Abstract
Aims: Epigenetic mechanisms, including histone post-translational modifications and DNA methylation, are implicated in the pathogenesis of diabetic nephropathy (DN), but the mediators are not well known. Moreover, although dyslipidemia contributes to DN, epigenetic changes triggered by lipids are unclear. In diabetes, increased expression of 12/15-lipoxygenase (12/15-LO) enhances oxidized lipids such as 12(S)-hydroxyeicosatetraenoic acid [12(S)-HETE], which promote oxidant stress, glomerular and mesangial cell (MC) dysfunction, and fibrosis, and mediate the actions of profibrotic growth factors. We hypothesized that 12/15-LO and its oxidized lipid products can regulate epigenetic mechanisms mediating profibrotic gene expression related to DN. Results: 12(S)-HETE increased profibrotic gene expression and enrichment of permissive histone lysine modifications at their promoters in MCs. 12(S)-HETE also increased protein levels of SET7, a histone H3 lysine 4 methyltransferase, and promoted its nuclear translocation and enrichment at profibrotic gene promoters. Furthermore, SET7 (Setd7) gene silencing inhibited 12(S)-HETE-induced profibrotic gene expression. 12/15-LO (Alox15) gene silencing or genetic knockout inhibited transforming growth factor-β1 (TGF-β1)-induced expression of Setd7 and profibrotic genes and histone modifications in MCs. Furthermore, 12/15-LO knockout in mice ameliorated key features of DN and abrogated increases in renal SET7 and profibrotic genes. Additionally, 12/15-LO siRNAs in vivo blocked increases in renal SET7 and profibrotic genes in diabetic mice. Innovation and Conclusion: These novel results demonstrate for the first time that 12/15-LO-derived oxidized lipids regulate histone modifications associated with profibrotic gene expression in MCs, and 12/15-LO can mediate similar actions of TGF-β1 and diabetes. Targeting 12/15-LO might be a useful strategy to inhibit key epigenetic mechanisms involved in DN. Antioxid. Redox Signal. 24, 361–375.
Introduction
Diabetic nephropathy (DN) is a major renal complication that can lead to end-stage kidney disease and increased mortality (13). Mesangial cell (MC) hypertrophy, fibrosis, and oxidant stress mediated by high glucose (HG) and downstream growth factors such as transforming growth factor-β (TGF-β) are key events in the progression of DN (16, 28, 46, 50, 63). TGF-β regulates the expression of several profibrotic and cell cycle genes through activation of key transcription factors such as Smad and E-box-binding proteins in MCs and other renal cells (18, 25, 29, 49). Emerging evidence shows that epigenetic mechanisms, such as chromatin histone post-translational modifications (PTMs) (24) and DNA methylation (14), play important roles in the regulation of profibrotic and inflammatory genes related to the pathogenesis of DN (44, 53). These studies demonstrated that TGF-β promotes changes in histone lysine modifications at profibrotic gene promoters and also mediates similar epigenetic changes induced by HG. These include enrichment of permissive histone modifications associated with transcription activation (histone H3 lysine-4 methylation [H3K4me] and histone H3 lysine-9 acetylation [H3K9Ac]), inhibition of repressive histone modifications (H3K9me), and increased expression, as well as enrichment of SET7 (Setd7), a key histone H3K4 methyltransferase, at profibrotic gene promoters in MCs (52, 62). Changes in certain histone modifications and corresponding writer enzymes (methyltransferases and acetylases) were also shown in kidneys and glomeruli from experimental models of DN (23, 43, 62). Treatment with histone deacetylase (HDAC) inhibitors blocked progression of DN in mice (1, 35). Furthermore, treatment of diabetic mice with the angiotensin II (Ang II) type I receptor (AT1R) blocker, losartan, ameliorated proteinuria and pathological gene expression in kidneys, but reversed only some of the diabetes-induced changes in various histone modifications (43). Together, these studies support a key role for epigenetic mechanisms in DN and suggest that inhibition of these mechanisms might improve the efficacy of currently available therapies. However, the precise mechanisms and mediators involved in epigenetic regulation of profibrotic genes by TGF-β and diabetes are still unclear.
Innovation.
12/15-Lipoxygenase (12/15-LO) plays key roles in fibrosis and hypertrophy induced by transforming growth factor beta1 (TGF-β1) in renal mesangial cells related to diabetic nephropathy. In this study we report the involvement of epigenetic mechanisms in these actions of 12/15-LO. We demonstrated that oxidized lipids derived from 12/15-LO can enhance active histone modifications and regulate H3K4-methyl transferase SET7 to promote fibrotic gene expression. 12/15-LO could also mediate similar actions of TGF-β1. Genetic knockout or siRNA targeting 12/15-LO inhibited renal glomerular dysfunction, fibrosis and SET7 expression in diabetic mice. Thus, targeting12/15-LO could be a novel approach to reverse epigenetic mechanisms mediating diabetic renal dysfunction.
Diabetes and obesity are associated with increased levels of oxidized lipids derived from the metabolism of polyunsaturated fatty acids (2, 8, 15, 32, 51). These oxidized lipids can promote oxidant stress, inflammation, fibrosis, and hypertrophy associated with vascular complications (8, 15, 32, 40, 45, 47, 57). However, it is not known if they can also regulate profibrotic genes via epigenetic mechanisms. Previous studies showed that increased expression and activation of 12/15-lipoxygenase (12/15-LO), the mouse homolog of human 15-LO, play an important role in increasing oxidized lipids in diabetes (8, 15, 32, 40, 47). 12/15-LO is a nonheme iron-containing enzyme that metabolizes arachidonic acid to generate oxidized lipids such as 12(S)-hydroxyeicosatetraenoic acid [12(S)-HETE] and 15-HETE (10, 33, 60). 12/15-LO has been implicated in several diabetic complications, including DN (8, 15, 32, 36, 40, 54, 57, 61). 12(S)-HETE levels are increased by HG and TGF-β in MCs in vitro and in experimental DN (15, 40, 57). 12/15-LO-derived 12(S)-HETE can promote hypertrophy and expression of profibrotic and proinflammatory genes, TGF-β, and AT1R in MCs (15, 40, 57–59). 12(S)-HETE regulates gene expression via CREB and Smad transcription factors and can augment similar actions of TGF-β in MCs (22, 40). 12/15-LO deficiency attenuated TGF-β signaling in MCs, further confirming a cross talk between 12/15-LO and TGF-β (21). Furthermore, administration of a cholesterol-tagged 12/15-LO siRNA ameliorated glomerular dysfunction and expression of renal TGF-β and profibrotic genes in diabetic mice (61). These results demonstrate the pathological role of the 12/15-LO pathway in the progression of DN via direct actions as well as cross talk with HG or diabetes-induced TGF-β.
However, it is unknown whether 12/15-LO activation and its oxidized lipid products such as 12(S)-HETE can enrich permissive histone modifications at profibrotic genes in MCs and whether 12/15-LO mediates similar epigenetic changes induced by TGF-β and diabetes. In this study, we demonstrated that 12(S)-HETE treatment directly induces enrichment of histone lysine modifications associated with transcriptional activation at profibrotic gene promoters and that it also regulates the expression of SET7 and promotes its nuclear translocation, as well as enrichment, at profibrotic gene promoters in MCs. Furthermore, using loss-of-function approaches with 12/15-LO knockout (LOKO) mice and siRNAs, we showed that 12/15-LO can mediate TGF-β-induced histone modifications in vitro in MCs and diabetes-induced renal changes in vivo in mouse models of DN.
