Abstract
Mutations in functionally constrained sites of the HIV envelope (Env) can affect entry efficiency and are potential targets for vaccine and drug design. We investigated Du151, a dual-infected individual with rapid disease progression. At her death 19 months postinfection (mpi), she was infected with a recombinant variant, which outgrew both parental viruses. We aimed to determine whether the recombinant virus had enhanced Env entry efficiency compared to the parental viruses and to identify the functional determinant. We generated 15 env clones at 1, 2, 8, and 19 mpi. Pseudovirus carrying a recombinant Env clone (PSV clone), C18 (19 mpi), had significantly higher entry efficiency compared to the parents, suggesting that the recombinant virus had enhanced fitness. To identify the functional determinant, we compared two recombinant PSV clones (C18 and C63)—differing in entry efficiency (2-fold) and by four and three amino acids in gp120 and gp41, respectively. The increased entry efficiency of a C18-gp41 PSV chimera indicated that the three amino acids in the C18 gp41 region were involved (K658, G671, and F717). Site-directed mutagenesis of the three amino acids of C63 showed that a single amino acid mutation, R658K, increased pseudovirion entry efficiency. The introduction of R658 into two PSV clones (C1 and C18) decreased their entry efficiency, suggesting that R658 carries a fitness cost. Thus, our data suggest that a recombinant virus emerged at 19 mpi with enhanced Env entry efficiency. Therefore, K658 in gp41 could in part be a contributing factor to the increased viral load and rapid disease progression of Du151.
Introduction
There is evidence to suggest that HIV-1 viral fitness increases during disease progression1–3 and a study comparing the relative fitness of viral isolates from long-term nonprogressors (LTNP) and rapid progressors demonstrated an association between disease progression and viral fitness.4 Several studies to date implicate the envelope (env) gene as playing a major role in the competitive ability of the virus.2,5–8 Dual infections, namely the infection of a single individual by two phylogenetically distinct variants, have been shown to be significantly associated with higher viral load at set point and rapid disease progression,9 particularly within the subtype C-infected Du cohort.10,11 However, recently, we have found this not to be the case in a second subtype C cohort from the same region (Centre for the AIDS Programme of Research in South Africa (CAPRISA 002).12
Dual-infected individuals harbor highly diverse viral populations providing an environment in which the virus has a much greater potential to evolve, by assembling through recombination, beneficial components from the different viral infections.13,14 Dual infections offer a unique opportunity to study phylogenetically distinct viral variants that evolve under the same selective pressures. Using intrasubtype C dual infection as a model system, we compared pseudovirus entry efficiency of Envelope (Env) clones representing viruses at 1, 2, 8, and 19 mpi to identify the regions or amino acid residues responsible for longitudinal changes in entry efficiency.
Du151 was identified as being coinfected at the first seropositive timepoint,10 and at the time of her death due to AIDS defining illness at 19 months mpi, she was infected with a recombinant variant, which outgrew both parental viruses. She was classified as a rapid progressor as her CD4 levels dropped to <200 cells/μl within 1.4 years and never controlled virus replication below 500,000 copies/ml.10 All of the study participants were antiretroviral (ARV) drug naive since treatment was not available in South Africa at the time of the study.
In a drug-naive patient, selective pressures on the virus are the result of both humoral and cellular immune responses5,8 directed toward the gag, pol, and env genes7,8 with immune evasion impacting HIV replicative fitness.15,16 As Env entry efficiency plays an important role in the overall replicative fitness of HIV2,5 and disease progression,17 we tracked the changes of Env function over the course of HIV infection of Du151.
Materials and Methods
A cohort of HIV-negative sex workers, including Du151, was recruited from five truck stops located along the main route between the port city of Durban and the commercial capital Johannesburg, South Africa. This cohort was established as part of a UNAIDS-funded Phase III vaginal microbicide trial, Col-149218 (ethics approval from the UCT Research Ethics Committee, number 137/95). To track the evolution of the variants infecting Du151, the C2–C3 region of env was amplified from plasma at 1, 2, 8, and 19 mpi by nested polymerase chain reaction (PCR) after limiting dilution of cDNA.11 The PCR products were cloned into the vector pGEM-T Easy vector using a TA cloning kit (Promega) and sequenced using an ABI PRISM dye terminator cycle-sequencing kit V3.1 (Applied Biosystems). Sequences were aligned using Clustal W (Bioedit) and neighbor-joining trees were constructed using the Kimura two-parameter model (Mega, version 5.1; Molecular Evolution Genetic Analysis).12 A heteroduplex tracking assay (HTA) was used to determine the relative frequency and fluctuation of the viral populations over the course of infection by combining population C2–C3 PCR products from each time point with a radiolabeled probe targeted to the C2–C3 region of the virus A identified at 1 mpi. Heteroduplex formation reactions were performed as described by Delwart and Gordon.19 The gels were dried and analyzed by using autoradiography with X-ray film followed by densitometry to determine the relative intensity of each of the bands.
