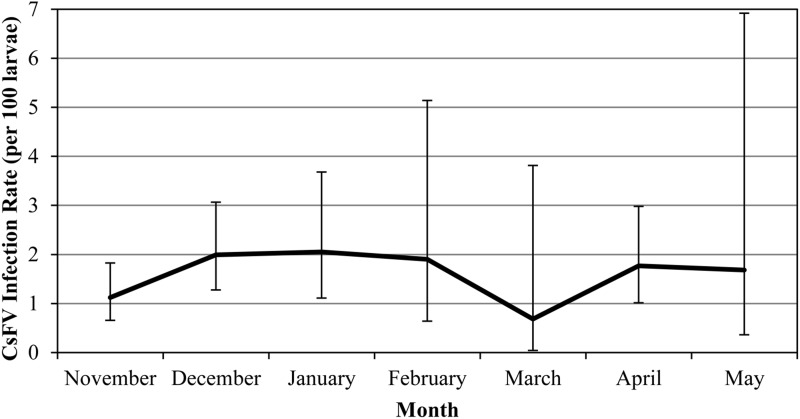
To investigate the overwintering portion of the annual virus cycle, pools of Cs. melanura larvae were collected from Chester, Connecticut (site CH23), during the collection period of November 6, 2012 to May 3, 2013. A total of 5822 mosquito larvae were processed as 144 pools for virus testing, and 69 of these pools tested positive for CsFV. The MLE-IR for all of the tested Cs. melanura larvae was 1.7 (per 100 larvae), and monthly infection rates were relatively stable throughout this time period, ranging from 0.7 to 2.1 (Fig. 5).
FIG. 5.
Monthly infection rates of Culiseta flavivirus (CsFV) in overwintering Cs. melanura larvae collected in Chester, Connecticut, between November of 2012 and May of 2013. Error bars indicate 95% confidence interval (CIs).
Genetic diversity of CsFV
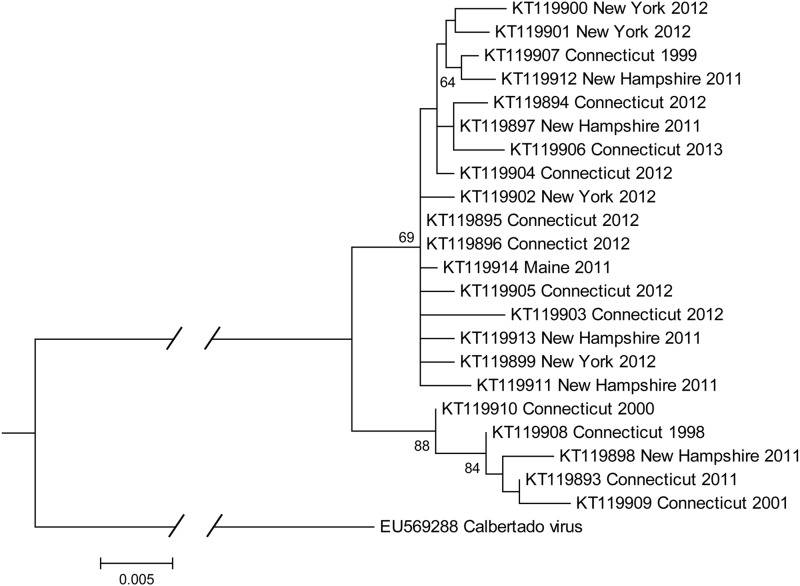
To evaluate the genetic relationships of CsFV strains circulating in the northeastern United States, we sequenced part of the NS5 gene (863 nucleotides) from 22 viral strains and compared these sequences by phylogenetic analysis (Fig. 6). We also included three CsFV strains that were detected retrospectively in historical pools of Cs. melanura collected during 1998–2001, in addition to more recent strains from Connecticut, Maine, New Hampshire, and New York. Mean nucleotide distances were 1.03% among all CsFV strains analyzed. Viruses segregated into two major clades by maximum likelihood analysis with 70% and 88% bootstrap support, but without distinct geographic or temporal patterns. Phylogenetic analysis indicates that viral genetic variation is limited in the northeastern United States and lacks obvious temporal and spatial definition.