Results
12(S)-HETE-induced profibrotic gene expression is associated with enhanced enrichment of promoter H3K9Ac and H3K4me in rat mesangial cell
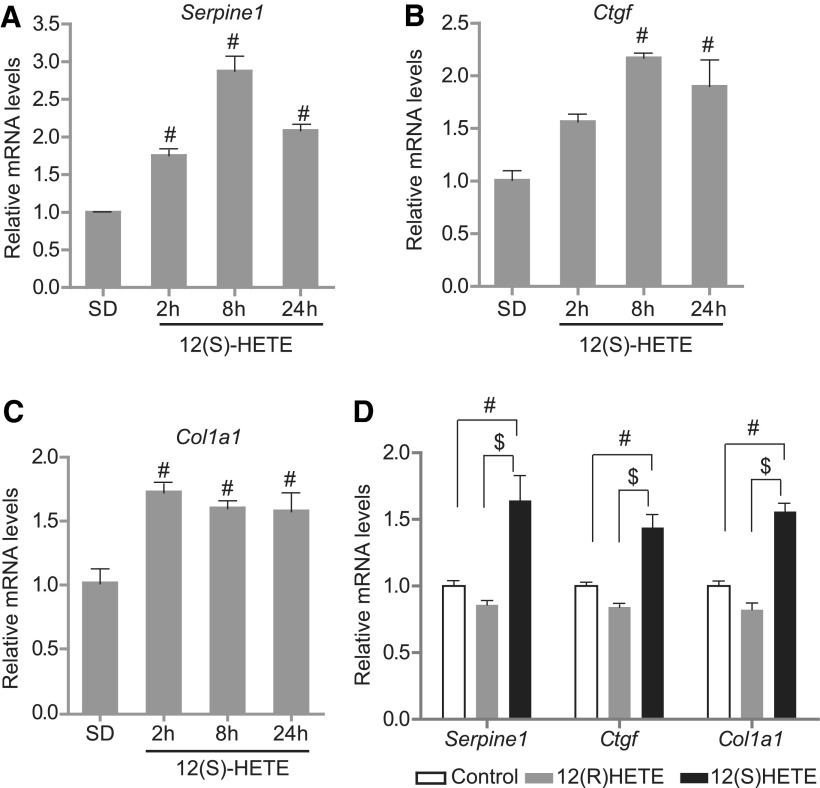
We first tested whether 12(S)-HETE-induced profibrotic gene expression in MCs is associated with alteration in key promoter histone modifications. We treated serum-depleted (SD) rat mesangial cells (RMC) with 12(S)-HETE (10−7 M) or vehicle (ethanol) and analyzed the expression of profibrotic genes using real-time quantitative polymerase chain reactions (RT-qPCRs) and histone modifications at their promoters using chromatin immunoprecipitation (ChIP) assays with specific antibodies. Results showed that 12(S)-HETE significantly upregulated Serpine1, Ctgf, and Col1a1 mRNAs from 2 to 24 h compared with vehicle controls (Fig. 1A–C). This effect was specific to 12(S)-HETE since its stereoisomer, 12(R)-HETE, which is not an LO metabolite, had no effect on the expression of these profibrotic genes (Fig. 1D). We have also confirmed increased levels of Serpine1 (plasminogen activator inhibitor type 1) protein levels after treatment with 12(S)-HETE by Western blotting (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars). These results confirmed that 12/15-LO products upregulate profibrotic gene expression in MCs related to the pathogenesis of DN.
FIG. 1.
12(S)-HETE induces profibrotic genes in RMCs. RMCs were SD and treated with vehicle ethanol or 12(S)-HETE (10−7 M) for the indicated time points, and expression of profibrotic genes, including Serpine1 (A), Ctgf (B), and Col1a1 (C), was determined by RT-qPCR. (D) SD RMCs were stimulated with vehicle, 12(S)-HETE (10−7 M), or 12(R)-HETE (10−7 M) for 2 h, and profibrotic gene expression was analyzed by RT-qPCR. Data represent mean ± SEM of three independent experiments. #p < 0.01 versus SD vehicle-treated control, $p < 0.01 versus 12(R)-HETE. 12(S)-HETE, 12(S)-hydroxyeicosatetraenoic acid; RMC, rat mesangial cell; RT-qPCR, real-time quantitative polymerase chain reaction; SD, serum depleted; SEM, standard error of the mean.
Furthermore, ChIP assays showed that 12(S)-HETE induced significant enrichment of permissive histone modifications, including H3K9Ac (Fig. 2A–C), H3K4me1 (Fig. 2D–F), and H3K4me3 (Fig. 2G–I), at the profibrotic gene promoters compared with controls (SD) in a time-dependent manner. However, no significant changes were seen at the cyclophilin A (CypA, Ppia) promoter, tested as a control for specificity (Supplementary Fig. S2A–C). 12(S)-HETE also had no effect on H3K4me2 (Supplementary Fig. S2D, E). These results suggest that the promoter enrichment of chromatin marks known to be associated with active gene expression, namely H3K9Ac, H3K4me1, and H3K4me3, may be involved in upregulation of profibrotic genes by oxidized lipids such as 12(S)-HETE in MCs.
FIG. 2.
12(S)-HETE induces permissive histone modifications (H3K9Ac and H3K4me) at profibrotic gene promoters. Bar graphs showing relative enrichment of permissive histone modifications at indicated gene promoters induced by 12(S)-HETE compared with SD controls at different time points. (A–C) H3K9Ac; (D–F) H3K4me1; (G–I) H3K4me3. RMCs were treated with 12(S)-HETE (10−7 M) for different time points and ChIP assays were performed with H3K9Ac, H3K4me1, and H3K4me3 antibodies. ChIP-enriched DNA was amplified by qPCR using primers spanning the promoter regions of indicated profibrotic genes. Results were normalized to input and expressed as fold over SD controls. Data represent mean ± SEM, #p < 0.01 versus SD, n = 3. ChIP, chromatin immunoprecipitation; H3K9Ac, histone H3 lysine-9 acetylation; H3K4me1, histone H3 lysine-4 monomethylation; H3K4me3, histone H3 lysine-4 trimethylation.
SET7 regulation by 12(S)-HETE in RMC
Because 12(S)-HETE enhanced H3K4me1 levels at profibrotic gene promoters, we next examined the role of the related methyltransferase SET7. SET7 regulates H3K4me1 in mammalian cells and evidence shows that its expression, nuclear translocation, and recruitment at key pathological genes are increased under diabetic conditions (9, 27, 37, 52). However, its regulation by oxidized lipids has not been studied. Our results showed that 12(S)-HETE treatment increased SET7 protein levels in MCs (Fig. 3A). Furthermore, immunoblotting of nuclear extracts (Fig. 3B) and immunofluorescence (IF) studies (Fig. 3C) showed that 12(S)-HETE enhanced SET7 nuclear translocation in RMC. ChIP assays showed that 12(S)-HETE also increased SET7 occupancy at the Serpine1 and Ctgf promoters in RMC (Fig. 3D, E). To further confirm the role of SET7, we transfected RMC with siRNAs targeting SET7 (siSET7) or control siNTC oligonucleotides and examined the effect of SET7 gene silencing on 12(S)-HETE-induced gene expression. Results showed that SET7 gene silencing (Fig. 3F) significantly inhibited 12(S)-HETE-induced profibrotic gene expression (Fig. 3G–I). Thus, for the first time, these data demonstrate direct regulation of SET7 expression and activation by oxidized lipids in mammalian cells, indicating a role for 12/15-LO activation in triggering epigenetic mechanisms involved in DN development.
FIG. 3.
Regulation of SET7 by 12(S)-HETE in MCs. (A) Quiescent RMCs were treated with 12(S)-HETE (10−7 M) and at indicated time points, SET7 protein levels in whole cell lysates were determined by immunoblotting with SET7 antibody. Actin was used as an internal control. Results are representative of two separate experiments. Intensity of SET7 bands was quantified using Bio-Rad GS-900 densitometer and expressed as fold over control (SD) cells (shown below each lane). Results are representative of two separate experiments. (B) Quiescent RMCs were treated with 12(S)-HETE (10−7 M), and at indicated time points, SET7 levels in nuclear fractions were determined by immunoblotting with SET7 antibody. Lamin B was used as an internal control. Intensity of nuclear SET7 bands was quantified using Bio-Rad GS-900 densitometer and expressed as fold over control (SD) cells (shown below each lane). Results are representative of two separate experiments. (C) RMCs were treated with 12(S)-HETE (10−7 M) or equal volume of vehicle (ethanol) for 6 h, and nuclear localization of SET7 was determined by immunofluorescence staining with SET7 antibody, followed by rabbit anti-mouse antibody conjugated with FITC (green) (upper panels). Blue color in lower panels indicates nuclear staining with DAPI. (D, E) RMCs were treated with 12(S)-HETE (10−7 M) for indicated time points and ChIP assays were performed with SET7 antibody to evaluate its enrichment at Serpine1 (D) and Ctgf promoters (E). ChIP DNA was analyzed by qPCR and results normalized to input are expressed as fold over SD controls. Data represent mean ± SEM. #p < 0.01 versus SD. n = 3. (F) SET7 gene silencing using siRNAs in RMCs. RMCs were transfected with siRNAs targeting SET7 (siSET7) or nontargeting control (siNTC) oligonucleotides. Gene expression was analyzed 96 h after transfection. Data represent mean ± SEM. *p < 0.05 versus siNTC. n = 3. (G–I) Effect of SET7 gene silencing on 12(S)-HETE-induced fibrotic genes. RMCs were transfected with siSET7 or siNTC and 96 h post-transfection, stimulated with vehicle (Ctrl) or 12(S)-HETE (10−7 M) for 8 h, and expression of indicated genes was analyzed by RT-qPCR. Results are expressed as fold over siNTC-Ctrl. Data represent mean ± SEM. *p < 0.05 versus siNTC-Ctrl and $p < 0.05 versus siSET7-Ctrl. n = 3. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
12/15-LO gene silencing attenuates TGF-β−induced SET7 and profibrotic gene expression in RMC
We have previously demonstrated that 12/15-LO and TGF-β regulate each other and that such cross talk between them augments gene expression involved in DN (22). Since evidence also shows that TGF-β-induced SET7 can mediate profibrotic gene expression (52), we next examined whether 12/15-LO regulates TGF-β-induced SET7 by using loss-of-function approaches. RMCs were transfected with siRNAs targeting 12-LO (si12-LO) or control siRNA (siNTC), and TGF-β (10 ng/ml, for 6 h)-induced gene expression was analyzed. Transfection with si12-LO significantly reduced 12/15-LO (Alox15) gene expression (Fig. 4A) and also inhibited both basal and TGF-β-induced Set7 (Setd7) mRNA expression (Fig. 4B) compared with siNTC-transfected RMCs. Furthermore, TGF-β-induced expression of Serpine1 and Ctgf was significantly attenuated in RMCs transfected with si12-LO (Fig. 4C, D). These results demonstrate that 12/15-LO can regulate TGF-β-induced SET7 and profibrotic gene expression.