Env cloning involved RNA extraction from either cell culture supernatants or plasma, RT-PCR using the Thermoscript RT-PCR System Kit (Invitrogen), limiting dilution of cDNA and then PCR of the env gene using primers Env 1A (5′ CAC CGG CTT AGG CAT CTC CTA TGG CAG GAA GAA 3′) and Env 1M (5′ TAG CCC TTC CAG TCC CCC CTT TTC TTT TA 3′) and Platinum Taq High Fidelity polymerase (Invitrogen, USA). The PCR product was ligated to the pCDNA3.1-TOPO vector (Invitrogen, USA). All clones were tested for function using a pseudovirion entry assay of TZM-bl cells20 and all functional env clones were sequenced.
Pseudovirus was prepared by cotransfecting HEK293T cells with the env clones and a subtype B HIV-1 backbone vector, pSG3.1Δenv (a gift from L. Morris, NICD; AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH; from Dr. John C. Kappes and Dr. Xiaoyun Wu) by using Polyfect Transfection Reagent (Qiagen, USA) at a ratio of 1:2.21 Cells were maintained in complete medium (10% fetal bovine serum, DMEM) for 48 h before the medium containing pseudovirions was harvested. Pseudovirions were lysed in 1% empigen/TBS and the p24 Gag concentration was determined using an ELISA (Vironostika HIV-1 Antigen kit, bioMérieux Clinical Diagnostics, France or Anti-HIV-1 p24-gag DIY ELISA kit, Aalto Biosystems, Ireland) and detected using the ELISA-Light Immunoassay kit with CDP-Star/SapphireII (Applied Biosystems, USA) on a GloMax-Multi Microplate Multimode Reader (Promega, USA; for the Aalto kit). TZM-bl cells (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH; from Dr. John C. Kappes, Dr. Xiaoyun Wu, and Tranzyme Inc.) were infected in triplicate with pseudovirus normalized for p24 Gag concentration (100 ng/ml) and 72 h postinfection luminescence (RLU) was measured using the Bright Glo Luciferase Assay System (Promega, USA).
Coreceptor tropism was determined by generating pseudovirus using a pNL4-lucR−E− backbone (NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: pNL4-3.Luc.R−.E− from Dr. Nathaniel Landau), normalizing pseudoviruses to 50 ng p24 and infecting U87-CD4-CCR5 and U87-CD4-CXCR4 in triplicate. The Env clones QHO22 and RPI23 were obtained from P. Moore (NICD, South Africa) and were used as positive controls for R5- and X4-tropic variants, respectively. Env chimeras were generated by digesting the env clones (C18 and C63) with BamHI and ClaI (Thermo Scientific/Fermentas, USA), which were located in the multiple cloning site of the pCDNA3.1-TOPO vector and the 3′ end of gp120 of both clones, respectively. Digestion and gel extraction of the DNA fragments followed by ligation swapped gp41 from one clone to the other and vice versa.
Specific amino acids were changed using a PCR-based strategy (modified Stratagene Quikchange II PCR-based protocol). PCR products were digested with DpnI (Thermo Scientific/Fermentas, USA), transformed into competent Escherichia coli JM109 cells (Promega, USA), and screened using restriction enzymes corresponding to the silent restriction sites introduced. The presence of the mutation, lack of any deleterious mutations, and preservation of the amino acid sequence were confirmed by sequencing of the env gene. Env incorporation into pseudoviral particles was measured by first ultracentrifugation of PSV through a sucrose cushion at 26,000×g for 2 h followed by Western blotting.
Results and Discussion
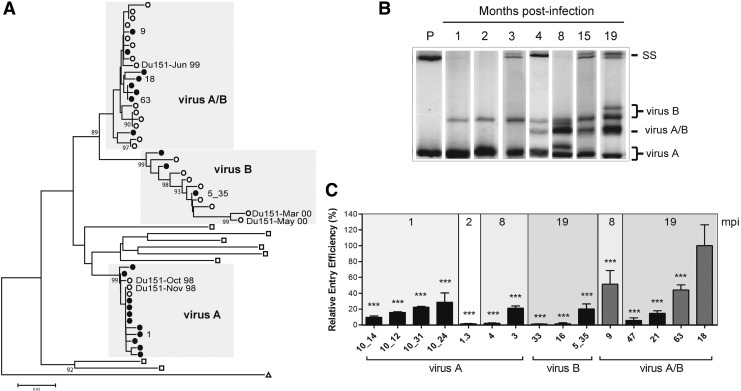
Sequence analysis of the env C2–C3 region amplified from plasma at 1, 2, 8, and 19 mpi demonstrated three distinct viral populations, with virus A and virus B present as phylogenetically distinct variants (Fig. 1A). We determined the relative abundance of these viral populations over the course of infection using HTA with a probe targeted to the C2–C3 region of virus A, 1 mpi. Mismatches between the probe and the PCR products resulted in heteroduplex formation resolving the viral population into variants with at least 3% diversity.24 Since the probe was identical to virus A, homoduplex formation indicated the level of virus A at each of the time points.
FIG. 1.