FIG. 6.
Maximum likelihood tree of Culiseta flavivirus (CsFV) strains based on partial NS5 nucleotide sequences. Labels specify GenBank accession number, geographic origin, and year of virus isolation of virus strains. Numbers at nodes indicate bootstrap support values > 60%.
Discussion
In this study, we describe a new ISF, termed CsFV, that was detected solely in Cs. melanura mosquitoes and found in all northeastern US states surveyed. CsFV represents the first ISF known to infect mosquitoes of the genus Culiseta, indicating that these single-host flaviviruses occur in diverse mosquito genera. CsFV was successfully isolated and visualized in mosquito cells, but failed to propagate in mammalian cell lines. The genome comprised a primary ORF encoding a viral polyprotein characteristic of the genus Flavivirus and a second overlapping ORF expressing a novel gene fifo due to a ribosomal frameshift that is common to other ISFs. Our phylogenetic analyses showed that CsFV represents a distinct evolutionary lineage that belongs to the classical ISF group.
CsFV was found to be very prevalent in northeastern US populations of Cs. melanura. The estimated MLE-IR for pools of adult females was 8.7–9.6 individuals per 100 mosquitoes collected in Connecticut, Maine, New Hampshire, and New York. This infection rate is at the lower end of the range of annual MLE-IRs calculated for CxFV in Cx. pipiens from Colorado, which ran from 7.2 to 46.2, but greater than ranges of annual infection rates of CxFV in Cx. tarsalis, 0.08–0.13, and of CLBOV in both Cx. pipiens (0–0.3) and Cx. tarsalis (0.3–4.0) from Colorado (Bolling et al. 2011), and similar to MLE-IRs for CxFV infection in Culex pools from Illinois (10.0–11.2) (Newman et al. 2011). The estimated CsFV infection rate falls well within the range of infection rates for other ISFs described in the literature.
To characterize spatial and temporal patterns of CsFV infection, Cs. melanura females were collected and tested from 30 sites in Connecticut between June and October, 2013. During the study period, weekly MLE-IRs per 100 mosquitoes ranged from 3.8 to 11.5 (Fig. 3). Although there were fluctuations in the MLE-IRs from week to week, overlapping 95% confidence intervals (CIs) indicate that the infection rate did not differ significantly during the study period. This differs from the weekly infection rates observed for another flavivirus, WNV, which cycles between bird and mosquito populations. In Connecticut, WNV is typically absent in mosquito populations until early July, at which point it begins to increase in prevalence before peaking in August, and then declining into early fall (Andreadis et al. 2004). This suggests that CsFV does not undergo seasonal amplification via horizontal transmission, as WNV does, and that a different mechanism acts to maintain CsFV in Cs. melanura populations.
Geographically, CsFV was found at all test sites in Connecticut with a sample size of >20 Cs. melanura per site (Fig. 4). The calculated MLE-IRs at these sites ranged between 3.4 and 50.9 infected individuals per 100 mosquitoes. Similar infection rates were found throughout the state, with the exception of site BB17 (MLE-IR = 50.9/100 mosquitoes), which was more than twice the next highest infection rate. We cannot explain this result either by a low sample size or by characteristics of the mosquito trapping site. BB17 is characterized as a woodland depression bordered by a forested wetland complex that is typical for this region. CsFV appears to be widespread and was found at all sites with a sufficient sample of more than 20 Cs. melanura mosquitoes.
Monthly MLE-IRs were calculated for Cs. melanura larvae collected from Chester, Connecticut, between November, 2012, and May, 2013 (Fig. 5). As with the MLE-IRs of CsFV in adults, the monthly larvae infection rates were very stable. For the most part, the infection rate per 100 larvae ranged from 1.1 to 2.1. The only month when the MLE-IR was outside of this range was March of 2013 (MLE-IR = 0.7). However, this anomaly is likely due to the small sample size for that month. A large snowstorm in February interfered with collection efforts, and only four pools containing 148 larvae were collected in March, of which one pool tested positive for CsFV. The infection rate for CsFV in overwintering larval Cs. melanura appears to be stable over the winter months. CsFV detection in larvae suggests that vertical transmission is occurring, and stable infection rates suggest that horizontal transmission is not occurring between larvae during the overwintering period. This reinforces observations of other ISFs, which indicate that vertical transmission is the major mode of transmission (Lutomiah et al. 2007, Bolling et al. 2012).