FIG. 4.
siRNAs targeting 12/15-LO attenuated TGF-β-induced profibrotic genes and SET7 expression. (A) RMCs were transfected with 0.3 μg of siRNA targeting 12/15-LO (si12-LO) or control (siNTC) oligonucleotides, and 48 h later, 12/15-LO (Alox15) mRNA expression was determined by RT-qPCR. 12/15-LO mRNA levels were expressed as fold over siNTC (mean ± SEM; #p < 0.01 vs. siNTC, unpaired t-test, n = 3). (B–D) RMCs were transfected with 0.3 μg of si12-LO or siNTC oligonucleotides, 48 h post-transfection, SD for 24 h, and then treated with TGF-β (10 ng/ml) for 6 h. Expression of Setd7 (B) and profibrotic genes (C, D) was determined by RT-qPCR. β-Actin was used as internal control. The results are expressed as fold over siNTC control (mean ± SEM, n = 3. #p < 0.01 vs. si-NTC or si12-LO, $p < 0.01 vs. siNTC+TGF-β). 12/15-LO, 12/15-lipoxygenase; TGF-β, transforming growth factor-β.
TGF-β-induced profibrotic gene expression and enrichment of key histone modifications at their promoters are attenuated in MCs derived from LOKO mice
To further confirm the role of 12/15-LO in TGF-β-induced epigenetic mechanisms, we tested cells from 12/15-LOKO mice (21). We treated MCs isolated from wild-type mice (WT-MC) and LOKO mice (LOKO-MC) with TGF-β (10 ng/ml) and analyzed the expression of TGF-β target genes. Results showed that TGF-β treatment significantly increased expression of profibrotic genes, Serpine1 and Ctgf, as well as Setd7 in WT-MC, but these increases were significantly attenuated in LOKO-MC (Fig. 5A–C). In contrast, Ppia expression was not affected in these treatments (Fig. 5D). Furthermore, ChIP assays revealed that TGF-β (10 ng/ml, for 2 h) increased H3K9Ac and H3K4me1 enrichment at the Serpine1, Ctgf, and Col1a1 promoters in WT-MC, but these events were significantly attenuated or completely blocked in LOKO-MC (Fig. 5E–J). These results demonstrate that the 12/15-LO pathway plays a mediatory role in TGF-β-induced epigenetic histone modifications associated with the upregulation of profibrotic gene expression in MCs.
FIG. 5.
TGF-β-induced profibrotic gene expression and permissive histone modifications at these promoters were attenuated in MCs from 12/15-LOKO mice. (A–D) TGF-β-induced profibrotic gene expression was attenuated in MMCs from LOKO mice. MCs cultured from WT and LOKO mice were SD for 24 h, and then treated with TGF-β (10 ng/ml) from 0.5 to 4 h. Expression of Serpine1 (A), Ctgf (B), Setd7 (C), and Ppia (D) was analyzed by RT-qPCR using β-actin as internal control. The results are expressed as fold over SD (mean ± SEM, n = 3. #p < 0.01 vs. SD). (E–J) TGF-β-induced enrichment of H3K9Ac (E–G) and H3K4me1 (H–J) at profibrotic gene promoters was attenuated in MCs from LOKO mice. Quiescent MCs from WT and LOKO mice were treated with TGF-β (10 ng/ml) for 2 h, ChIP assays were performed as described in Figure 2 with indicated antibodies, and ChIP-enriched DNA samples were amplified with primers specific to the profibrotic gene promoters. Results normalized to input DNA are expressed as fold enrichment over untreated WT MC (mean ± SEM, n = 3; *p < 0.05, #p < 0.01, $p < 0.01). LOKO, LO knockout; MMC, mouse mesangial cell; WT, wild-type.
12/15-LO deficiency in vivo attenuates increased expression of SET7 and profibrotic genes and ameliorates key features of DN in mice
Next, we examined the in vivo relevance of these results using WT and LOKO mice rendered diabetic by sequential injections of streptozotocin (STZ) as described earlier (61). In parallel, control mice were injected with citrate buffer. Control (no streptozotocin [NS]) and diabetic (STZ) mice were monitored up to 16 weeks. Diabetic WT as well as diabetic LOKO mice exhibited hyperglycemia, polyuria, and some weight loss, but no significant differences were observed in these and other physiological or biochemical parameters between the two groups (Tables 1 and 2). Diabetic WT and LOKO mice showed significant increases in proteinuria at 12 and 16 weeks relative to nondiabetic controls. However, the degree of proteinuria was relatively less in diabetic LOKO mice compared with diabetic WT mice at both time points (Table 2). Whereas the albumin-to-creatinine ratio (ACR) was significantly increased in diabetic WT mice at 12 and 16 weeks postdiabetes (Fig. 6A), in contrast, ACR was significantly increased only at 12 weeks, but not at 16 weeks, in diabetic LOKO mice (Fig. 6A). Diabetic WT mice exhibited kidney hypertrophy, including increased kidney-to-body weight ratio (Table 1) and mesangial expansion characterized by increased periodic acid-Schiff-positive matrix accumulation, as well as increased glomerular area, but these parameters were significantly ameliorated in diabetic LOKO mice compared with diabetic WT mice (Fig. 6B, C).
Table 1.
Physiological Parameters in Experimental Groups
| Parameter | WT+NS | WT+STZ | LOKO+NS | LOKO+STZ |
|---|---|---|---|---|
| Fasting glucose 1 week (mg/dl) | 144 ± 15.0 | 331 ± 46.4a | 156 ± 2.1 | 263 ± 58.2 |
| Fasting glucose 12 weeks (mg/dl) | 197 ± 13.5 | 558 ± 14.3b | 156 ± 6.8 | 439 ± 72.3c |
| Fasting glucose 16 weeks (mg/dl) | 140 ± 13.9 | 539 ± 28.5b | 155 ± 8.57 | 617 ± 58.4d |
| Serum creatinine 16 weeks (mg/dl) | 0.20 ± 0.01 | 0.14 ± 0.03 | 0.20 ± 0.01 | 0.17 ± 0.01 |
| Body weight 1 week (g) | 24.77 ± 0.82 | 22.57 ± 0.58 | 25.27 ± 2.07 | 25.47 ± 0.29 |
| Body weight 12 weeks (g) | 30.00 ± 1.53 | 25.67 ± 1.45 | 28.33 ± 2.96 | 28.00 ± 1.15 |
| Body weight 16 weeks (g) | 31.00 ± 1.53 | 26.00 ± 1.53 | 30.00 ± 3.21 | 29.33 ± 1.33 |
| Kidney weight 16 weeks (g) | 0.412 ± 0.013 | 0.471 ± 0.061 | 0.436 ± 0.082 | 0.515 ± 0.056 |
| K/B weight (%) | 1.330 ± 0.059 | 1.810 ± 0.076e | 1.456 ± 0.114 | 1.742 ± 0.054f,g |
Values are mean ± SE; n = 3 per group. C57BL/6 and LOKO mice were injected with STZ for 5 days. Nondiabetic groups of C57BL/6 and LOKO mice were injected with citrate buffer alone. Parameters were first measured at 1 week after establishment of hyperglycemia.
p < 0.05 versus WT+NS.
p < 0.001 versus WT+NS.
p < 0.01 versus LOKO+NS.
p < 0.001 versus LOKO+NS.
p < 0.01 versus WT+NS.
p < 0.05 versus LOKO+NS.
p < 0.05 versus WT+STZ.
LOKO, lipoxygenase knockout; NS, no streptozotocin; SE, standard error; STZ, streptozotocin; WT, wild-type.
Table 2.