Genotypic and phenotypic analysis of the viruses infecting Du151. (A) Neighbor-joining tree of the C2–C3 region of the env Du151 clones to demonstrate that strains A and B are phylogenetically distinct. The neighbor-joining tree was generated using Mega543 with HXB2-LAI-IIIB-BRU (△) and those from the South African CAPRISA subtype C cohort44 (□) as a reference. Sequences included functional env clones (●) and published Du151 sequences (◯).29 Functional env clones mentioned in the text are labeled. (B) The virus A env C2–C3 region was used as a radiolabeled probe to track the emergence of genetic diverse strains over the course of infection (1, 2, 3, 4, 8, 15, and 19 mpi) using HTA analysis. SS, single stranded bands; P, probe. The parent viruses (A and B) and the recombinant virus (A/B) are indicated. (C) The 15 functional env genes were used to generate pseudovirions and tested for differences in entry efficiency using TZM-bl cells. Results were plotted as the percent entry efficiency of clone C18. Virus A/B clones C9, C63, and C18 are indicated in gray. Statistical analysis was performed using a one-way ANOVA with Tukey's posttest (***p<0.001 compared to C18) using GraphPad Prism 5 software.
To confirm the identity of each band at each time point, the population PCR products were cloned and HTA was carried out after colony PCR and sequencing (data not shown). When representative clone sequences were phylogenetically analyzed, they grouped with either virus A, B, or recombinant A/B, confirming the banding pattern of each variant (data not shown). Two parent viral populations (viruses A and B) were detected at 1 mpi (80% and 20% of the total population, respectively), confirming that Du151 was coinfected. These viruses remained as two separate env populations until 4 mpi when a third variant (virus A/B) was identified as a recombinant population between the two parental infecting strains (Fig. 1B).
To confirm that virus A/B was a recombinant of the parental strains, full-length env genes were cloned and neighbor-joining trees of the C2–C3 region indicated that the clones either grouped with virus A, B, or A/B. Recombinant analysis, carried out using the Recombination Detection Programme (RDP),25 confirmed that virus A/B was a recombinant of the two parental strains carrying most of the virus B sequence, including gp41, although the majority of A/B recombinant variants carried the V3 loop from virus A. The A/B recombinant dominated at 8 mpi and at 19 mpi, contributing to 40% of the total population, estimated from the HTA banding pattern (Fig. 1B). From the sequence analysis, the recombinant virus A/B predominated at 19 mpi, representing 58% of the functional env clones at this time point.
Recombinant viruses might have a fitness advantage over other variants,3,13,26 and the outgrowth of virus A/B at 19 mpi suggested that it could be a contributing factor to the high viral load and thus rapid disease progression of Du151. As env has been demonstrated to be a determinant of viral replicative fitness and disease progression,2,5–8 we cloned 22 functional env representing viruses A, B, and A/B over the course of infection. Fifteen representative clones from the four time points: 1 (n=4), 2 (n=1), 8 (n=3), and 19 (n=7) mpi representing virus A (n=7), virus B (n=3), and recombinant virus (A/B) (n=5) were selected for further analysis (Fig. 1A). TZM-bl cells were infected with equivalent p24-normalized (ng) pseudovirus to compare the difference in entry efficiency between parent and recombinant clones.
One recombinant PSV clone present at 19 mpi (clone C18) had significantly higher entry efficiency compared to A and B viruses at all time points (Fig. 1C) and a 2-fold increase compared to a second recombinant PSV clone at 19 mpi (C63), suggesting that virus A/B had higher entry efficiency that contributed to its outgrowth. The sequence of env clone C18 clustered on a neighbor-joining tree with clones C9 and C63 (Fig. 1A), all of which were classified as the recombinant virus A/B and all had higher pseudovirion entry efficiency compared to PSV clones representing the parent strains A and B at all time points (Fig. 1C), suggesting that recombination contributed to generating a fitter virus.
As X4 tropism has been linked to increased disease progression27,28 and Coetzer et al.29 had previously indicated that Du151 variants became dual tropic at 19 mpi, the coreceptor tropism of the functional PSV Env clones were tested using U87-CD4-CCR5 and U87-CD4-CXCR4 cells and the Env sequences were analyzed using the online program PSSM. The outcome of the two methods both indicated that only a single functional Env clone (C5_35) was dual tropic at 19 mpi and that the recombinant Env clones with high entry efficiency (C18 and C63) did not utilize CXCR4 (data not shown). The entry efficiency of the PSV clone C5_35 was significantly lower compared to C18 (Fig. 1C) and based on PSSM results, of a total of 19 available gp120 sequences, including that of the functional Env clones, 11 (58%) were R5 tropic at 19 mpi, suggesting that the outgrowth of the recombinant variant was not due to coreceptor tropism.
Sequences of Env clones that differed in pseudovirion entry efficiency were compared to determine the region/amino acid residue that contributed to differences in entry (Table 1). The two A/B PSV clones, C18 and C63, present at 19 mpi with a 2-fold difference in entry efficiency differed by only seven amino acids—four in gp120 and three in gp41 (Fig. 1C and Table 1). To determine which region was responsible for the difference in entry efficiency, C18-gp120 and C18-gp41 chimeras were constructed (Table 1). The C18-gp41 chimeric PSV clone had 2-fold higher entry efficiency than its counterpart, similar to the wild-type (wt) clone (Fig. 2), indicating that the three gp41amino acid residues played a role in the enhanced entry efficiency of the PSV clone, C18.