An attempt was made to compare the MLE-IRs of overwintering larval Cs. melanura from Chester, Connecticut (1.7/100, 95% CI = 1.3–2.1), and adults trapped at the same site during the summer and fall of 2013 (4.0/100, 95% CI = 1.7–9.2). While a higher MLE-IR was calculated for the adult Cs. melanura, these differences were not significantly different on the basis of overlapping 95% CIs. Even if the larval and adult infection rates differed significantly, this is a difficult comparison to make because the larvae and adults were collected during different time periods. We attempted to collect Cs. melanura larvae during the fall of 2013, so that larvae and adults from the same location and time period could be compared, but this was prevented by extremely dry conditions.
Phylogenetic analyses were also performed to evaluate the genetic diversity and relationships of CsFV strains circulating in the northeastern United States (Fig. 6). Viral sequences proved to be highly conserved and grouped into two well-supported clades, but without evidence of spatial or temporal clustering. This indicates that CsFV has not undergone local isolation and differentiation among sampled locations in the northeastern United States. However, further testing of Cs. melanura from the mid-Atlantic and southeastern regions of the United States is needed to determine the prevalence of infection and phylogeography of this virus. CsFV could also potentially serve as a surrogate marker to identify geographically isolated Cs. melanura populations, given that this RNA virus likely mutates and differentiates faster than its mosquito host and appears to be stably maintained by vertical transmission. Future studies are warranted to evaluate the geographic range and population structure of CsFV.
In conclusion, we discovered and characterized CsFV that was found at high infection rates in Cs. melanura populations in the northeastern United States. Many of the findings, including the presence of the virus in larvae and the stable infection rates throughout the study period, suggest that CsFV is maintained in Cs. melanura populations through vertical transmission, and that it does not amplify through horizontal transmission. Studies need to be performed to determine whether the high prevalence of CsFV infection throughout Connecticut may be interfering with the transmission of WNV, a widespread flavivirus and human pathogen, through superinfection exclusion. Further studies are needed to determine the impact of CsFV on WNV infections in Cs. melanura.
Acknowledgments
We thank our support staff, specifically Angela Bransfield, John Shepard, and Michael Thomas, who provided technical assistance in the collection, identification, and testing of mosquitoes. This work was supported in part by grants from the Centers for Disease Control and Prevention (U50/CCU116806-01-1), the US Department of Agriculture Hatch Funds (CONH00773), the Multistate Research Project (NE1043), and the National Institutes of Health (AI067380).
Author Disclosure Statement
No competing financial interests exist.