Urinary Parameters in Experimental Groups
| Parameter | WT+NS | WT+STZ | LOKO+NS | LOKO+STZ |
|---|---|---|---|---|
| Urinary volume 1 week (ml) | 2.23 ± 0.38 | 19.83 ± 3.11a | 4.60 ± 0.35 | 12.50 ± 7.17 |
| Proteinuria 1 week (mg) | 23.68 ± 3.10 | 31.57 ± 2.96 | 22.76 ± 4.74 | 30.86 ± 4.03 |
| Urinary CR 1 week (mg/dl) | 14.71 ± 0.98 | 7.40 ± 0.86b | 11.07 ± 0.60 | 10.03 ± 2.15 |
| Urinary volume 12 weeks (ml) | 1.83 ± 0.88 | 27.67 ± 1.20b | 3.00 ± 0.81 | 28.67 ± 8.37c |
| Proteinuria 12 weeks (mg) | 22.45 ± 5.50 | 75.92 ± 9.61a | 24.79 ± 6.42 | 60.95 ± 10.04c |
| Urinary CR 12 weeks (mg/dl) | 13.95 ± 1.06 | 5.90 ± 0.33b | 12.49 ± 0.66 | 7.12 ± 1.50 |
| Urinary volume 16 weeks (ml) | 1.40 ± 0.64 | 35.33 ± 0.88b | 3.33 ± 0.38 | 22.67 ± 5.90c |
| Proteinuria 16 weeks (mg) | 27.88 ± 5.24 | 80.46 ± 13.54a | 21.48 ± 1.99 | 66.14 ± 11.14c |
| Urinary CR 16 weeks (mg/dl) | 17.77 ± 1.60 | 7.40 ± 0.86b | 10.59 ± 1.08 | 10.03 ± 2.15 |
Values are mean ± SE; n = 3 per group. C57BL/6 and LOKO mice were injected with STZ for 5 days. Nondiabetic groups of C57BL/6 and LOKO mice were injected with citrate buffer alone. Parameters were first measured at 1 week after establishment of hyperglycemia.
p < 0.01 versus WT+NS.
p < 0.001 versus WT+NS.
p < 0.05 versus LOKO+NS.
CR, creatinine.
FIG. 6.
Proteinuria, glomerular hypertrophy, and ECM deposition were less pronounced in diabetic LOKO mice. (A) Urine albumin/creatinine ratio (μg/mg) in urine samples from diabetic (STZ) and control (NS) WT and LOKO mice at 12 and 16 weeks postdiabetes. Biochemical assays were performed as described in the Materials and Methods section. *p < 0.05 versus LOKO+NS; #p < 0.01 versus WT+NS, Data represent mean ± SEM (n = 3). (B) Representative images of PAS staining and (C) quantification of glomerular area and mesangial expansion in kidneys from diabetic (STZ) and control (NS) WT and LOKO mice. Data represent mean ± SEM (n = 3). #p < 0.01 versus WT+NS or LOKO+NS, $p < 0.01 versus WT-STZ. (D) Representative images of indicated staining in kidney sections from WT and LOKO mice at 16 weeks postdiabetes. Images of H&E staining (1–4), PASM staining (5–8), Masson's trichrome staining (9–12), and TGF-β immunostaining in kidney sections from WT+NS (1, 5, 9, 13), WT+STZ (2, 6, 10, 14), LOKO+NS (3, 7, 11, 15), and LOKO+STZ mice (4, 8, 12, 16). (E, F) Quantification of Masson's trichrome-positive area and (E) and TGF-β-positive area (F) in the glomeruli using Image-Pro plus software and expressed as fold over WT+NS (n = 6). Values are mean ± SEM. #p < 0.01 versus WT+NS, $p < 0.01 versus WT-STZ. ECM, extracellular matrix; H&E, hematoxylin and eosin; NS, no streptozotocin; PAS, periodic acid-Schiff; PASM, periodic acid-silver methenamine; STZ, streptozotocin.
Furthermore, immunohistochemical (IHC) analysis of kidney sections revealed increased glomerular expansion (Fig. 6D, panel 2), extracellular matrix (ECM) accumulation and basement membrane thickening (Fig. 6D, panel 6), interstitial fibrosis and glomerulosclerosis (Fig. 6D, panel 10), and glomerular expression of TGF-β (Fig. 6D, panel 14) in diabetic WT mice (WT+STZ) versus controls (WT+NS). However, all these pathological features of DN were attenuated in the kidneys of diabetic LOKO mice (LOKO+STZ) compared with diabetic WT mice (16 weeks postdiabetes) (Fig. 6D, panels 4, 8, 12, and 16). Figure 6E and F shows quantitative analyses of trichrome staining (interstitial fibrosis) and TGF-β staining, respectively. Thus, key pathological characteristics of DN were clearly attenuated in diabetic LOKO mice, further supporting the notion that 12/15-LO deficiency in vivo confers renal protection during diabetes. These results are also consistent with the mRNA results in MCs derived from the LOKO mice (Figs. 4 and 5), demonstrating that the 12/15-LO gene knockout attenuates diabetes-induced TGF-β expression as well as associated mesangial matrix gene expression.
Diabetes increases renal expression of profibrotic genes and SET7 in WT mice, but these effects are attenuated in 12/15-LO-deficient mice
Next, we examined if diabetes-induced upregulation of profibrotic genes is associated with parallel increases in SET7 expression. RT-qPCR results showed that expression of several profibrotic genes was significantly increased in glomeruli from diabetic WT mice relative to nondiabetic WT controls, but these increases were significantly attenuated or abrogated in diabetic LOKO mice compared with diabetic WT mice (Fig. 7A). Furthermore, Setd7 mRNA levels were also significantly increased in kidneys of diabetic WT mice (cortex and glomeruli), but this effect was abrogated in diabetic LOKO mice (Fig. 7B). These changes in gene expression were further confirmed by Western blotting using protein samples from glomeruli (Fig. 7C) and renal cortex (Fig. 7D) of these mice. Some increase in basal protein levels observed in the LOKO mice, especially in the cortical samples, could be due to heterogeneity of these tissues. Overall, these results demonstrate that the 12/15LO pathway plays an important role in vivo in the regulation of SET7 and profibrotic genes in DN.
FIG. 7.
Increases in profibrotic genes and SET7 were attenuated in diabetic LOKO mice and diabetic mice treated with si12-LO. (A) Expression of profibrotic genes in glomeruli from diabetic (STZ) and control (NS) WT and LOKO mice. Kidneys from two mice were pooled to collect glomeruli from each group of mice, total RNA was extracted and profibrotic gene expression was determined by RT-qPCR. Profibrotic gene expression was normalized using β-actin and expressed as fold over nondiabetic control. Data represent mean ± SEM (n = 3). #p < 0.01 versus WT or LOKO, $p < 0.01 versus WT+STZ. (B) Total RNA isolated from renal cortex or pooled glomeruli was used to analyze expression of Setd7 mRNA by RT-qPCR from diabetic (STZ) and control (NS) WT and LOKO mice. Values are mean ± SEM (n = 3). *p < 0.05, $p < 0.01. (C, D) Total protein lysates from pooled glomeruli (C) and renal cortex (D) were immunoblotted with indicated antibodies. Results show that the increases in SET7 and profibrotic Serpine1 expression seen in diabetic WT mice are absent in diabetic LOKO mice compared with respective controls. (E) Expression of Setd7 mRNA in renal cortex from control nondiabetic mice (NS), control diabetic mice (STZ), diabetic mice injected with 12/15-LO siRNA (si12-LO+STZ), or control siRNA (mismatched oligos [MM], siMM+STZ). Total RNA from indicated mice was analyzed by RT-qPCR and Setd7 expression was expressed as fold over nondiabetic control (NS). Values are mean ± SEM (n = 4). *p < 0.05, #p < 0.01 versus NS. (F) Protein lysates from indicated mice were immunoblotted with SET7, Serpine1, and β-actin antibodies.
To further explore the functional role of 12/15-LO in the regulation of SET7 in DN, we examined SET7 expression in diabetic WT mice treated in vivo with cholesterol-tagged si12-LO or control mismatched siRNA (siMM). Our previous studies showed that treatment of diabetic mice with this siRNA ameliorated features of DN and inhibited profibrotic gene expression in vivo (61). Renal cortex samples from these 12-LO siRNA and control siRNA-treated mice (61) were analyzed by RT-qPCR and Western blotting. Results showed that Setd7 mRNA levels were increased in kidneys of diabetic mice (STZ) compared with nondiabetic controls (NS) (Fig. 7E), but these diabetes-induced increases in Setd7 expression were significantly ameliorated in diabetic mice treated with si12-LO (si12-LO+STZ) compared with nontargeting mismatched control oligos (siMM+STZ) (Fig. 7E). Furthermore, diabetes induced upregulation of SET7 and profibrotic Serpine1 protein levels were also ameliorated by si12-LO treatment (Fig. 7F). Because, si12-LO also ameliorated DN in these mice (61), these results indicate that 12/15-LO could be a potential therapeutic target to inhibit diabetes-induced epigenetic changes involved in profibrotic gene expression and DN progression.