Table 1.
Sequences of Du151 Envelope Clones, Chimeras, and Mutants Showing gp41 Amino Acid Residues That Differ Between Clones C63 and C18
| mpi | Clone ID | Abbreviation | Virus | Position 658 | Frequencya | Position 671 | Frequency | Position 717 | Frequency |
|---|---|---|---|---|---|---|---|---|---|
| 1–2 | Du151_1A_C1 | C1 | A | QQEKNEKDLLALD | 12/12 | SWKNLWNWFNITN | 12/12 | GYSPLSFQTLTPS | 12/12 |
| 8 | Du151_8A_C9 | C9 | A/B | ……K…… | 6/6 | ……N..D… | 6/6 | ……F…..N | 1/6 |
| 19 | Du151_19AB_C63 | C63 | A/B | …Q..R…… | 1/11 | K.Q…S…… | 10/11 | ……L…..N | 6/11 |
| 19 | Du151_19AB_C18 | C18 | A/B | …Q..K…… | 10/11 | K.Q…G…… | 1/11 | ……F…..N | 5/11 |
| 19 | C63-gp120/C18-gp41 | C18-gp41 | A/B | …Q..K…… | K.Q…G…… | ……F…..N | |||
| 19 | C18-gp120/C63-gp41 | C18-gp120 | A/B | …Q..R…… | K.Q…S…… | ……L…..N | |||
| 1–2 | Du151_1A_C1_K658R | C1-K658R | A | QQEKNERDLLALD | SWKNLWNWFNITN | GYSPLSFQTLTPS | |||
| 8 | Du151_8A_C9_ K658R | C9-K658R | A/B | ……R…… | ……N.D.. | ……F…..N | |||
| 19 | Du151_19AB_C18_ K658R | C18-K658R | A/B | …Q..R…… | K.Q…G…… | ……F…..N | |||
| 19 | Du151_19AB_C63_ R658K | C63-R658K | A/B | …Q..K…… | K.Q…S…… | ……L…..N | |||
| 19 | Du151_19AB_C63_ S671G | C63-S671G | A/B | …Q..R…… | K.Q…G…… | ……L…..N | |||
| 19 | Du151_19AB_C63_ L717F | C63-L717F | A/B | …Q..R…… | K.Q…S…… | ……F…..N |
The frequency of each amino acid (highlighted in bold) is indicated as the number of clone sequences carrying that specific residue relative to total clones at each time point. The 2F5 (ALDSWKN) and 4E10 (NWFNIT) epitopes are underlined.
FIG. 2.
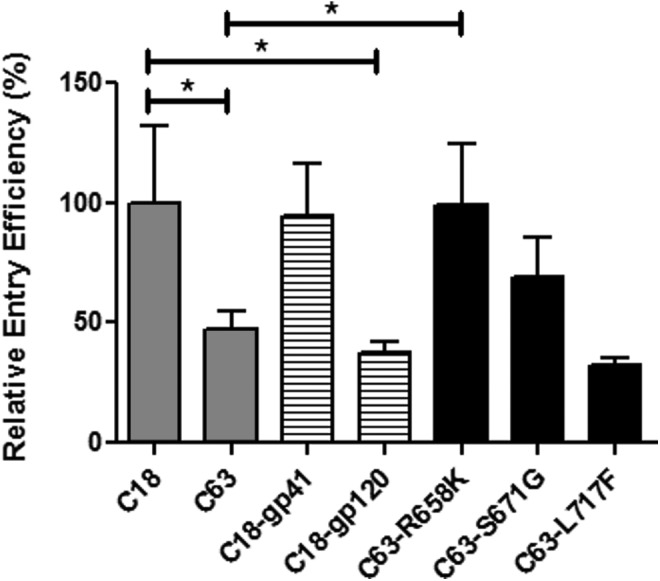
Identification of the determinant of Env entry efficiency. Comparison of entry efficiency between wt C18 and C63 (gray bars) with the chimeric Envs (C18-gp41 and C18-gp120, striped bars) and the site-directed mutants of C63 (R658K, S671G, and L717F, black bars). The entry efficiency of pseudoviruses normalized to 50 ng/ml p24 is displayed as a percentage of the entry efficiency of wt C18 (chimeras). This is a representative of three biological repeats with error bars indicating standard deviation of the mean of triplicate values. Statistical analysis was performed using an unpaired t-test (*p<0.05) with C63 and chimeras compared to C18 and site-directed mutants compared to C63 using GraphPad Prism 5 software.
