Abstract
Aims: The differentiation of embryonic stem (ES) cells into energetically efficient cardiomyocytes contributes to functional cardiac repair and is envisioned to ameliorate progressive degenerative cardiac diseases. Advanced cell maturation strategies are therefore needed to create abundant mature cardiomyocytes. In this study, we tested whether the redox-sensitive heme oxygenase-1/carbon monoxide (HO-1/CO) system, operating through mitochondrial biogenesis, acts as a mechanism for ES cell differentiation and cardiomyocyte maturation. Results: Manipulation of HO-1/CO to enhance mitochondrial biogenesis demonstrates a direct pathway to ES cell differentiation and maturation into beating cardiomyocytes that express adult structural markers. Targeted HO-1/CO interventions up- and downregulate specific cardiogenic transcription factors, transcription factor Gata4, homeobox protein Nkx-2.5, heart- and neural crest derivatives-expressed protein 1, and MEF2C. HO-1/CO overexpression increases cardiac gene expression for myosin regulatory light chain 2, atrial isoform, MLC2v, ANP, MHC-β, and sarcomere α-actinin and the major mitochondrial fusion regulators, mitofusin 2 and MICOS complex subunit Mic60. This promotes structural mitochondrial network expansion and maturation, thereby supporting energy provision for beating embryoid bodies. These effects are prevented by silencing HO-1 and by mitochondrial reactive oxygen species scavenging, while disruption of mitochondrial biogenesis and mitochondrial DNA depletion by loss of mitochondrial transcription factor A compromise infrastructure. This leads to failure of cardiomyocyte differentiation and maturation and contractile dysfunction. Innovation: The capacity to augment cardiomyogenesis via a defined mitochondrial pathway has unique therapeutic potential for targeting ES cell maturation in cardiac disease. Conclusion: Our findings establish the HO-1/CO system and redox regulation of mitochondrial biogenesis as essential factors in ES cell differentiation as well as in the subsequent maturation of these cells into functional cardiac cells. Antioxid. Redox Signal. 24, 345–360.
Introduction
Cell therapy holds unique promise in cardiovascular medicine for the prevention and remediation of diseases that destroy cardiomyocytes and lead to cardiomyopathies and congestive heart failure (31, 34, 52). The adult cardiomyocyte survives energetically through oxidative phosphorylation (24), and mitochondrial damage impairs cardiomyocyte survival and cardiac performance. An optimal complement of mitochondria is maintained by nuclear programming that is responsible for coordination of nuclear and mitochondrial-encoded mitochondrial genes. For example, the protein subunits of the mitochondrial electron transport complex (ETC) are encoded by both genomes (43). Moreover, nuclear-encoded transcriptional activator proteins such as the nuclear respiratory factors (NRFs) and peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), mitochondrial transcription factor A (Tfam), and DNA polymerase subunit gamma-1 (Polγ) indirectly and directly regulate mitochondrial DNA (mtDNA) replication and transcription (6, 16, 20).
Innovation.
The importance of heme oxygenase-1/carbon monoxide (HO-1/CO) in embryonic stem (ES) cell differentiation is shown by the requirement for the enzyme and for redox activation of mitochondrial biogenesis mediated by physiological levels of CO. HO-1/CO induction leads to mitochondrial reactive oxygen species generation and upregulation of proteins required for mitochondrial DNA replication before ES cell differentiation (2). The linkage to mitochondrial biogenesis is shown by loss-of-function experiments that interrupt the program and block the effectiveness of the CO messenger gas on differentiation. These findings put the HO-1/CO system and heme catabolism at a regulatory checkpoint in ES cell differentiation and cardiomyocyte maturation. Prior work suggesting that stem cell use, perhaps including marrow-derived cells, may contribute to repair of infarcts in excess of cell engraftment may indicate paracrine effects (26, 36, 45). Thus, agents that activate HO-1 in ES cell differentiation in vitro or in vivo may be expected to contribute to regenerative cell therapy in patients with advanced heart disease.
The embryonic deletion of Polγ or Tfam in mice causes mtDNA depletion and loss of mitochondrial function, leading to energy failure and death by days E8.5 (15) and E10.5 (20), respectively, in part because the energy needs of differentiated cells greatly exceed those of stem cells and must be met by adjustments in mitochondrial volume and phenotype (9). Well-differentiated cells exhibit high mtDNA copy number, distinctive mitochondrial morphology, and well-defined mitochondrial localization (8). Embryonic stem (ES) cells generally depend on glycolysis, and display low mtDNA copy number and low mitochondrial density (39, 46, 51).
During differentiation of ES cells, there are striking increases in mtDNA replication and nuclear- and mtDNA-encoded mitochondrial gene expression (22, 55) followed by mitochondrial proliferation through the activation of mitochondrial biogenesis (46). The implication is that mitochondria are necessary for aerobic maturation and possibly for terminal differentiation of mesenchymal stem cells (8), cardiac mesangioblasts (41), and ES cells (27). Cardiac differentiation is enhanced by certain small molecules, including retinoic acid (56), oxytocin (37), ascorbic acid (49), and nitric oxide donors (17, 33), but due to their low efficiencies, these are highly disadvantageous for therapeutic applications. Exploitable new approaches are actively being sought, and an effective strategy might be to bring inducible cell defense mechanisms to bear on the process of differentiation.
Mitochondrial biogenesis is regulated both by energy-dependent signals and by localized reactive oxygen species (ROS) production; both induce the transcriptional program to increase mitochondrial mass, particularly in aerobic organs such as the heart (18). The exact mitochondrial sensing and signaling of impending changes in cellular energetics during differentiation are poorly understood functionally, temporally, and spatially (32). This information gap creates a barrier for effective therapeutic use of various exogenous cardiac progenitor cells (CPCs) to improve bioenergetics and myocardial regeneration (5). For any pluripotent cell to adopt an adult contractile cardiomyocyte phenotype, however, it embodies a vast expansion of bioenergetic capacity during differentiation.
One critical myocardial defense for energy metabolism involves the inducible heme oxygenase-1 (HO-1; Hmox1) system, the rate-limiting step in heme catalysis (25). The enzyme eliminates heme, a potent pro-oxidant, and releases iron, biliverdin, and carbon monoxide (CO), which have important signaling and cytoprotective properties (54). Beyond heme clearance, the HO-1/CO system links mitochondrial redox status to the redox-active transcription factors, nuclear respiratory factor-1 (NRF-1) and nuclear factor erythroid 2-related factor 2 (Nfe2l2), which promote mitochondrial biogenesis (38, 48). Cardiac HO-1/CO induction stimulates gene expression for Tfam and opposes mitochondrial drug cardiotoxicity (47).
If modulation of mitochondrial biogenesis through the HO-1/CO system (4, 30) could be used to program ES cells onto a differentiation pathway, it may be possible to accelerate cardiomyocyte lineage development. It has been known that mitochondrial biogenesis is activated during the cell cycle (23), but no role has been identified for HO-1/CO. In this study, we demonstrate a critical regulatory step in ES cell embarkation to cardiomyocytes that is controlled by the HO-1/CO system through the mitochondrial biogenesis pathway. Mechanistically, HO-1/CO operates at a checkpoint involving CO, mitochondrial ROS, and mtDNA replication that are amenable to pharmacological intervention.
Results
CO-dependent loss of ES cell pluripotency
Undifferentiated mouse ES cells (E14) showed high expression of the pluripotency markers, transcription factor SOX-2 (Sox2) and POU domain, class 5, transcription factor 1 (Oct4). Stem cell differentiation into embryoid bodies (EBs) demonstrated a gradual loss of expression of these genes apparent by 72 h (Fig. 1A, B). After application of CO-releasing molecule (CORM)-2 at 25 μM (58), loss of pluripotent gene expression was accelerated by 72 h and there was dose-dependent repression of Sox2 and Oct4 by CORM-2 (Fig. 1C, D). These effects in E14 cells were confirmed in two other ES lines, mouse D3 and human HS-MEL-1 cells (Fig. 1E, F).
FIG. 1.