Discussion
The rapid rise in diabetes and obesity has led to significantly greater incidence and severity of DN. Since changes in the environment and lifestyles are likely key contributors, epigenetic mechanisms are increasingly implicated (12, 18). However, although dyslipidemia is involved in DN, the contribution of oxidized lipids to epigenetic mechanisms in DN has not been evaluated. In this study, we report for the first time that oxidized lipids such as 12(S)-HETE from 12/15-LO can directly promote epigenetic histone modification changes involved in profibrotic gene expression, at least in part via SET7, and that 12/15-LO also mediates similar effects elicited by TGF-β in MCs. Furthermore, 12/15-LO deficiency in mice ameliorates DN in vivo and inhibits diabetes-induced expression of TGF-β and profibrotic genes, as well as SET7. These results suggest an important role for 12/15-LO and its key lipid metabolites in regulating epigenetic mechanisms involved in the pathogenesis of DN.
We observed that the oxidized lipid 12(S)-HETE increased the enrichment of histone lysine modifications associated with active gene expression such as H3K9Ac and H3K4me at profibrotic gene promoters in MCs. This suggests that 12(S)-HETE can promote open chromatin formation accessible to transcription factors involved in the expression of these genes. We previously reported that 12(S)-HETE could regulate Smad activity (22). The ChIP primers we used in the study were near Smad binding sites, suggesting 12(S)-HETE might promote Smad-induced gene expression by acetylation-mediated chromatin relaxation.
Several lines of evidence support a key role for the 12/15-LO pathway of arachidonate metabolism in the pathogenesis of DN. 12/15-LO is highly expressed in the kidney and is induced under diabetic conditions, both in vitro in MCs and in vivo in type 1 and type 2 diabetes models (15, 40, 57, 58). 12/15-LO and its lipid products contribute to cellular hypertrophy and regulate the expression of profibrotic, proinflammatory, and growth factor receptor genes (11, 40, 58, 59, 61). Targeting 12/15-LO expression using cholesterol-tagged siRNAs alleviated ECM expansion, glomerular hypertrophy, and albuminuria in kidneys of type 1 diabetic mice (61). Another study showed that a chemical inhibitor of 12/15-LO ameliorated proteinuria and renal hypertrophy in type 2 diabetic rats (58). In the current study, we demonstrate for the first time that genetic knockdown of 12/15-LO in mice (LOKO mice) significantly attenuated proteinuria, glomerular hypertrophy, and fibrosis, providing further clear support for the in vivo involvement of 12/15-LO in DN. Reactive oxygen species (ROS) and lipid peroxidation are important in the pathogenesis of DN. Multiple factors related to DN are involved in the production of ROS, including hyperglycemia, advanced glycation end products, growth factors, and cytokines (4). 12/15-LO can also promote ROS production, lipid peroxidation (7), and superoxide anions (30). Several studies demonstrated that increased expression and activation of 12/15-LO lead to increased production of oxidized lipid products in kidneys of animals with DN (6, 15, 32, 40). Furthermore, 12/15-LO deficiency reduces oxidative stress in renal and vascular cells (21, 41), and increased expression of 12/15-LO and levels of 12(S)-HETE are associated with enhanced oxidative stress in prediabetic kidneys (6). Therefore, 12/15-LO is one of the key factors involved in ROS production and lipid peroxidation involved in the pathogenesis of DN.
Epigenetic changes in chromatin such as PTMs of histone tails, including lysine methylation and acetylation, have been demonstrated in regulation of genes associated with DN (23, 42, 43, 52, 62). We recently demonstrated that HG and TGF-β treatment of MCs led to increased levels of H3K9Ac and H3K4me (gene activation) and decreased levels of repressive H3K9me at promoters of profibrotic genes, as well increased expression of SET7 and its recruitment at profibrotic gene promoters (52, 62). Furthermore, a TGF-β antibody abolished HG-induced ECM gene expression and corresponding changes in histone modifications at their promoters (52). These observations suggest that targeting of TGF-β actions can reverse key epigenetic changes associated with diabetic complications. However, because targeting TGF-β itself can have adverse effects due to its important roles in immune cells, we explored key modulators and effectors of TGF-β-mediated epigenetic effects. We therefore examined 12/15-LO in this study because our previous studies demonstrated that a cross talk between these two key factors amplifies signal transduction cascades involved in hypertrophic and profibrotic gene expression associated with diabetic renal dysfunction (22). Most notably, in the current study, we found that 12/15-LO deficiency attenuated TGF-β-induced histone modifications (H3K9Ac and H3K4me1) at profibrotic gene promoters and SET7 expression. These new results suggest that 12/15-LO can mediate key epigenetic effects of TGF-β, leading to expression of profibrotic genes in MCs.
Our data showed increases in H3K4me1 at profibrotic promoters, suggesting a critical role of this modification in MCs. Evidence shows that SET7 mediates H3K4me1 associated with gene activation under diabetic conditions in monocytes, endothelial cells, and MCs and also cooperates with NF-κB (27, 37, 52). SET7 plays a role in gene transactivation by competing with HDACs to enhance H3K9Ac and prevent H3K9me (34, 56). In addition, SET7 can downregulate the repressive H3K9me3 methyltransferase activity of SUV39H1 (55). Furthermore, hyperglycemia could induce NF-κB (p65) gene expression in endothelial cells via increased H3K4me1 and decreased repressive modifications, H3K9me2 and H3K9me3 (3). Further studies showed that HG increased SET7 enrichment at the p65 promoter and its knockdown reduced H3K4me1 and attenuated increases in p65 expression. We previously showed that SET7 regulates TGF-β-induced profibrotic gene expression in MCs (52) and inflammatory gene expression in diabetic macrophages (27). Our current data also showed that 12(S)-HETE-induced increases in permissive histone PTMs and recruitment of SET7 correspond to increased expression of fibrotic genes, but not all the changes observed were sustained during the entire time course. These results demonstrate promoter and modification-specific differences in the formation of accessible chromatin needed for active gene expression. Furthermore, dynamic changes in chromatin remodeling arise from the cumulative effects and collaboration between several histone modifications. Thus, all histone modifications need not be altered simultaneously to sustain the chromatin relaxation involved in long-term gene expression. In addition, unlike the in vitro treatment with a single stimulus such as 12(S)-HETE, several pathological factors can act together in vivo, which can further amplify and sustain the alterations in chromatin structure involved in disease-associated gene expression. Altogether, these studies implicate SET7 as one of the key regulators of gene expression under diabetic conditions in diverse cell types involved in diabetic complications.
Interestingly, in the current study, we found that 12(S)-HETE directly increased SET7 protein levels and promoted its nuclear translocation, as well as occupancy, at the profibrotic gene promoters. Furthermore, TGF-β-induced SET7 expression was inhibited by 12-LO siRNA, and SET7 expression was attenuated in LOKO-MC, demonstrating a role for 12/15-LO and its oxidized lipid products in TGF-β-mediated SET7 expression. Evidence shows that nuclear translocation of SET7 can also be induced by HG in endothelial cells (37). Because, HG and diabetes can also increase 12/15-LO and 12(S)-HETE levels in MCs, SET7 may be a key target by which increased 12/15-LO and its products regulate pathologic genes involved in DN. Furthermore, similar mechanisms may be operative in endothelial cells and vascular smooth muscle cells, in which the 12/15-LO pathway is activated under diabetic conditions (26, 31, 45, 47), thereby contributing to multiple vascular complications. Thus, evidence shows that LO products and 12/15-LO can mediate low-density lipoprotein oxidation and are augmented in atherosclerosis, diabetes, and obesity and can mediate the development of diabetes and its key complications, including DN (8, 32, 33, 38).
The in vivo involvement of SET7 in diabetic complications is not fully clear. We speculated that there could be a cross talk between the TGF-β and 12/15-LO pathways in regulating SET7 expression in diabetic kidneys. In support of this, we observed that SET7 levels were enhanced in renal cortices and glomeruli from STZ-injected diabetic mice relative to nondiabetic mice, along with increases in TGF-β, 12/15-LO, and profibrotic genes. Importantly, 12/15-LO gene silencing by cholesterol-tagged siRNA injection or genetic knockdown (LOKO mice) significantly abrogated these diabetes-induced effects. A recent study showed increased expression of SET7 in kidneys from type 2 diabetic db/db mice, where it was implicated in monocyte chemoattractant protein-1 (MCP-1) expression (5), further highlighting the importance of SET7 in DN pathogenesis.
In summary, our study with in vitro and in vivo models demonstrates that the 12/15-LO pathway is involved in diabetes and TGF-β-associated signaling, histone modifications, and SET7 regulation associated with the sustained expression of multiple profibrotic genes involved in the progression of DN (Fig. 8). Apart from its role in HG and TGF-β effects, 12/15-LO also regulates AT1R expression and mediates Ang II effects (32, 40, 45, 59). 12/15-LO may therefore also modulate histone modifications mediated by Ang II under diabetic conditions. Thus, 12/15-LO could be a key common mediator of epigenetic mechanisms induced by multiple factors involved in DN pathogenesis. Therefore, inhibitors of the 12/15-LO pathway might offer novel therapeutic opportunities for DN by targeting these epigenetic changes. This has translational significance because epigenetic mechanisms may be key factors responsible for the relative inefficacy of traditional drugs used for DN, such as Ang II-converting enzyme inhibitors and AT1R blockers (48), and also for metabolic memory, the phenomenon in which periods of poor glycemic control can lead to continued sustained progression of diabetic complications such as DN, despite subsequent good glycemic control (20, 44).