References
- Andreadis TG, Anderson JF, Vossbrinck CR, Main AJ. Epidemiology of West Nile virus in Connecticut: A five-year analysis of mosquito data 1999–2003. Vector Borne Zoonotic Dis 2004; 4:360–378 [DOI] [PubMed] [Google Scholar]
- Andreadis TG, Thomas MC, Shepard JJ. Identification Guide to the Mosquitoes of Connecticut. Bulletin of The Connecticut Agricultural Experiment Station, No. 966, 2005:1–173 [Google Scholar]
- Biggerstaff BL. PooledInfRate, Version 3.0: A Microsoft Excel Add-In to Compute Prevalence Estimates from Pooled Samples. Centers for Disease Control and Prevention, Fort Collins, CO, 2006 [Google Scholar]
- Blitvich BJ, Firth AE. Insect-specific flaviviruses: A systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organization. Viruses 2015; 7:1927–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitvich BJ, Lin M, Dorman KS, Soto V, et al. Genomic sequence and phylogenetic analysis of Culex flavivirus, an insect-specific flavivirus, isolated from Culex pipiens (Diptera: Culicidae) in Iowa. J Med Entomol 2009; 46:934–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolling BG, Eisen L, Moore CG, Blair CD. Insect-specific flaviviruses from Culex mosquitoes in Colorado, with evidence of vertical transmission. Am J Trop Med Hyg 2011; 85:169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolling BG, Olea-Popelka FJ, Eisen L, Moore CG, et al. Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology 2012; 427:90–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540–552 [DOI] [PubMed] [Google Scholar]
- Cook S, Moureau G, Harbach RE, Mukwaya L, et al. Isolation of a novel species of flavivirus and a new strain of Culex flavivirus (Flaviviridae) from a natural mosquito population in Uganda. J Gen Virol 2009; 90:2669–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook S, Moureau G, Kitchen A, Gould EA, et al. Molecular evolution of the insect-specific flaviviruses. J Gen Virol 2012; 93:223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree MB, Sang RC, Stollar V, Dunster LM, et al. Genetic and phenotypic characterization of the newly described insect flavivirus, Kamiti River virus. Arch Virol 2003; 148:1095–1118 [DOI] [PubMed] [Google Scholar]
- Crockett RK, Burkhalter K, Mead D, Kelly R, et al. Culex flavivirus and West Nile virus in Culex quinquefasciatus populations in the southeastern United States. J Med Entomol 2012; 49:165–174 [DOI] [PubMed] [Google Scholar]
- Farfan-Ale JA, Lorono-Pino MA, Garcia-Rejon JE, Soto V, et al. Detection of flaviviruses and orthobunyaviruses in mosquitoes in the Yucatan Peninsula of Mexico in 2008. Vector Borne Zoonotic Dis 2010; 10:777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AE, Blitvich BJ, Wills NM, Miller CL, et al. Evidence for ribosomal frameshifting and a novel overlapping gene in the genomes of insect-specfic flaviviruses. Virology 2010; 399:153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 2011; 29:644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimley PM. Togaviridae and Flaviviridae. In: Adams JR, Bonami JR, eds. Atlas of Invertebrate Viruses. Boca Raton, FL: CRC Press, 1991:461–497. [Google Scholar]
- Grubaugh ND, Smith DR, Brackney DE, Bosco-Lauth AM, et al. Experimental evolution of an RNA virus in wild birds: evidence for host-dependent impacts on population structure and competitive fitness. PLoS Pathog 2015; 11:e1004874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddow AD, Guzman H, Popov VL, Wood TG, et al. First isolation of Aedes flavivirus in the Western Hemisphere and evidence of vertical transmission in the mosquito Aedes (Stegomyia) albopictus (Diptera: Culicidae). Virology 2013; 440:134–139 [DOI] [PubMed] [Google Scholar]
- Hobson-Peters J, Yam AW, Lu JW, Setoh YX, et al. A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS One 2013; 8:e56534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino K, Isawa H, Tsuda Y, Yano K, et al. Genetic characterization of a new insect flavivirus isolated from Culex pipiens mosquito in Japan. Virology 2007; 359:405–414 [DOI] [PubMed] [Google Scholar]
- Hoshino K, Isawa H, Tsuda Y, Sawabe K, et al. Isolation and characterization of a new insect flavivirus from Aedes albopictus and Aedes flavopictus mosquitoes in Japan. Virology 2009; 391:119–129 [DOI] [PubMed] [Google Scholar]