Oct4 and Sox2 mRNA expression levels in ES cells. (A, B) The mRNA levels for Oct3/4 and Sox2 pluripotency genes in E14 cells in the absence of LIF decline between 0 and 72 h where 0 is control value. Loss of Oct 3/4 and Sox2 mRNA expression is accelerated in E14 cells exposed to CORM-2 (25 μM) after LIF withdrawal, but not iCORM. (C, D) E14 cells demonstrate CORM-2 concentration-dependent loss of Oct3/4 and Sox2 mRNA expression at 72 h after dose escalation. (E, F) CORM-2 accelerates loss of Oct4 and Sox2 mRNA in human MEL-1 and mouse mD3 ES cells (at 72 h after 25 μM CORM-2). Values are mean ± SD of 3–4 independent experiments. Asterisks indicate significant differences from control (Concentration or Time 0 by ANOVA; p < 0.05). ANOVA, analysis of variance; CORM-2, CO-releasing molecule; ES, embryonic stem; iCORM, inactivated CORM; LIF, leukemia inhibitory factor; SD, standard deviation. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
To compare changes in pluripotency with mitochondrial mass, we tracked nuclear Oct4 localization, HO-1 protein induction, and mitochondrial mass. HO-1 protein was induced in control EBs by day 7 (data not shown), while CORM-2 treatment enhanced early HO-1 expression (by day 3) compared with controls (Fig. 2A). For Oct4 and mitochondrial mass (using MitoTracker green), we performed immunofluorescence microscopy under control and CORM-2-treated conditions (Fig. 2B[a–c]). During early differentiation (day 3), nuclear Oct4 was present in all conditions, but less nuclear staining (puncta) was observed in EBs treated with CORM-2 (Fig. 2B[d–f]). Oct4 nuclear staining was strongly attenuated at days 7–9. To evaluate HO-1, the enzyme was silenced and the effect of CORM-2 was determined on Oct4 expression. HO-1 silencing delayed the loss of Oct4 (Fig. 2B[g–i]), an effect only marginally reversed by CORM-2 (Fig. 2B[j–l]). Images of untreated control cells are shown in Figure 2B(m–o).
FIG. 2.
HO-1 induction and loss of pluripotency in ES cells. (A) Control ES cells show increases in HO-1 mRNA levels after exposure to CORM-2 (25 μM), while iCORM has no effect. Cells transfected with Sh-HO-1 show 70–80% suppression of HO-1 mRNA relative to control. Sh-vector does not affect HO-1 expression and the cells behave like controls. Data analyzed by T test and presented as mean ± SD of three separate experiments. (*p < 0.05 iCORM compared with CORM-2). (B) E14 cells were stained for mitochondria and loss of OCT3/4 on days 3, 7, and 9. Mitochondria are stained with MitoTracker (green), nuclei with DAPI (blue), and anti-OCT4 (red). (a–c) E14 cells treated with iCORM. Nuclear Oct4 appears pink. As the nucleus loses OCT4, mitochondrial proliferation is initiated. (d–f) E14 cells treated with CORM-2 (25 μM) display complete loss of nuclear OCT4 by day 7. (g–i) E14 cells after transfection with ShHO-1 show delayed nuclear OCT4 loss. (j–l) E14 cells transfected with Sh-HO-1 and treated with CORM-2 show persistence of nuclear OCT4 at day 7. (m–o) Images of untreated control cells for comparison. Images acquired at 400×. (C) E14 cells cultured in hanging drops for 2 days were plated onto gelatin-coated microtiter plates and treated at days 2, 5, and 7 with iCORM or CORM-2 (25 μM). Adherent EBs were monitored daily by inverted microscopy for spontaneous beating. Results are percentage of beating EBs and represent mean ± SD of five determinations. Control cells began beating by day 7. CORM-2 significantly increased the number of EB beating foci at days 7–13. Cells transduced with Sh-HO-1 showed few beating foci and could not be rescued by CORM-2. (*p < 0.05 HO-1-silenced CORM-2 vs. CORM-2; **p < 0.05 WT CORM-2 vs. iCORM on the same days). DAPI, 4′,6-diamidino-2-phenylindole; EBs, embryoid bodies; HO-1/CO, heme oxygenase-1/carbon monoxide; Oct/3/4, POU domain, class 5, transcription factor 1; WT, wild-type.
To distinguish serum-driven cardiac differentiation, E14 cells were cultured in hanging drops and the emerging EBs placed individually into tissue culture plates, and cardiac development was monitored by examination of rhythmically contracting focal areas. Spontaneously beating focal areas within individual differentiated EBs were observed from day 7, with >55% of EBs containing beating areas by the end of the experiment (Fig. 2C). In CORM-2 (25 μM)-treated EBs, beating areas reached 80% by day 13. Contrastingly, HO-1 silencing almost completely abrogated the appearance of beating areas and was not improved by CORM-2 treatment (Fig. 2C).
ES cell differentiation
Mouse ES cells were able to develop into structural and functional cardiomyocytes. In the hanging drop stage, EB aggregates in control media or in media plus CORM-2 had a similar morphological appearance and size for the first 2 days (not shown), indicating no basic perturbations of EB formation by the compound. For cardiac differentiation, changes in nuclear expression of critical transcription factors, homeobox protein Nkx-2.5 (Nkx2.5) and transcription factor Gata4 (GATA4), were monitored by immunochemical staining (Fig. 3A, B). Nkx2.5 and GATA4 nuclear staining was more widespread in CORM-2- than in inactivated CORM (iCORM)-treated EBs (Fig. 3A, B[a, b], respectively). HO-1-silenced cells showed rare nuclei that stained for Nkx2.5 and GATA4 even after CORM-2 (Fig. 3A, B[c, d], respectively). Immunoblot analysis indicated increases in EB nuclear Nkx2.5, GATA4, Mef2c, and in particular heart- and neural crest derivatives-expressed protein 1 (Hand1) proteins and their inhibition with shHO-1 (Fig. 3C). These transcription factors are characteristically activated in well-differentiated cardiomyocytes, and CORM-2 was especially effective for Hand1, a noncanonical E-box transcription factor required for cardiac morphogenesis. Immunoblot analysis also documented the extent to which CORM-2 stimulated EB maturation into cardiomyocytes. The responses included increased expression of cardiac MHC6, MLC-2a, MLC-2v, and troponin I (cTnI) proteins consistent with the contractile myocyte (Fig. 3D, upper panel). CORM-2 also progressively increased cardiac structural marker levels for α-actinin, connexin 43 (Cxn43), and brain natriuretic peptide (BNP) (Fig. 3C, lower panel).
FIG. 3.
HO-1 activation drives E14 cell differentiation into beating cardiac cells. (A) Cardiac GATA4 (red) localization to nuclei stained with DAPI (blue). GATA4-positive nuclei are pink. (a) Control cells treated with iCORM, (b) Control cells treated with CORM-2 (25 μM), (c) Sh-HO-1-transfected cells, (d) Sh-HO-1-transfected cells treated with CORM-2. Nuclear accumulation of GATA4 is inhibited by Sh-HO-1 in iCORM and CORM-2-treated cells. Images acquired at 200×. (B) Cardiac Nkx2.5 transcription factor expression and localization by immunofluorescence staining in control and HO-1-silenced EBs (day 13). E14 cells were differentiated in the absence or presence of CORM-2. Nkx2.5 (red) localization to nucleus is pink; nuclei stained with DAPI (blue). (a) Control cells plus iCORM, (b) Controls plus CORM-2 (25 μM), (c) ShHO-1-transfected cells, (d) Sh-HO-1-transfected cells treated with CORM-2. Nuclear localization is enriched in cells treated with CORM-2. Images acquired at 200×. (C) Histograms of cardiac lineage nuclear proteins, Nkx2.5, GATA4, Mef2c, and Hand1, relative to Lamin B loading control. Lineage marker expression is enhanced by CORM-2 (25 μM) and inhibited by Sh-HO-1. Gel shows a representative immunoblot for Hand1 versus Lamin B. (D) Upper panel, Western blots for cardiac MHC6, MLC-2a, MLC-2v, and cTnI proteins after EB differentiation into cardiac lineage (day 13). Lower panel, immunoblot analysis showing CORM-2 enhancement of the expression of the structural cardiac proteins, α-actinin, Cxn43, and BNP, at day 13 compared with day 0. Stripped blots were stained for β-actin as the loading control. (E) Histogram quantification of (D). Values are mean ± SD of three independent experiments (*p < 0.05). Hand1, heart- and neural crest derivatives-expressed protein 1; Gata4, transcription factor Gata4; MLC2a, myosin regulatory light chain 2, atrial isoform; Nkx2.5, homeobox protein Nkx-2.5.
Mitochondrial expansion promotes cardiac differentiation
Mitochondrial expansion is a key step in differentiation that involves mtDNA transcription, replication, protein synthesis, and activation of oxidative phosphorylation (10). Since HO-1/CO regulates mitochondrial proliferation (48), we considered that mitochondrial pathway activation might induce ES cell differentiation. Using mitochondrial content monitoring by fluorescence microscopy and fluorescence staining of citrate synthase (CS), we found that CORM-2 (25 μM) significantly augmented EB mitochondrial mass (Fig. 4A) and CORM-2 treatment significantly increased mtDNA content in differentiating EBs by mtDNA measurements (Fig. 4B). Since mtDNA replication depends on nuclear NRF-1, PGC-1α coactivator, and imported mitochondrial factors such as Polγ and Tfam (43), these four transcripts were checked in EBs (Fig. 4C–F) and increased mRNA levels were found for NRF-1 and PGC-1α by day 3 and for Polγ and Tfam by day 7, coinciding with the peak change in mtDNA content (Fig. 4B). This also increased mitochondrial transcriptional activity as shown by enhanced expression of mtDNA-encoded subunit II of complex IV (cytochrome c oxidase subunit 2 [COII]; Fig. 4G).
FIG. 4.