The three gp41 amino acid differences between C63 and C18 comprise an R/K at position 658 (HXB2 numbering), an S/G at position 671, and an L/F at position 717 (Table 1). We determined the frequency of the amino acid residues at each of the positions in gp41 to identify evidence of selection over time (Table 1). There was a preference for lysine at position 658 and a transition from asparagine to serine at position 671 at 19 mpi, except with C18, which carried a glycine. Phenylalanine was the preferred residue at position 717 with leucine appearing to be under positive selection. These frequencies reflected those in subtype C sequences (n=1,459) from South Africa (www.hiv.lanl.gov). At position 658, lysine was predominant (87.5%) and arginine was present in 4.2% of sequences, suggesting positive selection for lysine. At position 671, asparagine predominated (62%) with serine in 32.5% and glycine present in two sequences (0.1%). Lastly, phenylalanine at position 717 was present in the majority of sequences (86.9%) with leucine in 14%.
To identify the functional determinant, we altered the three gp41 amino acids in C63 to the corresponding C18 residue using site-directed mutagenesis to generate C63-R658K, C63-S671G, and C63-L717F (Table 1). We performed a pseudovirion single round of infection assay on the three mutants to determine whether any one of the amino acids would rescue C63's low entry efficiency and raise it to that of C18 (Fig. 2). Mutant PSV clones C63-S671G and C63-L717F had no significant increase in entry efficiency compared to wt C63, whereas PSV clone C63-R658K had 2-fold increased entry (p=0.0268) and displayed activity similar to wt C18. This result suggests that K658 was responsible for the increased entry efficiency of the PSV clone C18 compared to C63.
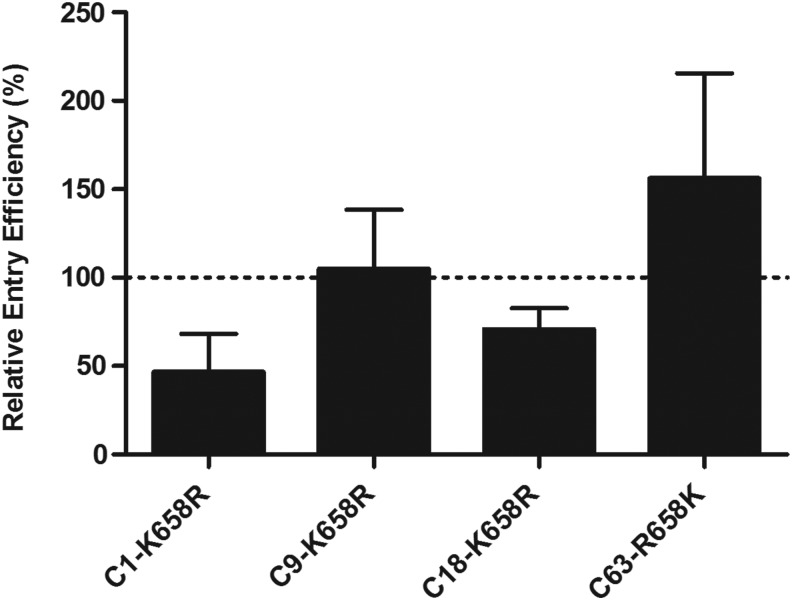
We next determined the importance of lysine at position 658 in Env entry efficiency over time and across clones with high genetic diversity compared with C18 (C9: 3.9% and C1: 6.8%). We introduced the R658 mutation into clones C1 (virus A, 1 mpi), C9 (virus A/B, 8 mpi), and C18 (virus A/B, 19 mpi) (Table 1, Fig. 3)—generating C1-K658R, C9-K658R, and C18-K658R, respectively—and compared their entry efficiency to wt. The K/R transition resulted in an observable, but not significant, decrease in viral entry in the pseudovirion assay compared to the wt C1 and C18 PSV clones (p=0.225 and p=0.0567, respectively), as expected if arginine at this position came with a fitness cost. K658R resulted in a 2-fold change in entry efficiency, which might not be sufficient to impact disease progression despite the association between Env function, replicative fitness,7 and disease progression.4 However, the entry efficiency of C18 was approximately 10-fold higher than clones at 1 mpi, suggesting that the enhanced entry of C18 could be due to K658 within the context of other amino acid changes.
FIG. 3.
Effect of K658 on entry efficiency. Comparison of entry efficiency of mutated clones from 1 mpi (C1-K658R), 8 mpi (C9-K658R), and 19 mpi (C18-K658R). The entry efficiency was determined using pseudoviruses normalized to 50 ng/ml p24 and displayed as a percentage of the entry efficiency of wt clones C1, C18, and C9. This is the mean of three biological repeats. Statistical analysis was performed using a paired t-test (with each mutant compared to the wt clone) using GraphPad Prism 5 software.
Brockman et al.30 showed that the well-known Gag TW10 T242N cytotoxic T lymphocyte (CTL) escape mutation, associated with slowed disease progression of B5801/B57-positive individuals, attenuated viral replication less than 2-fold in primary cells and this effect was reversed due to the presence of compensatory mutations. Although Gag is more structurally constrained than Env, the 10-fold increase in Env entry efficiency could have contributed to the rapid disease progression of Du151. Contrary to C18 and C1, K658R did not lower the entry efficiency of C9, supporting the suggestion that the effect of K658 on entry efficiency is dependent on the context of other amino acids.