- Huhtamo E, Putkuri N, Kurkela S, Manni T, et al. Characterization of a novel flavivirus from mosquitoes in northern europe that is related to mosquito-borne flaviviruses of the tropics. J Virol 2009; 83:9532–9540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junglen S, Kopp A, Kurth A, Pauli G, et al. A new flavivirus and a new vector: Characterization of a novel flavivirus isolated from uranotaenia mosquitoes from a tropical rain forest. J Virol 2009; 83:4462–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012; 28:1647–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney JL, Solberg OD, Langevin SA, Brault AC. Characterization of a novel insect-specific flavivirus from Brazil: Potential for inhibition of infection of arthropod cells with medically important flaviviruses. J Gen Virol 2014; 95:2796–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent RJ, Crabtree MB, Miller BR. Transmission of West Nile virus by Culex quinquefasciatus say infected with Culex Flavivirus Izabal. PLoS Negl Trop Dis 2010; 4:e671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Guzman H, Bueno R, Dennett JA, et al. Characterization of Culex flavivirus (Flaviviridae) strains isolated from mosquitoes in the United States and Trinidad. Virology 2009; 386:154–159 [DOI] [PubMed] [Google Scholar]
- Kolodziejek J, Pachler K, Bin H, Mendelson E, et al. Barkedji virus, a novel mosquito- borne flavivirus identified in Culex perexiguus mosquitoes, Israel, 2011. J Gen Virol 2013; 94:2449–2457 [DOI] [PubMed] [Google Scholar]
- Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, et al. Phylogeny of the genus Flavivirus. J Virol 1998; 72:73–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Grubaugh ND, Kondig JP, Turell MJ, et al. Isolation and genomic characterization of Chaoyang virus strain ROK144 from Aedes vexans nipponii from the Republic of Korea. Virology 2013; 435:220–224 [DOI] [PubMed] [Google Scholar]
- Lee WP, Stromberg MP, Ward A, Stewart C, et al. MOSAIK: A hash-based algorithm for accurate next-generation sequencing short-read mapping. PLoS One 2014; 9:e90581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutomiah JJ, Mwandawiro C, Magambo J, Sang RC. Infection and vertical transmission of Kamiti River virus in laboratory bred Aedes aegypti mosquitoes. J Insect Sci 2007; 7:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado DC, Mondini A, dos Santos Santana V, Yonamine PT, et al. First identification of Culex flavivirus (Flaviviridae) in Brazil. Intervirology 2012; 55:475–483 [DOI] [PubMed] [Google Scholar]
- Morales-Betoulle ME, Monzon Pineda ML, Sosa SM, Panella N, et al. Culex flavivirus isolates from mosquitoes in Guatemala. J Med Entomol 2008; 45:1187–1190 [DOI] [PubMed] [Google Scholar]
- Newman CM, Cerutti F, Anderson TK, Hamer GL, et al. Culex flavivirus and West Nile virus mosquito coinfection and positive ecological association in Chicago, United States. Vector Borne Zoonotic Dis 2011; 11:1099–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara-Nagoya M, Yamauchi T, Watanabe M, Hasegawa S, et al. Ecological and genetic analyses of the complete genomes of Culex flavivirus strains isolated from Culex tritaeniorhynchus and Culex pipiens (Diptera: Culicidae) group mosquitoes. J Med Entomol 2013; 50:300–309 [DOI] [PubMed] [Google Scholar]
- Randolph VB, Hardy JL. Phenotypes of St Louis encephalitis virus mutants produced in persistently infected mosquito cell cultures. J Gen Virol 1988; 69:2199–2207 [DOI] [PubMed] [Google Scholar]
- Saiyasombat R, Bolling BG, Brault AC, Bartholomay LC, et al. Evidence of efficient transovarial transmission of Culex flavivirus by Culex pipiens (Diptera: Culicidae). J Med Entomol 2011; 48:1031–1038 [DOI] [PubMed] [Google Scholar]
- Scott TW, Weaver SC. Eastern equine encephalomyelitis virus: Epidemiology and evolution of mosquito transmission. Adv Virus Res 1989; 37:277–328 [DOI] [PubMed] [Google Scholar]
- Stollar V, Thomas VL. An agent in the Aedes aegypti cell line (Peleg) which causes fusion of Aedes albopictus cells. Virology 1975; 64:367–377 [DOI] [PubMed] [Google Scholar]
- Tyler S, Bolling BG, Blair CD, Brault AC, et al. Distribution and phylogenetic comparisons of a novel mosquito flavivirus sequence present in Culex tarsalis mosquitoes from western Canada with viruses isolated in California and Colorado. Am J Trop Med Hyg 2011; 85:162–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou G, Zhang B, Lim PY, Yuan Z, et al. Exclusion of West Nile virus superinfection through RNA replication. J Virol 2009; 83:11765–11776 [DOI] [PMC free article] [PubMed] [Google Scholar]