Mitochondrial biogenesis promotes cardiac differentiation in EBs. (A) Mitochondrial density in differentiating EBs imaged at D13 using CS immunofluorescence (green). Nuclei stained blue with DAPI. (a) Control cells plus iCORM, (b) Control cells plus CORM-2 (25 μM), (c) ShHO-1-transfected cells, (d) ShHO-1-transfected cells plus CORM-2. Mitochondrial density is most pronounced in CORM-2-treated cells, while ShHO-1-transfected cells show lowest mitochondrial density. Lower histogram is quantification of fluorescence intensity. Images acquired at 200×. (B) Changes in mtDNA copy number during spontaneous and CORM-2-induced differentiation in murine EBs. (C–E) Mitochondrial gene expression and EB cell differentiation. During differentiation, mRNA levels for upstream mtDNA regulators (NRF-1 and PGC-1α) and downstream mtDNA transcription/replication regulators (Polγ and Tfam) increase (F). Gene expression normalized to 18S and expressed as fold change compared with day 0. (G) Mitochondrial COII gene expression by qPCR. Data are fold changes compared with day 0. Significant differences between iCORM and CORM-2 treated cells (*p < 0.05). Values are mean ± SD of three independent experiments. COII, cytochrome c oxidase subunit 2; CS, citrate synthase; mtDNA, mitochondrial DNA; NRF-1, nuclear respiratory factor-1; PGC-1α, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; Polγ, DNA polymerase subunit gamma-1; qPCR, quantitative real-time RT-PCR; Tfam, mitochondrial transcription factor A.
Maturation of ES cells via redox signals
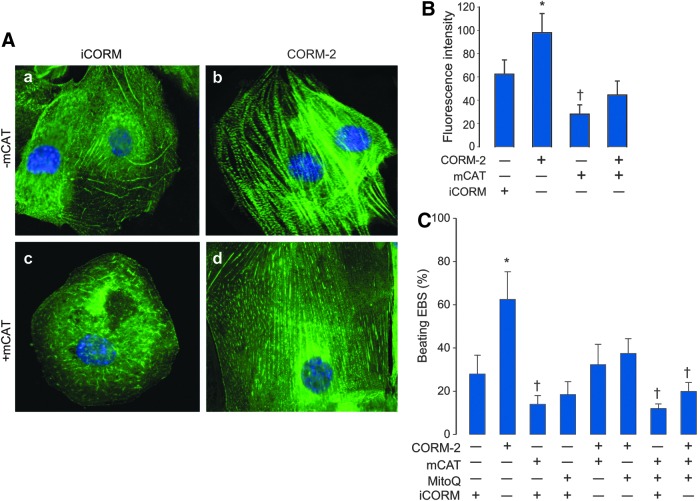
The role of mitochondrial ROS production as a maturation signal was evaluated using transfection with mitochondrial-targeted catalase (mCAT). With immunofluorescence for sarcomeric α-actinin, we demonstrated a characteristic pattern of banded continuous filaments in differentiating cells that became more pronounced in CORM-2-treated cells (Fig. 5A[a, c]). This supported the ability of ES cells to generate cardiomyocyte structures and their augmentation by CORM-2. EB cell differentiation was impaired by mCAT transfection as indicated by poorly defined cardiac-specific α-actinin filament staining (Fig. 5A[b, d]). The α-actinin fluorescence signal increased significantly after CORM-2, and mCAT reduced both the basal and CORM-2 effects (Fig. 5B, C). This implicated mitochondrial redox signals in the cardiac differentiation of ES cells (7, 11), and this mechanism was further explored using the mitochondrial-targeted antioxidant, mito-ubiquinone (MitoQ). The emergence of beating foci was quantified as above through day 13 in wild-type and mCAT-transfected EBs plus MitoQ with or without CORM-2. Both MitoQ and mCAT transduction significantly reduced the percentage of beating EBs and in concert they greatly inhibited the CORM-2 effect (Fig. 5C). These data together support that changes in mitochondrial redox state induced by CORM-2 are necessary for EB differentiation.
FIG. 5.
Cellular redox state and differentiation of mouse ES cells into cardiomyocytes. EBs were developed by hanging drop and harvested at different times of cardiac differentiation. (A) For differentiation markers, day 13 EBs were digested and single-cell suspensions subjected to immunofluorescence staining of cardiac myofibrils with cardiac anti-α-sarcomeric actinin (green). Nuclei are stained blue (DAPI). (a) iCORM-treated cells, (b) CORM-2-treated cells (25 μM), (c) Cells transfected with mCAT on day 3 and collected at day 13, (d) Cells transfected with mCAT and then treated with CORM-2 (25 μM). CORM-2-treated cells exhibit a classical striated appearance of mature cardiomyocytes. Final magnification 500×. (B) Histogram for quantification of α-sarcomeric actinin fluorescence intensity in (A). (C) ES cells were differentiated and then transfected with mCAT or treated with MitoQ (0.3 μM), then exposed to CORM-2 (25 μM). The number of beating foci was counted on day 13. Values are mean ± SD of four experiments from independent batches of cells. (*p < 0.05 > control; †p < 0.05 < control by ANOVA). mCAT, mitochondrial catalase; MitoQ, mito-ubiquinone.
Tfam deficiency and ES cell differentiation
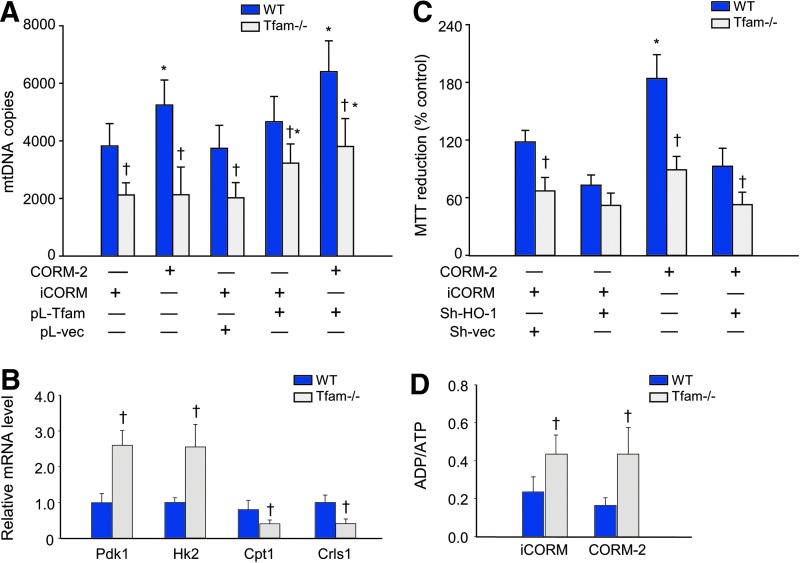
The transition from noncontractile ES cells into actively beating cardiomyocytes requires an efficient metabolic and electromechanical infrastructure (12, 35). Moreover, Tfam is indispensable to mtDNA transcription and to the packaging and stabilization of mtDNA during mitochondrial biogenesis (2, 13). To determine if mitochondrial biogenesis is required for ES cell differentiation, we used Tfam ablation (Fig. 6A). In Tfam−/− cells, HO-1 gene upregulation by CORM-2 was still weakly present (Fig. 6B), but differentiation into beating cardiomyocytes was blocked as was the CORM-2 effect on it (Fig. 6C). The impact of loss of Tfam on differentiation was indicated by low α-actinin, MLC-2a, and cTnI protein levels compared with controls even after CORM-2 treatment (Fig. 6D). Additional evaluation of mitochondrial function and expression of mitochondrial ETC complex and assembly factors in Tfam−/− ES cells showed that NADH dehydrogenase I alpha subcomplex 1 of complex I (Ndufa1), as well as complex I assembly factor 4 (Ndufaf4) were downregulated (Fig. 6E). We also found depletion of complex IV protein COI and COII expression (Fig. 6E), confirming the functional effectiveness of Tfam ablation. The effects shown in Figure 6D and E are quantified in Figure 6F.
FIG. 6.