The three amino acid changes at sites 658, 671, and 717 lie within the C-terminal heptad repeat (HR-2), the membrane proximal external region (MPER), and the cytoplasmic tail, respectively, of gp41, which are involved in fusion of the viral and cellular membranes during entry.31 Sivaraman et al.32 investigating two isolates from a longitudinal study of a rapid progressor with different fusogenic abilities, mapped the variation to the HR-2 region of gp41 known to be involved in membrane fusion.33,34 The R658K transition in our Du151 clones mapped to this region, suggesting that lysine at position 658 could play an important role in the fusogenic capacity of Env. When we compared the fusion capacity of the clones using a HEK293T cell-TZM-bl cell fusion assay similar to Sakamoto et al.,35 there was no difference between C18, C63, C63-R658K, and C18-K658R (data not shown). As expression and processing in env-transfected HEK293T cells could affect the fusion assay, we compared gp160 and gp120 levels by Western blot (data not shown). There was no difference in expression levels of gp160 and gp120 between the Env clones. Therefore, K658 might not play a role in viral–host membrane fusion despite its effect on Env entry efficiency.
As the entry efficiency assay relies on the incorporation of Env into viral particles and the cell–cell fusion assay is dependent on expression levels, it is possible that K658 could play a role in Env incorporation during viral budding. It has been suggested that both the cytoplasmic tail and the transmembrane domain of gp41 mediate Env incorporation into viral particles.36,37 To test whether R658 affected Env incorporation, C18, C63, and C63-R658K PSVs were concentrated and Western blotting was used to compare gp160 and gp120 levels after normalizing with p24. There was a slight, observable increase in incorporation of C18 gp120 compared to C63 (1.2-fold), possibly due to differences in cleavage of gp160 into gp120 (1.2-fold) (Fig. 4). C63-R658K also had only 20% higher gp120 incorporation relative to C63. Bachrach et al.38 showed that a slight increase in Env incorporation (2-fold) resulted in a 100-fold increase in infectivity. However, this association was dependent on the total number of Env incorporated because when Env density was further increased to 16-fold there was no concomitant increase in infectivity. Therefore, we cannot conclude that the slight increase in incorporation associated with the presence of lysine at position 658 is sufficient for the 2-fold difference in entry efficiency of C18 and C63.
FIG. 4.
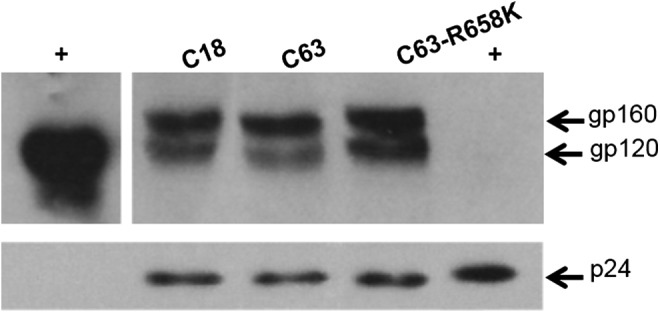
Pseudovirion incorporation of Env. Incorporation of Env was measured by ultracentrifugation of C18, C63, and C63-R658K pseudoviruses followed by Western blotting after normalizing p24 levels. The Western blot was probed with sheep anti-gp120 and rabbit ARP432 raised against gp120 and p24, respectively. The positive control (+) of gp120 was HIV-1 gp120 CM Envelope protein (Cat. #2968) and p24 was HIV-1 p24 IIB (Cat. #12028) (NIH Aids Reagent Program).
Another two longitudinal studies focused on the effect of mutations in the 2F5 and 4E10 epitopes of the MPER,39,40 which are adjacent to positions 658 and 671. In a comparative study of subtype C sequences from slow progressors and progressors, Archary et al.39 demonstrated changes in the 2F5 epitope from ALDKWQN to ALDSWKN from slow progressor to progressors where the primary residues that determine sensitivity to 2F5 are DKW.33,41 They suggest that there is a loss of this antibody site in the progressors. Interestingly, we observe the opposite in Du151 clonal sequences. The ALDSWKN motif is present in sequences at early time points (1–2 and 8 mpi) and switches to the 2F5-sensitive motif ALDKWQN at 19 mpi (Table 1). In addition, the N671S mutation described by Archary et al.39 in the 4E10 epitope (NWFNIT) corresponds to what we observed between clonal sequences at early time points and clone C63, where this changes to G in C18 (Table 1).