FIG. 8.
Model for the role of the 12/15-LO pathway in HG or TGF-β-induced histone modifications and gene expression. The 12/15-LO pathway and its oxidized lipid metabolites such as 12(S)-HETE induce dynamic changes in permissive histone modifications and promote SET7 nuclear translocation, as well as its occupancy, at target gene promoters such as Serpine1 and Ctgf. These events play important roles in mediating HG or TGF-β-induced epigenetic changes involved in profibrotic gene expression. Deficiency of 12/15-LO attenuates these responses. Thus, cross talk between 12/15-LO and TGF-β pathways can lead to sustained expression of profibrotic genes involved in the progression of DN. DN, diabetic nephropathy; HG, high glucose. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Materials and Methods
Materials
12(S)-Hydroxyeicosa-5Z, 8Z, 10E, 14Z-tetraenoic acid [12(S)-HETE], and its stereoisomer, 12(R)-HETE, were from Biomol (Plymouth Meeting, PA); pan-specific TGF-β1 antibody (MAB1835) and recombinant human TGF-β1 (240-B) were obtained from R&D Systems (Minneapolis, MN); lamin B1 (9087) antibody and horseradish peroxidase-conjugated secondary antibodies were from Cell Signaling (Beverly, MA); anti-SET7 (ab14820), anti-H3K9 acetylation (ab10812), anti-H3 monomethyl K4 (ab8895), anti-H3 dimethylK4 (ab32356), and anti-H3 trimethyl K4 (ab8580) were from Abcam (Cambridge, MA); purified mouse anti-PAI-1 (612024) was from BD Biosciences (San Jose, CA); normal rabbit IgG (PP64B) and normal mouse IgG (12-371) were from Upstate Biotechnology, and β-actin antibody was from Sigma (St. Louis, MO). SET7 ON-TARGET plus Smartpool siRNA (J-059399[09–12]), leukocyte-type 12-LO ON-TARGET plus Smartpool siRNA (J-083898[09-12]), and silencer nontargeting control siRNA #1 (D-001810-01-05) were from Thermo Scientific (Waltham, MA); RNA-STAT60 reagent was from Tel-Test (Friendswood, TX) and RNeasy columns were from Qiagen (Valencia, CA). Reverse transcriptase kits and SYBR Green PCR Master Mix kits were from Applied Biosystems (Foster City, CA). Sequences of the qPCR primers used in this study are listed in Table 3.
Table 3.
Primer Sequences Used in This Study
| Primer | Forward primer | Reverse primer |
|---|---|---|
| cDNA primers | ||
| Serpine1 (R) | cggaCTTCTTCAAGCTCTTCCG | TGAAATAGAGGGCGTTCACCAG |
| Col1a1 (R) | TGGTGCTCCTGGTATTGCTG | cggaCGTTTTCCTTCTTCTCCG |
| Ctgf (R/m) | cagctCCGAGAAGGGTCAAGCTG | AACAGGCGCTCCACTCTGTG |
| Setd7 (R) | ACAGAAGAAGGGAAGCCACA | CGGACTCATAAGGGTCTGGA |
| Actb (R/m) | CTGCCCTGGCTCCTAGCAC | cggacGCAGCTCAGTAACAGTCCG |
| Serpine1 (m) | GACGCCTTCATTTGGACGAA | CGGACCTTTTCCCTTCAAGAGTCCG |
| Col1a1 (m) | CGGATAGCAGATTGAGAACATCCG | CGGCTGAGTAGGGAACACACA |
| Col4a1 (m) | CACGAGCTTCCCTGGTAGTCGTG | GGACAACCTTTCCTGCCTCA |
| Tgfb1 (m) | AGGAAGGACCTGGGTTGGAAG | CGTCTCGACCCACGTAGTAGACG |
| Rplp0 (m) | GCCCTGCACTCTCGCTTTCT | CAACTGGGCACCGAGGCAACAGTTG |
| Setd7 (m) | GGGAAGTTCATTGACGGAGA | AGGGTCTGGAAGGAGAGCAT |
| ChIP primers | ||
| Serpine1 pro (R) | cggcTATACCAGATGTGGGCcG | GACGACCGACCAGCCAAAG |
| Col1a1 pro (R) | GGCTGGAGAAAGGTGGGTCT | CCCAGGTATGCAGGGTAGGA |
| Ctgf pro (R) | ATCAGGAAGGGTGCGAAGAG | TCCACATTCCTCCGTCTGAA |
| Ppia pro (R) | TATCTGCACTGCCAAGACTGAGTG | CTTCTTGCTGGTCTTGCCATTCC |
| Serpine1 pro (m) | cggcCTTTATACCAGATGTGAGCcG | TCCAAACACCAGGCTTTGTAGG |
| Col1a1 pro (m) | TGGACTCCTTTCCCTTCCTT | CTGGGCCCCTTTTATACCAT |
| Ctgf pro (m) | AGCCAGAACTGGCAAAGAGA | ACACTTTAGCTCGCCAGGAA |
ChIP, chromatin immunoprecipitation; m, mouse; R, rat.
Cell culture
Primary cultures of RMCs from Sprague-Dawley rats, and mouse mesangial cells (MMCs) from genetic control mice (C57BL/6, WT) and leukocyte-type 12/15-LOKO mice (Strain B6. 129S2-Alox15tm1Fun; Jackson Laboratories, Bar Harbor, ME) were prepared as described previously (21, 52). Briefly, kidney cortices from mice (8–10 weeks old) or rats (250–300 g) were minced and the glomeruli were isolated by differential sieving. Then, glomeruli were digested with collagenase (Sigma Chemical Co., Inc., St. Louis, MO, USA) for 15 minutes and cultured in T-25 culture flasks (Nunc, Rochester, NY, USA) in RPMI 1640 medium supplemented with 15% fetal calf serum (FCS) (Omega Scientific, Co., Tarzana, CA, USA) and 20 μg/mL insulin (Sigma Chemical Co.) at 37°C under 5% CO2 for about a week to 10 days until outgrowths of spindle-like mesangial cells were observed. Then mesangial cells were cultured for a further 48 hours in the presence of 50 mmol/L D-valine containing medium to exclude fibroblasts. Purity of MC was tested by staining cells plated on glass chamber slides with monoclonal Thy1.1 antibody. MCs were maintained in RPMI 1640 supplemented with L-glutamine (2.5 mM), HEPES (7 mM), Penicillin/streptomycin and 10% fetal calf serum. Cells between 5 to 12 passages were used in all the studies.
Transient transfection
MCs were placed in 60-mm dishes upto 75% confluency, then cotransfected with 0.3 μg of 12-LO ON-TARGETplus Smartpool siRNA (si12-LO) or Silencer Negative Control #1 siRNA (siNTC) using Nucleofection reagent as described earlier (17). After the transfected cells recovered, they were placed in serum-free RPMI 1640 medium containing 0.2% bovine serum albumin for 24 h, treated with or without TGF-β (10 ng/ml), and processed for RNA extraction at the indicated time periods. In SET7 gene silencing experiments, RMCs were transfected using Smartpool siSET7 (siSET7) or siNTC (20 nM) using RNAiMAX (Lifetechnologies, Carlsbad, CA). After 2 days, cells were retransfected with siSET7 or siNTC, and 48 h after the second transfection, cells were treated with vehicle or 12(S)-HETE. Total RNA was extracted and gene expression was determined by RT-qPCR.
RNA isolation and RT-qPCR
Total RNA was isolated from MCs, renal cortices, and glomeruli using RNA-STAT60 reagent and RNeasy columns according to the manufacturer's instructions. Total RNA (0.5–1 μg) was used for cDNA synthesis using Gene Amp RNA PCR kits (Applied Biosystems) and qPCRs were performed in triplicate in a final volume of 20 μl with an ABI 7500 real-time PCR thermal cycler (Applied Biosystems) using specific gene primers as described previously (52,61). Dissociation curves were run to detect nonspecific amplification and to confirm amplification of a single product in each reaction. Data was analyzed by 2−ΔΔCt method using Actb, Ppia or Rplp0 genes as internal controls (52,61). Results were expressed as fold over control.
Western blots
Protein lysates from MCs, glomeruli, and renal cortex tissues were prepared by using 1.5× sodium dodecyl sulfate (SDS) sample buffer and immunoblotting was performed as described previously (61, 62). Nuclear extracts were prepared using the NE-PER kit (Cat#78833; Thermo Scientific) according to the manufacturer's protocols.