Disruption of differentiation in Tfam-deficient ES cells. (A, B) Tfam and HO-1 mRNA expression by qPCR and Tfam protein by immunoblot in WT and Tfam−/− cell EBs at day 7. Asterisks indicate significant differences in iCORM versus CORM-2, and crosses are WT versus Tfam−/− cells (p < 0.05) by T-test. CORM-2 is ineffective in Tfam−/− cells. Values are mean ± SD of three independent experiments. (C) Tfam-deficient ES cells cannot differentiate into beating cardiomyocytes and CORM-2 cannot rescue them. Differences between iCORM and CORM-2 treatment and WT and Tfam−/− EBs are significant (p < 0.05). N = 3–4 experiments per group. (D) Western analysis of α-actinin, MLC-2a, and cTnI protein expression in Tfam−/− cells compared with WT cells (tubulin control). (E) Immunoblot analysis of mitochondrial ETC member proteins, COI and COII, and assembly factors, Ndufa1 and Ndufaf4 (porin control). (F) Histogram for quantification of protein in (D, E). Two-way ANOVA was used for data analysis. Values are mean ± SD of n = 4 independent batches of cells (*p < 0.05 WT iCORM vs. CORM-2; †p < 0.05 WT vs. Tfam−/−). ETC, electron transport complex. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
The loss of mitochondrial biogenesis as predicted also blocked ES cell differentiation and maturation into functional cardiomyocytes. Tfam−/− ES cells had low mtDNA content, which was restorable by Tfam transfection (Fig. 7A). Tfam deficiency also reprogrammed the ES cell metabolically for glycolysis as reflected by upregulation of pyruvate dehydrogenase kinase isozyme 1, mitochondrial (Pdk1), and hexokinase 2 (HK2). In contrast, the loss of Tfam decreased cardiolipin synthase (Crls) and carnitine O-palmitoyltransferase 1 (Cpt1) expression (Fig. 7B). CORM-2 was ineffective in preventing these effects as in Figure 7B (negative data not shown). Confirmation that Tfam deficiency lowered EB metabolic activity was done by MTT assay (Fig. 7C), which showed that a loss of Tfam decreased MTT reduction and was not improved by CORM-2 treatment. The net energetic effect was a lower average ATP content and a higher ADP/ATP ratio revealing a relative energy insufficiency in differentiation (Fig. 7D).
FIG. 7.
Tfam depletion decreases mtDNA and alters ES cell metabolic phenotype. (A) Decrease in mtDNA copy number as a result of Tfam deficiency is rescued by transfection of Tfam into Tfam−/− ES cells. (B) Metabolism‐related Pdk1, HK2, Crls, and Cpt1 mRNA expression. (C) Histogram for functional MTT reduction in WT control and Tfam−/− cells at day 13. CORM-2 enhances the metabolic activity (MTT reduction) in EB controls, but had no effect on Tfam−/− cells. (D) ADP/ATP ratio in Tfam-deficient and WT control cells treated with CORM-2. Values are mean ± SD of n = 3 batches of cells (*p < 0.05 iCORM vs. CORM-2; †p < 0.05 WT vs. Tfam−/− by two-way ANOVA). Cpt1, carnitine O-palmitoyltransferase 1; CRLS, cardiolipin synthase; HK2, hexokinase 2; Pdk1, pyruvate dehydrogenase kinase isozyme 1, mitochondrial. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Maturation of the mitochondrial network during cardiogenesis
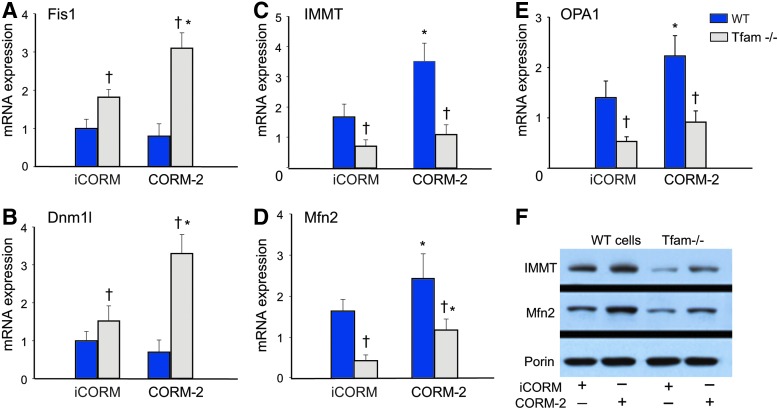
The dependence of cardiac differentiation and maturation on mitochondrial number, phenotype, and structural organization hence depends on genes involved in mitochondrial network formation (8). In tracking mRNA levels for selected genes involved in network formation and maintenance through day 13 in wild-type (WT) and Tfam-deficient cells, we found greatly increased fission (fragmentation) protein 1 (mitochondrial fission 1 protein [Fis1]) after loss of Tfam, while CORM-2, which had no effect on Fis1 levels in WT cells, increased Fis1 in Tfam−/− cells (Fig. 8A). The mRNA responses for Dynamin-1-like protein (Dnm1l) were similar both after Tfam deletion and after CORM-2 treatment (Fig. 8B). In contrast, mRNA levels for the crista junction maturing mitofilin protein (MICOS complex subunit Mic60 [Immt]) decreased significantly in Tfam−/− cells and, after CORM-2, increased significantly only in differentiating WT control cells (Fig. 8C). Similarly, mRNA levels for mitofusin 2 (Mfn2), an essential transmembrane GTPase that mediates mitochondrial fusion, and optic atrophy type 1 (dynamin-like 120 kDa protein, mitochondrial [Opa1]) were downregulated in Tfam−/− cells and upregulated only in WT controls by CORM-2 (Fig. 8D, E). The Immt and Mfn2 protein levels paralleled the changes of their mRNA levels (Fig. 8F). This pattern of changes in these four proteins indicates that Tfam deficiency blocks mitochondrial network formation and that CORM-2 can accelerate mitochondrial network formation only in mtDNA replication-sufficient cells.
FIG. 8.
EB mitochondrial network maturation during cardiac differentiation. (A, B) The expression of genes involved in mitochondrial fission and membrane remodeling increases only in Tfam−/− cells (Dnm1l and Fis1). (C, D) Expression of genes involved in mitochondrial fusion and cristae maturation (Immt and Mfn2) in control and Tfam−/− cells at day 13 of differentiation. (E) OPA1 cristae protein increases with CORM-2 treatment only in WT cells. (F) Immt and Mfn2 protein expression in control and Tfam−/− cells at day 13 of differentiation. Asterisks indicate iCORM versus CORM-2 (*p < 0.05). Crosses (†) indicate Tfam−/− compared with WT cells (p < 0.05). Values are mean ± SD of three independent experiments. Dnm1l, dynamin-1-like protein; Fis1, mitochondrial fission 1 protein; Mfn2, mitofusin 2; Opa1, dynamin-like 120 kDa protein, mitochondrial. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
H9c2 cardiomyoblast differentiation
To confirm that HO-1/CO signaling is integral to cardiac differentiation, we used adult myocardioblasts (H9c2 cells) and checked their ability to differentiate morphologically by shape elongation and multinucleation. HO-1 mRNA expression was induced in these cells by CORM-2 (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars), which also modulated morphological characteristics such as elongation and cell fusion, as well as proliferation tracked by changes in cell cycle marker, cyclin-dependent kinase 2 (CDK2), mRNA levels (Supplementary Fig. S1A). In HO-1-silencing experiments, myogenin and the myosin regulatory light chain 2 (Myl2) contractile apparatus protein were upregulated by CORM-2 in an HO-1-dependent manner (Supplementary Fig. S1A). The implication is that the HO-1/CO system promotes cell cycle arrest in preparation for the synchronous expansion of myogenic differentiation. H9c2 cell differentiation was accompanied by mitochondrial network development and maturation, especially after CORM-2 administration (Supplementary Fig. S1B). Orderly emergence of the mitochondrial network tracked by Mfn2 staining indicated disruption of cardiomyocyte differentiation steps by HO-1 silencing, leaving behind a discontinuous mitochondrial network reminiscent of fusion impairment. Mitochondrial expansion and network formation were accelerated by CORM-2 as shown by increased expression of inner membrane, mitofilin (Immt), and outer membrane, Mfn2, which was absent after HO-1 silencing (Supplementary Fig. S1C).
To visualize the myocytes, we used sarcomeric α-actinin immunofluorescence staining to locate increases in actin filament assembly with clear sarcomere cross-striation and increased cell size. This was more pronounced in CORM-2-treated cells (Supplementary Fig. S2A). Early transduction with sh-HO-1 blocked these responses, and α-actinin in those cells appeared as unassembled or disarrayed filaments or aggregates (Supplementary Fig. S2A).
CPCs in adult mouse heart
Because the cellular work strongly supported a regulatory role for HO-1 induction, and specifically for endogenous CO production in cardiomyocyte differentiation, we sought in vivo evidence of relevance in mice. As CO also appeared to be a key pharmacological mechanism of action of CORM-2, which was unavailable in large enough quantities for in vivo studies, we induced the HO-1/CO system in the mouse heart using inhaled CO (47) and checked for evidence of CPC expansion ex vivo (Supplementary Fig. S3). After 3 days of intermittent CO breathing, the mouse hearts contained new clusters of Sca1+ cells, consistent with expansion of the progenitor cell population (Supplementary Fig. S3A–C). CO also increased the cardiomyocyte lineage commitment as indicated by increases in Nkx2.5 protein expression (Supplementary Fig. S3D–F; p < 0.05).
Discussion
These studies demonstrate that the cell-protective HO-1/CO system, which promotes energy homeostasis in the heart and other tissues (40), also helps regulate the differentiation of murine ES into functional cardiac cells. The data show temporal and spatial connections between changes in HO-1 gain and loss of function and corresponding changes in mitochondrial mass. In pluripotent cells, the expression of cardiac-specific structural genes is preceded by mitochondrial biogenesis, and the emergence of the morphological contractile phenotype is blocked by HO-1 silencing and stimulated by a novel CO-releasing molecule. Moreover, the HO-1/CO system promotes differentiation through mitochondrial ROS production, which is required in the ES cell transition into the cardiac lineage. Mitochondrial-targeted antioxidants prevent ES cell differentiation, while Tfam knockdown blocks it even after CO. This means that mitochondrial proliferation and maturation are definitive mechanisms to actually drive cardiomyocyte phenotype.