Zhang et al.40 compared subtype B sequences in drug-naive slow progressors and progressors and found that N671S and T676S mutations in the 4E10 epitope correlated with disease progression. However, this change does not seem to impact Env entry efficiency since we did not observe significant differences in the pseudovirion entry efficiency of PSV clones C63 and C63-S671G (Fig. 2). In agreement with these earlier studies, our data suggest that residues in gp41 are under positive selection during the course of infection. Moreover, the changes in PSV entry efficiency after site-directed mutagenesis highlight the role of this region in Env function (Fig. 2), which emphasizes the need for further investigation into the association between Env biological determinants and disease progression to inform vaccine design. Vaccines that target Env functional determinants might cause a loss of replicative fitness and lower viral loads, and thus lower the probability of HIV transmission.42
In this work, we sought to identify the functional determinant responsible for increased pseudovirion entry efficiency of Env clones isolated from the dual-infected individual, Du151. We located a residue within gp41 that was responsible for changes in entry efficiency of three of four Env clones tested, suggesting that sequence variation between the Env clones could compensate for the fitness cost of arginine at position 658. However, the presence of lysine at this position most likely contributed to the increased entry efficiency of PSV C18 and the outgrowth of the recombinant virus A/B at this stage of infection, suggesting that this increase in entry efficiency may have impacted viral loads and possibly led to increased disease progression of Du151.
GenBank Accession Numbers
The GenBank accession numbers for the 15 env sequences cloned in this study are KF146938.1–KF146952.
Acknowledgments
This work was funded by CAPRISA, the South African AIDS Vaccine Initiative (SAAVI), the Poliomyelitis Research Foundation (PRF), and the Medical Research Council (MRC). We thank Prof. Lynn Morris (National Institute for Communicable Diseases) for her generous gifts of pSG3.1Δenv and Du151 clones. Z.W. was funded by the Sydney Brenner Trust. K.G. was funded by the National Research Foundation and the Claude Leon Foundation.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Connor RI. and Ho DD: Human immunodeficiency virus type 1 variants with increased replicative capacity develop during the asymptomatic stage before disease progression. J Virol 1994;68(7):4400–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Troyer RM, Collins KR, Abraha A, et al. : Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. J Virol 2005;79(14):9006–9018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tebit DM, Nankya I, Arts EJ, and Gao Y: HIV diversity, recombination and disease progression: How does fitness “fit” into the puzzle? AIDS Rev 2010;9(2):75–87 [PubMed] [Google Scholar]
- 4.Quiñones-Mateu M, Ball S, Marozsan AJ, et al. : A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J Virol 2000;74(19):9222–9233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rangel HR, Weber J, Chakraborty B, et al. : Role of the human immunodeficiency virus type 1 envelope gene in viral fitness. J Virol 2003;77(16):9069–9073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Travers SAA, O'Connell MJ, McCormack GP, and McInerney JO: Evidence for heterogeneous selective pressures in the evolution of the env gene in different human immunodeficiency virus type 1 subtypes. J Virol 2005;79(3):1836–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ball SC, Abraha A, Collins KR, et al. : Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J Virol 2003;77(2):1021–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marozsan AJ, Moore DM, Lobritz MA, et al. : Differences in the fitness of two diverse wild-type human immunodeficiency virus type 1 isolates are related to the efficiency of cell binding and entry. J Virol 2005;79(11):7121–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sagar M, Lavreys L, Baeten JM, et al. : Infection with multiple human immunodeficiency virus type 1 variants is associated with faster disease progression. J Virol 2003;77(23):12921–12926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottlieb GS, Nickle DC, Jensen Ma, et al. : Dual HIV-1 infection associated with rapid disease progression. Lancet 2004;363(9409):619–622 [DOI] [PubMed] [Google Scholar]
- 11.Grobler J, Gray CM, Rademeyer C, et al. : Incidence of HIV-1 dual infection and its association with increased viral load set point in a cohort of HIV-1 subtype C-infected female sex workers. J Infect Dis 2004;190(7):1355–1359 [DOI] [PubMed] [Google Scholar]
- 12.Woodman Z, Mlisana K, Treurnicht F, et al. : Decreased incidence of dual infections in South African subtype C-infected women compared to a cohort ten years earlier. AIDS Res Hum Retroviruses 2011;27(11):1167–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S-L, Mittler JE, Nickle DC, et al. : Selection for human immunodeficiency virus type 1 recombinants in a patient with rapid progression to AIDS. J Virol 2002;76(21):10674–10684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramirez BC, Simon-Loriere E, Galetto R, and Negroni M: Implications of recombination for HIV diversity. Virus Res 2008;134(1–2):64–73 [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Picado J, Prado JG, Fry EE, et al. : Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol 2006;80(7):3617–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song H, Pavlicek JW, Cai F, et al. : Impact of immune escape mutations on HIV-1 fitness in the context of the cognate transmitted/founder genome. Retrovirology 2012;9(1):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lassen KG, Lobritz MA, Bailey JR, et al. : Elite suppressor-derived HIV-1 envelope glycoproteins exhibit reduced entry efficiency and kinetics. PLoS Pathog 2009;5(4):e1000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Damme L, Ramjee G, Alary M, et al. : Effectiveness of COL-1492, a nonoxynol-9 vaginal gel, on HIV-1 transmission in female sex workers: A randomised controlled trial. Lancet 2002;360(9338):971–977 [DOI] [PubMed] [Google Scholar]