ChIP assays
Quiescent RMCs were treated with 12(S)-HETE for the indicated time points, and then fixed with 1% formaldehyde for 10 min at room temperature, followed by neutralization with 125 mM glycine for 5 min. Fixed cells were washed three times with cold phosphate-buffered saline supplemented with protease inhibitors and lysed using 1% SDS lysis buffer supplemented with protease inhibitors. Cell lysates were sonicated until DNA was fragmented up to 200–1000 bp. Chromatin immunoprecipitation (ChIP) assays with the relevant antibodies indicated above under the Materials section were performed using commercially available kits (EMD Millipore, New York, NY) as described previously (52). Cell lysates were diluted with ChIP dilution buffer to reduce SDS concentration to < 0.1%, precleared by incubating with Protein A/G agarose beads for an hour. Then, lysates were centrifuged and supernatants were incubated with indicated antibodies (1–5 μg) overnight at 4°C. Next day, protein A/G-coupled agarose beads were added and 2 h later beads were collected by centrifugation. Beads were washed sequentially with low-salt buffer, high-salt buffer, LiCl buffer, and TE buffer at 4°C. Washed beads were incubated with elution buffer (1% SDS in 0.1 M NaHCO3) at room temperature for 15 min to elute immune complexes. DNA-protein cross links were removed by incubation at 65°C for 4 h. Then, ChIP-DNA was extracted by digestion with proteinase K (50 μg/ml) for 30 min, followed by extraction with phenol–chloroform and precipitation of DNA from the aqueous layer using two volumes of ethanol at −20°C overnight. An aliquot of the cell lysate was used to isolate total input DNA. Rabbit or mouse IgG was used as antibody controls. Using qPCR, we confirmed the enrichment (15- to 12,000-fold enrichment) of histone modifications at gene promoters relative to IgG samples demonstrating specificity of ChIP assays. Furthermore, Western blots also showed that these antibodies detected specific bands corresponding to histone proteins in RMC lysates. Input DNA samples as well as antibody-enriched ChIP DNA samples were analyzed by qPCR using primers corresponding to sequences within the promoter regions near Smad binding sites for profibrotic genes (Table 3). Data were analyzed using the 2−ΔΔCt method and normalized as described (52). Results are expressed as fold over respective control samples.
Immunofluorescence
Confluent RMCs were placed on glass slides, SD, and treated with 12(S)-HETE (10−7 M), followed by IF as described before (27). Cellular localization of stained protein was observed under a fluorescent microscope (Olympus, Tokyo, Japan). To detect the nuclei, cells were stained with 0.2% Hoechst 33342 (Molecular Probe, Eugene, OR).
Mouse models of DN
All animal studies were conducted using protocols approved by the Institutional Research Animal Care and Use Committee (IACUC). The animals were housed in a temperature-controlled room and given free access to water and standard laboratory chow. To induce type 1 diabetes, about 8-week-old WT and LOKO mice (n = 6) were injected with 50 mg/kg of STZ intraperitoneally for 5 consecutive days as described earlier (39). Nondiabetic control mice were injected with citrate buffer (NS). All mice were euthanized at 16 weeks after development of diabetes (blood glucose levels >350 mg/dl in STZ versus 140 mg/dl in control mice). Blood samples were collected before euthanization. Individual mice were placed in metabolic cages to collect 24-h urine samples during the first, 12, and 16 weeks postdiabetes. Renal cortical tissues were saved for Western blots and qPCRs. Glomeruli were isolated from cortical tissues by sequential sieving as described before (62) and pooled from two mice in each group. Body weight and blood glucose were checked monthly, and kidneys were weighed after the animals were euthanized. Treatment of diabetic mice with the cholesterol-tagged siRNAs targeting 12/15-LO or control siRNAs was described earlier (61). Briefly, diabetes was induced by injecting STZ (50 mg/kg) for 5 days in 8-week-old DBA/2J mice. Diabetic mice (glucose >300 mg/dl) were subcutaneously injected with normal saline or cholesterol-tagged 12-LO siRNAs or mismatched siRNAs (MM oligos) for 7 weeks (400 μg each, twice/week). As controls, nondiabetic mice were injected with normal saline (61). After 7 weeks, mice were euthanized and kidneys were collected for further processing.
Urinary protein and albumin assay
Urine samples (24 h) were diluted 5× with Milli Q distilled water and the amount of protein (in milligrams) was determined using detergent-compatible protein assay kits (Bio-Rad, Inc., Hercules, CA) as described previously (39). Urinary albumin concentration was measured with an indirect competitive ELISA kit (Exocell, Philadelphia, PA) according to the manufacturer's instructions.
Histology and morphology analyses
Renal cortices were fixed immediately after harvesting in 10% neutral buffered formalin solution and submitted to the Anatomic Pathology Core at the City of Hope Medical Center to obtain paraffin-embedded sections. To evaluate diabetes-associated renal pathological changes, sections were processed for hematoxylin and eosin, periodic acid-silver methenamine, and Masson's trichrome staining. Images were captured using an Olympus DP72 Microscope Digital Camera and processed with DP2-BSW software. The areas of glomeruli and mesangial expansion were quantified using Image-Pro (version 5.1; Media Cybernetics, Silver Spring, MD).
Immunohistochemical staining
IHC staining was performed as described (19, 61) using TGF-β1 antibody (1:100 dilution) for 0.5 h at room temperature. Twenty glomeruli in each section were examined using light microscopy at a magnification of 400×, and stained positive area per glomerulus was quantified using Image-Pro software.
Statistical analyses
Data are expressed as mean ± standard error of the mean of multiple experiments. Paired Student's t-tests were used to compare two groups or ANOVA with Dunnet's post-test for multiple groups using PRISM software (Graph Pad, San Diego, CA). Statistical significance was detected at the 0.05 level.
Supplementary Material
Abbreviations Used
- 12(S)-HETE
12(S)-hydroxyeicosatetraenoic acid
- 12/15-LO
12/15-lipoxygenase
- ACR
albumin-to-creatinine ratio
- Ang II
angiotensin II
- AT1R
angiotensin II type I receptor
- ChIP
chromatin immunoprecipitation
- CR
creatinine
- DN
diabetic nephropathy
- ECM
extracellular matrix
- H&E
hematoxylin and eosin
- H3K4me1
histone H3 lysine-4 monomethylation
- H3K4me3
histone H3 lysine-4 trimethylation
- H3K9Ac
histone H3 lysine-9 acetylation
- HDAC
histone deacetylase
- HG
high glucose
- IF
immunofluorescence
- IHC
immunohistochemical
- LOKO
LO knockout
- MC
mesangial cell
- MCP-1
monocyte chemoattractant protein-1
- MMC
mouse mesangial cells
- NF-κB
nuclear factor of kappa light polypeptide gene enhancer in B-cells
- NS
no streptozotocin
- PAS
periodic acid-Schiff
- PASM
periodic acid-silver methenamine
- PTMs
post-translational modifications
- RMC
rat mesangial cell
- ROS
reactive oxygen species
- RT-qPCR
real-time quantitative polymerase chain reaction
- SD
serum depleted
- SDS
sodium dodecyl sulfate
- SE
standard error
- SEM
standard error of the mean
- SET7
SET domain containing (lysine methyltransferase) 7
- si12-LO
siRNA targeting 12/15-LO
- siMM
control nontargeting siRNA
- siNTC
nontargeting siRNA
- STZ
streptozotocin
- TGF-β1
transforming growth factor-β1
- WT
wild-type
Acknowledgments
The authors gratefully acknowledge funding from the National Institutes of Health (grants R01 DK058191 and R01 DK081705) to R.N. Research reported in this publication included work performed in the Anatomic Pathology Core and the Animal Research Core supported by the National Cancer Institute of the National Institutes of Health under award number P30CA33572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Advani A, Huang Q, Thai K, Advani SL, White KE, Kelly DJ, Yuen DA, Connelly KA, Marsden PA, and Gilbert RE. Long-term administration of the histone deacetylase inhibitor vorinostat attenuates renal injury in experimental diabetes through an endothelial nitric oxide synthase-dependent mechanism. Am J Pathol 178: 2205–2214, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonipillai I, Nadler J, Vu EJ, Bughi S, Natarajan R, and Horton R. A 12-lipoxygenase product, 12-hydroxyeicosatetraenoic acid, is increased in diabetics with incipient and early renal disease. J Clin Endocrinol Metab 81: 1940–1945, 1996 [DOI] [PubMed] [Google Scholar]