Studies on the role of the HO-1/CO system in cardiac regeneration have proposed the facilitation of CPC mobilization by improving their migration, proliferation, and/or viability (19). Similarly, HO-1 induction in transplanted mesenchymal stem cells (MSCs) augments their regenerative potential in infarcted or ischemic myocardium. These salutary effects are attributable, in part, to improved cell viability or to a better balance between proangiogenic growth factor production and proinflammatory cytokine production (50, 53), but data have so far been lacking on precise molecular pathways and rates of MSC differentiation. Moreover, induction of the mitochondrial signaling pathway through HO-1 operates in connection with Akt/PKB (48), which has a known role in the proliferation of cardiomyocytes derived from human ES cells (28).
The differentiation of ES cells along the cardiac lineage creates a characteristic gene expression profile that includes the early expression of a network of mesodermal cardiac transcription factors such as Nkx2.5, Gata4, and Mef2c, which activate gene expression for structural proteins, MHC, Mlc2v, and troponin I (3). In this study, the early upregulation of the Nkx2.5 cardiac transcription factor and the repression of OCT4, followed by increases in mitochondrial mass, were greatly facilitated by CORM-2. HO-1 induction mirrored loss of OCT4 and was linked to accelerated mitochondrial network formation, thereby exposing the heme and energy requirements of terminal differentiation. HO-1 silencing markedly decreases nuclear Nkx2.5 and Gata4 expression and localization in EB outgrowths, which delays and decreases the number of self-contracting foci. CORM-2 had no ability to rescue the contractile phenotype in HO-1-silenced cells, thereby indicating that HO-1/CO, in this setting, is necessary for the development of contractile activity. Expansion of the mitochondrial population for terminal differentiation in EBs was accompanied by increases in α-actinin, MHC, Mlc2v, and troponin T proteins and was facilitated by CO, further implicating HO-1/CO in regulation of cardiomyogenesis.
The origin of cardiomyocytes from ES cells is well described empirically, but a lack of complete lineage definitions has made it difficult to devise interventions involving oxidant/antioxidant balance (1, 21, 49). Accordingly, the influence of mitochondrial redox state on cardiomyogenesis is known only insofar as what selected antioxidants inhibit, while mitochondrial ROS facilitates cardiac differentiation (42). In this study, we report that mitochondrial ROS production occurring early in cardiomyocyte differentiation promotes and enhances differentiation, while the CO molecule acting as a lynchpin is driving expansion of the mitochondrial population. ES cell transfection with mCAT prevented α-actinin upregulation and the development of contractile function, while augmentation of differentiation by CORM-2 was abrogated by mCAT and mitoQ, thereby underscoring the importance of sensing the mitochondrial redox state.
Although certain redox-active compounds at high levels, such as ascorbate, tend to promote cardiac differentiation, postcommitment the molecule has no additional effect (49). The capacity of CO to induce mitochondrial biogenesis by slowing the ETC and enhancing mitochondrial ROS leak rate not only activates antioxidant response genes but certain cardiac-specific genes as well (38, 47, 48). Our work supports the operation of this mechanism because mitochondrial-targeted antioxidants block the CORM-2 response and inhibit cardiomyocyte maturation.
The mitochondrial actions of the HO-1/CO system provided our focal point because the differentiation steps for the cardiac lineage eventually give rise to a compatible bioenergetic state (10, 41). Typically, energy demand dynamically regulates mitochondrial number and functional capacity in the cardiomyocyte, and as the rate of respiration increases, so does the cell's mitochondrial density. Thus, key proteins that facilitate and coordinate mtDNA transcription and replication, including NRF-1 and PGC-1α, tracked with Polγ and Tfam early in differentiation and led to subsequent expression of mitochondrial-encoded proteins. This is reminiscent of a conditioning effect on aerobic differentiation of cardiomyocytes analogous to the transition from fetal to neonatal life where cardiac metabolism switches away from glucose as the preferred substrate for ATP generation (44, 46).
The dependence of undifferentiated ES cells on glycolysis and the low respiration rates of such highly proliferative cells are reflected by low mtDNA copy number and low mitochondrial-encoded COII levels (44). In murine ES cells, Tfam deficiency provided evidence of depressed mitochondrial function and reversion to glycolysis as demonstrated by high HK2 and Pdk1 expression. By forcing reentry into glycolysis, stem maintenance and differentiation are negatively impacted, and retransfection of Tfam−/− cells with Tfam vector improves mtDNA levels and allows return of aerobic metabolism and differentiation. That mitochondrial biogenesis per se influences cardiac differentiation implies that the organelle directs the ES cell toward a phenotype capable of maintaining an efficient energetic infrastructure (8). Pluripotency regulators are known to influence energy metabolism and the balance between glycolysis and oxidative metabolism (10); hence, aerobic metabolism regulates not only cell fate and self-renewal (8) but also the capacity for differentiation.
Disruption of HO-1 interrupted mitochondrial network organization and compromised the development of energetic infrastructure accompanied by improper sarcomere formation, poor beating area assembly, and defective contractility. Although CORM-2 was found to be quite potent in WT cells, in the absence of HO-1, CORM-2 did not rescue differentiation, implying that HO-1 is essential and that our pharmacological CO release profile did not fully capture the physiological regulatory processes involving HO-1. These processes would include the enzyme's roles in heme turnover and iron handling, which are topics for further investigation.
Several molecular prerequisites underlying the generation of energetically competent cardiomyocytes have been identified in the transition of stem cells to heart cell progenitors. For instance, two critical fusion-facilitating genes (Mfn2 and Immt) were increased in differentiating ES cells, but mitochondrial fission proteins did not change. The former proteins are required for crista junction maintenance and inner membrane architecture, allowing contact sites to form on the outer membrane. Thus, mitochondrial network fusion and expansion in connection with respiratory capacity are able to support a cardiac phenotype (10). Coordination of maturation with mitochondrial biogenesis highlights the specificity of mitochondrial to nuclear retrograde communication for cardiomyogenesis and cardiac differentiation. Accordingly, final differentiation occurs after the mitochondria are functional, and interference with completion of biogenesis by Tfam knockdown also interrupts differentiation and degrades EB beating.
Similarly, in differentiated rat H9c2 embryonic cardiomyoblasts, the HO-1/CO system has a key role in the expression of cardiac gene products and sarcomere structures in a developmentally regulated manner. Both lines express molecular cardiac markers and maintain the ability to differentiate into multinucleated myotubes (38). In H9c2 cells, CORM-2 upregulates the myogenin transcription factor, which promotes muscle-specific target gene expression during muscle differentiation and participates in cell cycle exit. Contractile apparatus proteins such as Myl2 are upregulated by myogenic differentiation, a hallmark of the differentiated phenotype of mature myogenic cells. Myl2 mRNA expression is also enhanced by CORM-2 and prevented by HO-1 silencing. Thus, the HO-1/CO system in both cell lines drives ES cell differentiation toward the functional cardiomyocyte by a redox-dependent mechanism of activation of mitochondrial biogenesis.
Finally, the relevance of these cellular events is seen by expansion of the pool of cardiac progenitors in the mouse heart after brief exposures to CO by inhalation in vivo. The level and duration of these exposures are insufficient to recruit hypoxic mechanisms (48), but sufficient to increase the number of Sca1 and Nkx2.5-expressing cells in the heart. This finding implies significant biological potential for cardiac tissue rescue using induction of the HO-1/CO system in certain heart diseases.
Materials and Methods
Reagents
All chemicals and reagents were purchased from Sigma or Molecular Probes (Invitrogen,) unless otherwise indicated. A water-soluble CO-releasing molecule, CORM-2, synthesized and characterized at the University of Zurich (57, 58) was used to produce endogenous levels of CO. This well-characterized agent binds to B12 receptors and releases two molecules of CO in ∼20 min along with one of soluble ReO4, which is nontoxic and is excreted in vivo. We dissipated the prodrug in phosphate-buffered saline (PBS) for 1 h before applying it to cells as iCORM. Dose ranging studies were performed in murine ES and rat H9c2 cells to identify CORM-2 concentrations that did not cause lactate dehydrogenase release or cell death from CO toxicity.
Cell culture
Mouse ES cells (E14-TG2a; American Type Culture Collection, ATCC) were grown in gelatin-coated plastic tissue culture flasks and maintained in Iscove's modified Dulbecco's medium (Life Technologies; # 12440053) containing 15% newborn calf serum (Atlanta Biologicals), nonessential amino acids, and nucleosides. To prevent differentiation, 2-mercaptoethanol (100 μM) and leukemia inhibitory factor (LIF 1000 U/ml; EMD Millipore; #ESG1106) were added to the media. To induce differentiation, a hanging drop method was used (14).