- 19.Delwart EL. and Gordon CJ: Tracking changes in HIV-1 envelope quasispecies using DNA heteroduplex analysis. Methods 1997;12(4):348–354 [DOI] [PubMed] [Google Scholar]
- 20.Montefiori DC: Measuring HIV neutralization in a luciferase reporter gene assay. In: HIV Protocols (Prasad VR, Kalpana GV, eds.). Humana Press, Totowa, NJ, 2009, pp. 395–405 [DOI] [PubMed] [Google Scholar]
- 21.Montefiori D: Preparation and titration of HIV-1 Env-pseudovirus [Internet]. Cited March 24, 2013. Available from www.hiv.lanl.gov/content/nab-reference-strains/html/Protocol-Preparation-and-Titration-of-HIV-1-Env-Pseuodoviruses-July-2008.pdf
- 22.Li M, Gao F, Mascola JR, et al. : Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 2005;79(16):10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cilliers T, Moore P, Coetzer M, and Morris L: In vitro generation of HIV type 1 subtype C isolates resistant to enfuvirtide. AIDS Res Hum Retroviruses 2005;21(9):776–783 [DOI] [PubMed] [Google Scholar]
- 24.Resch W, Parkin N, Stuelke EL, et al. : A multiple-site-specific heteroduplex tracking assay as a tool for the study of viral population dynamics. Proc Natl Acad Sci USA 2001;98(1):176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin DP, Lemey P, Lott M, et al. : RDP3: A flexible and fast computer program for analyzing recombination. Bioinformatics 2010;26(19):2462–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van der Kuyl AC. and Cornelissen M: Identifying HIV-1 dual infections. Retrovirology 2007;4(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goetz MB, Leduc R, Kostman JR, et al. : Relationship between HIV coreceptor tropism and disease progression in persons with untreated chronic HIV infection. J Acquir Immune Defic Syndr 2009;50(3):259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clementi M. and Lazzarin A: Human immunodeficiency virus type 1 fitness and tropism: Concept, quantification, and clinical relevance. Clin Microbiol Infect 2010;16(10):1532–1538 [DOI] [PubMed] [Google Scholar]
- 29.Coetzer M, Cilliers T, Papathanasopoulos M, et al. : Longitudinal analysis of HIV type 1 subtype C envelope sequences from South Africa. AIDS Res Hum Retroviruses 2007;23(2):316–321 [DOI] [PubMed] [Google Scholar]
- 30.Brockman MA, Schneidewind A, Lahaie M, et al. : Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 gag alter capsid interactions with cyclophilin. J Virol 2007;81(22):12608–12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montero M, van Houten NE, Wang X, and Scott JK: The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: Dominant site of antibody neutralization and target for vaccine design. Microbiol Mol Biol Rev 2008;72(1):54–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sivaraman V, Zhang L, Meissner EG, et al. : The heptad repeat 2 domain is a major determinant for enhanced human immunodeficiency virus type 1 (HIV-1) fusion and pathogenicity of a highly pathogenic HIV-1 Env. J Virol 2009;83(22):11715–11725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunter E: gp41, a multifunctional protein involved in HIV entry and pathogenesis [Internet]. 1997;55–73. Available from www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/1997/partIII/gp41.pdf
- 34.Chan DC, Fass D, Berger JM, and Kim PS: Core structure of gp41 from the HIV envelope glycoprotein. Cell 1997;89(2):263–273 [DOI] [PubMed] [Google Scholar]
- 35.Sakamoto T, Ushijima H, Okitsu S, et al. : Establishment of an HIV cell–cell fusion assay by using two genetically modified HeLa cell lines and reporter gene. J Virol Methods 2003;114(2):159–166 [DOI] [PubMed] [Google Scholar]
- 36.Murakami T. and Freed EO: Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J Virol 2000;74(8):3548–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yue L, Shang L, and Hunter E: Truncation of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein defines elements required for fusion, incorporation, and infectivity. J Virol 2009;83(22):11588–11598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachrach E, Dreja H, Linb Y, et al. : Effects of virion surface gp120 density on infection by HIV-1 and viral production by infected cells. Virology 2005;332:418–429 [DOI] [PubMed] [Google Scholar]
- 39.Archary D, Gordon ML, Green TN, et al. : HIV-1 subtype C envelope characteristics associated with divergent rates of chronic disease progression. Retrovirology 2010;7:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X-L, Han X-X, Dai D, et al. : Association of variations of NAb 2F5 and 4E10 epitopes and disease progression in Chinese antiretroviral treatment-naïve patients infected with HIV-1 clade B′. Chin Med J (Engl) 2010;123(23):3406–3411 [PubMed] [Google Scholar]
- 41.Zwick MB, Jensen R, Church S, et al. : Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J Virol 2005;79(2):1252–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zwick MB: The membrane-proximal external region of HIV-1 gp41: A vaccine target worth exploring. AIDS 2005;19(16):1725–1737 [DOI] [PubMed] [Google Scholar]
- 43.Tamura K, Peterson D, Peterson N, et al. : MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28(10):2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abrahams M-R, Anderson Ja, Giorgi EE, et al. : Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-poisson distribution of transmitted variants. J Virol 2009;83(8):3556–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]