On day 0, ES cells were cultivated without LIF in hanging drops (30 μl) to form EBs (∼1000 cells per drop) for 2 days, then held in suspension. On day 4, the EBs were independently plated on gelatin-coated 96-well plates, single-culture dishes, or slides and grown in differentiation medium without LIF. This consisted of Dulbecco's modified Eagle's medium (DMEM), 15% fetal calf serum, 0.1 mM mercaptoethanol, and 1% nonessential amino acids. For CORM2, the drug was prepared in differentiation medium (5–25 μM final concentrations) and was changed every 1–2 days. Rhythmically beating aggregates in EB outgrowths were observed daily by light microscopy to record morphology and number of beating cardiomyocytes. The ADP/ATP ratio was determined using ApoSENSOR™ (# K255-200; BioVision, Inc.) with the manufacturer's instructions.
Two other ES cell lines were used in confirmatory studies, the mESC line, D3 (ATCC), and hESC line, MEL-1 (#SCC020; Chemicon). Feeder-free mES D3 cells were cultured on Nunclon surface dishes. The MEL-1 line was maintained on mitotically inactivated mouse embryonic fibroblasts in gelatin-coated culture dishes as recommended by the supplier (Chemicon). All cells were maintained at 37°C with 5% CO2 in DMEM (Gibco), supplemented with 15% heat-inactivated fetal bovine serum (FBS; Hyclone), GIBCO reagents 0.5 mM β-mercaptoethanol, 2 mM l-glutamine, 0.1 mM MEM nonessential amino acid, and 5000 U/ml penicillin/streptomycin, and for MEL-1 cells, insulin–transferrin–selenium (ITS # 41400-045; Life Technologies) is added instead of LIF.
Cell transfection
Transfections were performed in six-well plates at 70–80% confluence with mCAT vector or HuSH shRNA (pGFPCRS) vector containing HO-1 oligonucleotide sequence for optimal suppression of HO-1 (#TR30018; Origene), using FuGENE-HD (#E2311; Promega). After 24–48 h, the cells were subjected to drug selection at either 500 μg/ml G418 (#10131035; Life Technologies) or 4 μg/ml puromycin (#A1113803; Life Technologies) to assess transfection stability. These procedures did not result in measurable cell toxicity or cell death. Antibiotic-resistant colonies were expanded and the transfected cells harvested for EB formation or further measurements. To confirm mCAT expression or HO-1 knockdown, quantitative real-time RT-PCR (qPCR) was used.
MTT assay
Active mitochondrial mass was determined by the reduction of yellow MTT (#11465007001; Sigma/Roche) to the purple formazan by mitochondrial enzymes. ES cells plated at 10,000 per well were analyzed by adding 10 μl of MTT reagent to each well. After incubation in standard or differentiation media at 37°C for 4 h, MTT was removed and the precipitate solubilized at room temperature. Absorbance was determined at 540 nm by a microplate reader (Molecular Devices). MTT reduction was defined as [100 × (absorbance of treated cells) / (absorbance of control cells)].
CRE recombination
For Cre recombinase-stimulated deletion of loxP sites in ES E14/Tfam/loxp derivative cells [generated in our laboratory after (13)], ∼1 million cells were electroporated with 50 μg pBS185 (Cre recombinase expression vector; Life Technologies) using 225 V and 950 μF or 230 V and 500 μF parameters. Cells were replated on ES medium, and 48 h after electroporation, stably transfected cells were selected using G418 started at 300 μg/ml. Seven to nine days later, colonies were counted (to determine the frequency of G418R colonies), isolated in 96-well plates, and expanded in 24-well dishes before freezing. G418 selection was maintained throughout the expansion. Fractions of these clones were put on gelatin-coated plates and tested functionally for loss of puromycin effect or used to extract nucleic acids. A single ES cell clone carrying Tfam deletion was replated for clonal expansion in fresh ES medium containing 1000 U/ml LIF. After expansion, the cells were gradually differentiated as above. For rescue experiment, cells were transfected with pLenti-Tfam vector (#MR203015L2; Origene).
Rat H9c2 cells
Rat cardiomyoblast-derived H9c2 cells (ATCC CRL1446) were grown in Dulbecco's minimal essential medium containing 10% FBS and 1% (v/v) antibiotic mixture at 37°C in a humidified atmosphere containing 5% CO2. Cells were induced to differentiate toward the cardiac-like phenotype of Menard et al. (29). Following decreases in growth factor concentration, myoblasts rapidly become multinucleated myotubes; therefore, subculturing was performed preconfluence to prevent spontaneous transdifferentiation into a skeletal phenotype. Differentiation was induced by culturing myoblasts in DMEM supplemented with 1% fetal calf serum. CORM-2 (25 μM) was added at days 1 and 3, and media replaced every 2 days. H9c2 cardiomyoblasts were transfected with mCAT vector (48) or Sh-HO-1 (Origene) using FuGENE-HD (Roche). Cells transfected with pcDNA3 vector lacking the insert were the controls. Transfected cells were differentiated as above.
RNA extraction
Total cellular RNA was extracted using the RNAqueous-4PCR kit (Ambion) according to the manufacturer's instructions. Samples were treated with DNase I (4 U) for 2 h at 37°C, then inactivated with DNase. The RNA was reverse transcribed using the Reverse Transcription System (Promega).
Real-time qPCR
cDNA was prepared with a high-capacity cDNA archive kit (Applied Biosystems), according to the manufacturer's suggestions. Real-time qPCR primers for HO-1, NRF-1, Polγ, PGC-1α, Tfam, COII, Oct4, Sox2, CS, CRLS-1, Dnm1, Fis1, Pdk1, HK2, CPt1, CDK2, MLC2 (Myl7) and MLC2v, myogenin, and 18S from Applied Biosystems were used with the manufacturer's protocol. All reactions were conducted on StepOnePlus for 40 cycles. The results were analyzed using the 2−ΔΔCt method after being normalized to the level of 18S in each sample.
mtDNA copy number
Total cellular DNA was extracted using the Sigma DNA isolation kit. mtDNA copy number was quantified with real-time PCR on the StepOnePlus Sequence Detector System (AB Applied Biosystems). Primers were designed for cytochrome b (Cyt b) with ABI Probe Design software (Applied Biosystem) and amplifications performed on 10 ng total mtDNA using PCR primers (cyt b-s and cyt b-as). One copy of linearized pGEMT-cyt b vector (47) was used for standard mtDNA quantification. The cyt b probe, 5′ FAM-ttcctccacgaaacaggatcaaa-TAMRA 3′, contained FAM (6-carboxyfluorescein) at the 5′ end as a fluorescent reporter dye and, at the 3′ end, the TAMRA (6-carboxy-tetramethylrhodamine) as a quencher dye selected from a highly conserved region of mouse cyt b gene. Serial dilutions of 105–10 copies of standard cyt b plasmid were prepared for a standard curve. Samples were tested for mtDNA at 1:100 and 1:1000 dilutions. Samples were analyzed in triplicate, and mtDNA copy number/ng DNA determined relative to the standards known mtDNA copies per dilution.
Immunoblot analysis
Cell lysates were prepared from cell preparations (H9c2 or E14 cells) and equal amounts of protein (20–30 μg) separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Membranes were incubated with validated polyclonal rabbit antibodies against COI, COII (#PA5-26688, # A-6404; Life Technologies), myosin regulatory light chain 2, atrial isoform (sc-34488), MLC2v (sc-34490), Mfn2 (sc-50331), Immt (sc-79636), Mef2c (sc-365862), Hand1(sc-374296), Cxn43 (sc-6560), LaminB (sc-374015), BNP (sc-271185), α-actinin(sc-15335), cTnI(sc-20642), porin (sc-390996), MHC6(sc-168676), and OPA1 (sc-30573), all from Santa Cruz. Secondary antibody was 1:5000 horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse immunoglobulin G (Santa Cruz), as appropriate. Blots were developed with ECL, quantified on digitized images in the mid-dynamic range, and expressed relative to Lamin B, tubulin, β-actin, or porin.
Immunohistochemical imaging
Live cells on single slide chambers were incubated in media containing 250 nM MitoTracker green (Molecular Probes, Invitrogen). After 30 min, this was followed by three washes in media and fixation with 4% formaldehyde in 1× PBS. Fixed cells were permeabilized with 0.2% Triton X-100 and immunostained by first digesting EBs with 3 mg/ml collagenase II (Gibco) for 15 min at 37°C. Cells were resuspended in differentiation media, plated onto gelatin-coated single-chamber slides, and incubated overnight at 37°C + 5% CO2. ES cells were fixed in 2% formaldehyde (Sigma) for 1 h, permeabilized for 30 min in 1% (v/v) Triton X-100, placed in blocking solution for 30 min, and incubated with antibody to CS (1:100; Molecular Probes), OCT3/4 (1:100; Santa Cruz), sacomeric α-actinin (Abcam), Gata 4, Mfn2 (Santa Cruz), or Nxk2.5 (Cell Signaling) in PBS containing 1% bovine serum albumin for 2 h at room temperature. After three washes, the cells were incubated with Alexa Fluor secondary antibodies, followed by a nuclear probe (4′,6-diamidino-2-phenylindole; Invitrogen).
Confocal and fluorescence imaging
Fluorescent images were obtained with a Laser Scanning Microscope 510 (Carl Zeiss Microimaging) and analyzed using a Zeiss Laser Scanning Microscope Image Browser or MetaMorph® software (Universal Imaging Corporation). ES cells were visualized on a Hamamatsu Digital Camera C4742-95. Image data were collected using NIS-element 3.1 software (Nikon). Negative controls were used to determine exposure times and prevent false positives.
Mouse studies
Mouse exposures were preapproved by the Duke IACUC. Authentic CO gas was administered to 10- to 12-week-old male B57L6 mice (Jackson) in special plastic chambers at 200 ppm CO in air for 1 h to increase the carboxyhemoglobin level to ∼20% without tissue hypoxia (22). Mice received 1 h of CO per day or room air for three consecutive days and the hearts harvested at day 4. The hearts were fixed in 10% formalin, paraffin embedded, and sectioned. For immunofluorescence techniques, the sections were costained with Sca1 (Abcam; # Ab51317) and CS (Cell Signaling; #14309) or Nkx2.5 (Cell Signaling; #8792) and CS. The positive cells in ventricles and atria were counted and expressed as a percentage of number of nuclei per field (10 random fields per slide and five slides per group) using NIS element 3.2 software.
Statistics
Grouped data are expressed as mean ± standard deviation. Each time point analysis contained at least three samples from three to four experiments per group. Differences between iCORM and CORM-2 were determined by T test, while time-dependent comparisons were made by two-way analysis of variance, followed by Tukey's post-test analysis performed using Sigma Stat and Sigma Plot software (Systat Software, Inc.). p < 0.05 was considered significant, and the statistical tests identified in the figure legends as appropriate.
Supplementary Material
Abbreviations Used
- ANOVA
analysis of variance
- BNP
brain natriuretic peptide
- CDK2
cyclin-dependent kinase 2
- COII
cytochrome c oxidase subunit 2
- CORM-2
CO-releasing molecule
- CPC
cardiac progenitor cell
- Cpt1
carnitine O-palmitoyltransferase 1
- CRLS-1
cardiolipin synthase
- CS
citrate synthase
- cTnI
cardiac troponin I
- Cxn43
connexin 43
- Cyt b
cytochrome b
- DAPI
4′,6-diamidino-2-phenylindole
- DMEM
Dulbecco's modified Eagle's medium
- Dnm1l
dynamin-1-like protein
- ES
embryonic stem
- ETC
electron transport complex
- FBS
fetal bovine serum
- Fis1
mitochondrial fission 1 protein
- Gata4
transcription factor Gata4
- Hand1
heart- and neural crest derivatives-expressed protein 1
- HK2
hexokinase 2
- HO-1/CO
heme oxygenase-1/carbon monoxide
- iCORM
inactivated CORM
- Immt
MICOS complex subunit Mic60
- LIF
leukemia inhibitory factor
- mCAT
mitochondrial catalase
- Mfn2
mitofusin 2
- MitoQ
mito-ubiquinone
- MLC2a
myosin regulatory light chain 2, atrial isoform
- mtDNA
mitochondrial DNA
- Myl2
myosin regulatory light chain 2
- Nkx2.5
homeobox protein Nkx-2.5
- NRF-1
nuclear respiratory factor-1
- Nrf2
nuclear factor erythroid 2-related factor 2
- NRFs
nuclear respiratory factors
- Oct/3/4
POU domain, class 5, transcription factor 1
- Opa1
dynamin-like 120 kDa protein, mitochondrial
- PBS
phosphate-buffered saline
- Pdk1
pyruvate dehydrogenase kinase isozyme 1, mitochondrial
- PGC-1α
peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- Polγ
DNA polymerase subunit gamma-1
- qPCR
quantitative real-time RT-PCR
- ROS
reactive oxygen species
- SD
standard deviation
- Sox2
transcription factor SOX-2
- Tfam
mitochondrial transcription factor A
Acknowledgments
The authors thank Martha Salinas, Lynn Tatro, and Eli Colman for technical assistance. This work was supported by NHLBI: P01HL108801 04.
Author Contributions
H.B.S.: Conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, and final approval of manuscript. C.A.P.: Conception and design, assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript, and financial support. F.Z.: Conception and design, provision of study material, data analysis and interpretation, and final approval of manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Akazawa H. and Komuro I. Roles of cardiac transcription factors in cardiac hypertrophy. Circ Res 92: 1079–1088, 2003 [DOI] [PubMed] [Google Scholar]
- 2.Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, and Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res 31: 1640–1645, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behfar A, Perez-Terzic C, Faustino RS, Arrell DK, Hodgson DM, Yamada S, Puceat M, Niederlander N, Alekseev AE, Zingman LV, and Terzic A. Cardiopoietic programming of embryonic stem cells for tumor-free heart repair. J Exp Med 204: 405–420, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boheler KR, Czyz J, Tweedie D, Yang HT, Anisimov SV, and Wobus AM. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ Res 91: 189–201, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Bolli R, Chugh AR, D'Amario D, Loughran JH, Stoddard MF, Ikram S, Beache GM, Wagner SG, Leri A, Hosoda T, Sanada F, Elmore JB, Goichberg P, Cappetta D, Solankhi NK, Fahsah I, Rokosh DG, Slaughter MS, Kajstura J, and Anversa P. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet 378: 1847–1857, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Bonawitz ND, Clayton DA, and Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell 24: 813–825, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Buggisch M, Ateghang B, Ruhe C, Strobel C, Lange S, Wartenberg M, and Sauer H. Stimulation of ES-cell-derived cardiomyogenesis and neonatal cardiac cell proliferation by reactive oxygen species and NADPH oxidase. J Cell Sci 120: 885–894, 2007 [DOI] [PubMed] [Google Scholar]
- 8.Chen CT, Shih YR, Kuo TK, Lee OK, and Wei YH. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells 26: 960–968, 2008 [DOI] [PubMed] [Google Scholar]
- 9.Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park Do J, Park KS, and Lee HK. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem Biophys Res Commun 348: 1472–1478, 2006 [DOI] [PubMed] [Google Scholar]
- 10.Chung S, Dzeja PP, Faustino RS, Perez-Terzic C, Behfar A, and Terzic A. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat Clin Pract Cardiovasc Med 4 Suppl 1: S60–S67, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crespo FL, Sobrado VR, Gomez L, Cervera AM, and McCreath KJ. Mitochondrial reactive oxygen species mediate cardiomyocyte formation from embryonic stem cells in high glucose. Stem Cells 28: 1132–1142, 2010 [DOI] [PubMed] [Google Scholar]
- 12.Dzeja PP. and Terzic A. Phosphotransfer networks and cellular energetics. J Exp Biol 206: 2039–2047, 2003 [DOI] [PubMed] [Google Scholar]
- 13.Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, and Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet 13: 935–944, 2004 [DOI] [PubMed] [Google Scholar]
- 14.Gissel C, Doss MX, Hippler-Altenburg R, Hescheler J, and Sachinidis A. Generation and characterization of cardiomyocytes under serum-free conditions. Methods Mol Biol 330: 191–219, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Hance N, Ekstrand MI, and Trifunovic A. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum Mol Genet 14: 1775–1783, 2005 [DOI] [PubMed] [Google Scholar]
- 16.Jeong-Yu S. and Clayton DA. Regulation and function of the mitochondrial genome. J Inherit Metab Dis 19: 443–451, 1996 [DOI] [PubMed] [Google Scholar]
- 17.Kanno S, Kim PK, Sallam K, Lei J, Billiar TR, and Shears LL, 2nd., Nitric oxide facilitates cardiomyogenesis in mouse embryonic stem cells. Proc Natl Acad Sci U S A 101: 12277–12281, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karamanlidis G, Bautista-Hernandez V, Fynn-Thompson F, Del Nido P, and Tian R. Impaired mitochondrial biogenesis precedes heart failure in right ventricular hypertrophy in congenital heart disease. Circ Heart Fail 4: 707–713, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lakkisto P, Kyto V, Forsten H, Siren JM, Segersvard H, Voipio-Pulkki LM, Laine M, Pulkki K, and Tikkanen I. Heme oxygenase-1 and carbon monoxide promote neovascularization after myocardial infarction by modulating the expression of HIF-1alpha, SDF-1alpha and VEGF-B. Eur J Pharmacol 635: 156–164, 2010 [DOI] [PubMed] [Google Scholar]
- 20.Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, and Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet 18: 231–236, 1998 [DOI] [PubMed] [Google Scholar]
- 21.Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, Platoshyn O, Yuan JX, Evans S, and Chien KR. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature 433: 647–653, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leary SC, Battersby BJ, Hansford RG, and Moyes CD. Interactions between bioenergetics and mitochondrial biogenesis. Biochim Biophys Acta 1365: 522–530, 1998 [DOI] [PubMed] [Google Scholar]
- 23.Lee S, Kim S, Sun X, Lee JH, and Cho H. Cell cycle-dependent mitochondrial biogenesis and dynamics in mammalian cells. Biochem Biophys Res Commun 2007 [DOI] [PubMed] [Google Scholar]
- 24.Liu T. and O'Rourke B. Enhancing mitochondrial Ca2+ uptake in myocytes from failing hearts restores energy supply and demand matching. Circ Res 103: 279–288, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maines MD. Heme oxygenase: function, multiplicity, regulatory mechanisms, and clinical applications. FASEB J 2: 2557–2568, 1988 [PubMed] [Google Scholar]
- 26.Malecki M, Sabo C, Putzer E, Stampe C, Foorohar A, Quach C, Beauchaine M, Tombokan X, and Anderson M. Recruitment and retention of human autologous CD34+ CD117+ CD133+ bone marrow stem cells to infarcted myocardium followed by directed vasculogenesis: novel strategy for cardiac regeneration. Mol Cell Ther 1: pii , 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandal S, Lindgren AG, Srivastava AS, Clark AT, and Banerjee U. Mitochondrial function controls proliferation and early differentiation potential of embryonic stem cells. Stem Cells 29: 486–495, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDevitt TC, Laflamme MA, and Murry CE. Proliferation of cardiomyocytes derived from human embryonic stem cells is mediated via the IGF/PI 3-kinase/Akt signaling pathway. J Mol Cell Cardiol 39: 865–873, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menard C, Pupier S, Mornet D, Kitzmann M, Nargeot J, and Lory P. Modulation of L-type calcium channel expression during retinoic acid-induced differentiation of H9C2 cardiac cells. J Biol Chem 274: 29063–29070, 1999 [DOI] [PubMed] [Google Scholar]
- 30.Miller-Hance WC, LaCorbiere M, Fuller SJ, Evans SM, Lyons G, Schmidt C, Robbins J, and Chien KR. In vitro chamber specification during embryonic stem cell cardiogenesis. Expression of the ventricular myosin light chain-2 gene is independent of heart tube formation. J Biol Chem 268: 25244–25252, 1993 [PubMed] [Google Scholar]
- 31.Montaigne D, Hurt C, and Neviere R. Mitochondria death/survival signaling pathways in cardiotoxicity induced by anthracyclines and anticancer-targeted therapies. Biochem Res Int 2012: 951539, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moyes CD, Battersby BJ, and Leary SC. Regulation of muscle mitochondrial design. J Exp Biol 201: 299–307, 1998 [PubMed] [Google Scholar]
- 33.Mujoo K, Sharin VG, Bryan NS, Krumenacker JS, Sloan C, Parveen S, Nikonoff LE, Kots AY, and Murad F. Role of nitric oxide signaling components in differentiation of embryonic stem cells into myocardial cells. Proc Natl Acad Sci U S A 105: 18924–18929, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murry CE, Field LJ, and Menasche P. Cell-based cardiac repair: reflections at the 10-year point. Circulation 112: 3174–3183, 2005 [DOI] [PubMed] [Google Scholar]
- 35.Naya FJ, Black BL, Wu H, Bassel-Duby R, Richardson JA, Hill JA, and Olson EN. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med 8: 1303–1309, 2002 [DOI] [PubMed] [Google Scholar]
- 36.Oldroyd KG, Berry C, and Bartunek J. Myocardial repair and regeneration: bone marrow or cardiac stem cells? Mol Ther 20: 1102–1105, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paquin J, Danalache BA, Jankowski M, McCann SM, and Gutkowska J. Oxytocin induces differentiation of P19 embryonic stem cells to cardiomyocytes. Proc Natl Acad Sci U S A 99: 9550–9555, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piantadosi CA, Carraway MS, Babiker A, and Suliman HB. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via Nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circ Res 103: 1232–1240, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivolta MN. and Holley MC. Asymmetric segregation of mitochondria and mortalin correlates with the multi-lineage potential of inner ear sensory cell progenitors in vitro. Brain Res Dev Brain Res 133: 49–56, 2002 [DOI] [PubMed] [Google Scholar]
- 40.Ryter SW, Alam J, and Choi AM. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol Rev 86: 583–650, 2006 [DOI] [PubMed] [Google Scholar]
- 41.San Martin N, Cervera AM, Cordova C, Covarello D, McCreath KJ, and Galvez BG. Mitochondria determine the differentiation potential of cardiac mesoangioblasts. Stem Cells 29: 1064–1074, 2011 [DOI] [PubMed] [Google Scholar]
- 42.Sauer H, Rahimi G, Hescheler J, and Wartenberg M. Role of reactive oxygen species and phosphatidylinositol 3-kinase in cardiomyocyte differentiation of embryonic stem cells. FEBS Lett 476: 218–223, 2000 [DOI] [PubMed] [Google Scholar]
- 43.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev 88: 611–638, 2008 [DOI] [PubMed] [Google Scholar]
- 44.Spitkovsky D, Sasse P, Kolossov E, Bottinger C, Fleischmann BK, Hescheler J, and Wiesner RJ. Activity of complex III of the mitochondrial electron transport chain is essential for early heart muscle cell differentiation. FASEB J 18: 1300–1302, 2004 [DOI] [PubMed] [Google Scholar]
- 45.Sreejit P. and Verma RS. Natural ECM as biomaterial for scaffold based cardiac regeneration using adult bone marrow derived stem cells. Stem Cell Rev 9: 158–171, 2013 [DOI] [PubMed] [Google Scholar]
- 46.St. John JC, Ramalho-Santos J, Gray HL, Petrosko P, Rawe VY, Navara CS, Simerly CR, and Schatten GP. The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. Cloning Stem Cells 7: 141–153, 2005 [DOI] [PubMed] [Google Scholar]
- 47.Suliman HB, Carraway MS, Ali AS, Reynolds CM, Welty-Wolf KE, and Piantadosi CA. The CO/HO system reverses inhibition of mitochondrial biogenesis and prevents murine doxorubicin cardiomyopathy. J Clin Invest 117: 3730–3741, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suliman HB, Carraway MS, Tatro LG, and Piantadosi CA. A new activating role for CO in cardiac mitochondrial biogenesis. J Cell Sci 120: 299–308, 2007 [DOI] [PubMed] [Google Scholar]
- 49.Takahashi T, Lord B, Schulze PC, Fryer RM, Sarang SS, Gullans SR, and Lee RT. Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation 107: 1912–1916, 2003 [DOI] [PubMed] [Google Scholar]
- 50.Tang YL, Tang Y, Zhang YC, Qian K, Shen L, and Phillips MI. Improved graft mesenchymal stem cell survival in ischemic heart with a hypoxia-regulated heme oxygenase-1 vector. J Am Coll Cardiol 46: 1339–1350, 2005 [DOI] [PubMed] [Google Scholar]
- 51.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, and Jones JM. Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147, 1998 [DOI] [PubMed] [Google Scholar]
- 52.Tilemann L, Ishikawa K, Weber T, and Hajjar RJ. Gene therapy for heart failure. Circ Res 110: 777–793, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsubokawa T, Yagi K, Nakanishi C, Zuka M, Nohara A, Ino H, Fujino N, Konno T, Kawashiri MA, Ishibashi-Ueda H, Nagaya N, and Yamagishi M. Impact of anti-apoptotic and anti-oxidative effects of bone marrow mesenchymal stem cells with transient overexpression of heme oxygenase-1 on myocardial ischemia. Am J Physiol Heart Circ Physiol 298: H1320–H1329, 2010 [DOI] [PubMed] [Google Scholar]
- 54.Wagener FA, Volk HD, Willis D, Abraham NG, Soares MP, Adema GJ, and Figdor CG. Different faces of the heme-heme oxygenase system in inflammation. Pharmacol Rev 55: 551–571, 2003 [DOI] [PubMed] [Google Scholar]
- 55.Williams RS. Mitochondrial gene expression in mammalian striated muscle. Evidence that variation in gene dosage is the major regulatory event. J Biol Chem 261: 12390–12394, 1986 [PubMed] [Google Scholar]
- 56.Wobus AM, Kaomei G, Shan J, Wellner MC, Rohwedel J, Ji G, Fleischmann B, Katus HA, Hescheler J, and Franz WM. Retinoic acid accelerates embryonic stem cell-derived cardiac differentiation and enhances development of ventricular cardiomyocytes. J Mol Cell Cardiol 29: 1525–1539, 1997 [DOI] [PubMed] [Google Scholar]
- 57.Zobi F. and Blacque O. Reactivity of 17 e− complex [Re(II)Br4(CO)2]2- with bridging aromatic ligands. Characterization and CO-releasing properties. Dalton Trans 40: 4994–5001, 2011 [DOI] [PubMed] [Google Scholar]
- 58.Zobi F, Blacque O, Jacobs RA, Schaub MC, and Bogdanova AY. 17 e(−) rhenium dicarbonyl CO-releasing molecules on a cobalamin scaffold for biological application. Dalton Trans 41: 370–378, 2012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.