- 3.Brasacchio D, Okabe J, Tikellis C, Balcerczyk A, George P, Baker EK, Calkin AC, Brownlee M, Cooper ME, and El-Osta A. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes 58: 1229–1236, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature 414: 813–820, 2001 [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Guo Y, Zeng W, Huang L, Pang Q, Nie L, Mu J, Yuan F, and Feng B. ER stress triggers MCP-1 expression through SET7/9-induced histone methylation in the kidneys of db/db mice. Am J Physiol Renal Physiol 306: F916–F925, 2014 [DOI] [PubMed] [Google Scholar]
- 6.Chua S, Jr., Li Y, Liu SM, Liu R, Chan KT, Martino J, Zheng Z, Susztak K, D'Agati VD, and Gharavi AG. A susceptibility gene for kidney disease in an obese mouse model of type II diabetes maps to chromosome 8. Kidney Int 78: 453–462, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cyrus T, Pratico D, Zhao L, Witztum JL, Rader DJ, Rokach J, FitzGerald GA, and Funk CD. Absence of 12/15-lipoxygenase expression decreases lipid peroxidation and atherogenesis in apolipoprotein e-deficient mice. Circulation 103: 2277–2282, 2001 [DOI] [PubMed] [Google Scholar]
- 8.Dobrian AD, Lieb DC, Cole BK, Taylor-Fishwick DA, Chakrabarti SK, and Nadler JL. Functional and pathological roles of the 12- and 15-lipoxygenases. Prog Lipid Res 50: 115–131, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, Cooper ME, and Brownlee M. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 205: 2409–2417, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funk CD. The molecular biology of mammalian lipoxygenases and the quest for eicosanoid functions using lipoxygenase-deficient mice. Biochim Biophys Acta 1304: 65–84, 1996 [DOI] [PubMed] [Google Scholar]
- 11.Guo QY, Miao LN, Li B, Ma FZ, Liu N, Cai L, and Xu ZG. Role of 12-lipoxygenase in decreasing P-cadherin and increasing angiotensin II type 1 receptor expression according to glomerular size in type 2 diabetic rats. Am J Physiol Endocrinol Metab 300: E708–E716, 2011 [DOI] [PubMed] [Google Scholar]
- 12.Jirtle RL. and Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet 8: 253–262, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones CA, Krolewski AS, Rogus J, Xue JL, Collins A, and Warram JH. Epidemic of end-stage renal disease in people with diabetes in the United States population: do we know the cause? Kidney Int 67: 1684–1691, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492, 2012 [DOI] [PubMed] [Google Scholar]
- 15.Kang SW, Adler SG, Nast CC, LaPage J, Gu JL, Nadler JL, and Natarajan R. 12-Lipoxygenase is increased in glucose-stimulated mesangial cells and in experimental diabetic nephropathy. Kidney Int 59: 1354–1362, 2001 [DOI] [PubMed] [Google Scholar]
- 16.Kanwar YS, Sun L, Xie P, Liu FY, and Chen S. A glimpse of various pathogenetic mechanisms of diabetic nephropathy. Annu Rev Pathol 6: 395–423, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato M, Dang V, Wang M, Park JT, Deshpande S, Kadam S, Mardiros A, Zhan Y, Oettgen P, Putta S, Yuan H, Lanting L, and Natarajan R. TGF-beta induces acetylation of chromatin and of Ets-1 to alleviate repression of miR-192 in diabetic nephropathy. Sci Signal 6: ra43, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato M. and Natarajan R. Diabetic nephropathy—emerging epigenetic mechanisms. Nat Rev Nephrol 10: 517–530, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato M, Yuan H, Xu ZG, Lanting L, Li SL, Wang M, Hu MC, Reddy MA, and Natarajan R. Role of the Akt/FoxO3a pathway in TGF-beta1-mediated mesangial cell dysfunction: a novel mechanism related to diabetic kidney disease. J Am Soc Nephrol 17: 3325–3335, 2006 [DOI] [PubMed] [Google Scholar]
- 20.Keating ST. and El-Osta A. Glycemic memories and the epigenetic component of diabetic nephropathy. Curr Diab Rep 13: 574–581, 2013 [DOI] [PubMed] [Google Scholar]
- 21.Kim YS, Reddy MA, Lanting L, Adler SG, and Natarajan R. Differential behavior of mesangial cells derived from 12/15-lipoxygenase knockout mice relative to control mice. Kidney Int 64: 1702–1714, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Kim YS, Xu ZG, Reddy MA, Li SL, Lanting L, Sharma K, Adler SG, and Natarajan R. Novel interactions between TGF-{beta}1 actions and the 12/15-lipoxygenase pathway in mesangial cells. J Am Soc Nephrol 16: 352–362, 2005 [DOI] [PubMed] [Google Scholar]
- 23.Komers R, Mar D, Denisenko O, Xu B, Oyama TT, and Bomsztyk K. Epigenetic changes in renal genes dysregulated in mouse and rat models of type 1 diabetes. Lab Invest 93: 543–552, 2013 [DOI] [PubMed] [Google Scholar]
- 24.Kouzarides T. Chromatin modifications and their function. Cell 128: 693–705, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Kutz SM, Higgins CE, Samarakoon R, Higgins SP, Allen RR, Qi L, and Higgins PJ. TGF-beta 1-induced PAI-1 expression is E box/USF-dependent and requires EGFR signaling. Exp Cell Res 312: 1093–1105, 2006 [DOI] [PubMed] [Google Scholar]
- 26.Li SL, Reddy MA, Cai Q, Meng L, Yuan H, Lanting L, and Natarajan R. Enhanced proatherogenic responses in macrophages and vascular smooth muscle cells derived from diabetic db/db mice. Diabetes 55: 2611–2619, 2006 [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Reddy MA, Miao F, Shanmugam N, Yee JK, Hawkins D, Ren B, and Natarajan R. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem 283: 26771–26781, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loeffler I. and Wolf G. Transforming growth factor-beta and the progression of renal disease. Nephrol Dial Transplant 29 Suppl 1: i37–i45, 2014 [DOI] [PubMed] [Google Scholar]
- 29.Miyazono K, ten Dijke P, and Heldin CH. TGF-beta signaling by Smad proteins. Adv Immunol 75: 115–157, 2000 [DOI] [PubMed] [Google Scholar]
- 30.Murea M, Park JK, Sharma S, Kato H, Gruenwald A, Niranjan T, Si H, Thomas DB, Pullman JM, Melamed ML, and Susztak K. Expression of Notch pathway proteins correlates with albuminuria, glomerulosclerosis, and renal function. Kidney Int 78: 514–522, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natarajan R. and Nadler JL. Lipoxygenases and lipid signaling in vascular cells in diabetes. Front Biosci 8: s783–s795, 2003 [DOI] [PubMed] [Google Scholar]
- 32.Natarajan R. and Nadler JL. Lipid inflammatory mediators in diabetic vascular disease. Arterioscler Thromb Vasc Biol 24: 1542–1548, 2004 [DOI] [PubMed] [Google Scholar]
- 33.Natarajan R. and Reddy MA. HETEs/EETs in renal glomerular and epithelial cell functions. Curr Opin Pharmacol 3: 198–203, 2003 [DOI] [PubMed] [Google Scholar]
- 34.Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, and Reinberg D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev 16: 479–489, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noh H, Oh EY, Seo JY, Yu MR, Kim YO, Ha H, and Lee HB. Histone deacetylase-2 is a key regulator of diabetes- and transforming growth factor-beta1-induced renal injury. Am J Physiol Renal Physiol 297: F729–F739, 2009 [DOI] [PubMed] [Google Scholar]
- 36.Obrosova IG, Stavniichuk R, Drel VR, Shevalye H, Vareniuk I, Nadler JL, and Schmidt RE. Different roles of 12/15-lipoxygenase in diabetic large and small fiber peripheral and autonomic neuropathies. Am J Pathol 177: 1436–1447, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okabe J, Orlowski C, Balcerczyk A, Tikellis C, Thomas MC, Cooper ME, and El-Osta A. Distinguishing hyperglycemic changes by set7 in vascular endothelial cells. Circ Res 110: 1067–1076, 2012 [DOI] [PubMed] [Google Scholar]
- 38.Parthasarathy S. and Santanam N. Mechanisms of oxidation, antioxidants, and atherosclerosis. Curr Opin Lipidol 5: 371–375, 1994 [DOI] [PubMed] [Google Scholar]
- 39.Putta S, Lanting L, Sun G, Lawson G, Kato M, and Natarajan R. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol 23: 458–469, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reddy MA, Adler SG, Kim YS, Lanting L, Rossi J, Kang SW, Nadler JL, Shahed A, and Natarajan R. Interaction of MAPK and 12-lipoxygenase pathways in growth and matrix protein expression in mesangial cells. Am J Physiol Renal Physiol 283: F985–F994, 2002 [DOI] [PubMed] [Google Scholar]
- 41.Reddy MA, Kim YS, Lanting L, and Natarajan R. Reduced growth factor responses in vascular smooth muscle cells derived from 12/15-lipoxygenase-deficient mice. Hypertension 41: 1294–1300, 2003 [DOI] [PubMed] [Google Scholar]
- 42.Reddy MA. and Natarajan R. Epigenetics in diabetic kidney disease. J Am Soc Nephrol 22: 2182–2185, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reddy MA, Sumanth P, Lanting L, Yuan H, Wang M, Mar D, Alpers CE, Bomsztyk K, and Natarajan R. Losartan reverses permissive epigenetic changes in renal glomeruli of diabetic db/db mice. Kidney Int 85: 362–373, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy MA, Tak Park J, and Natarajan R. Epigenetic modifications in the pathogenesis of diabetic nephropathy. Semin Nephrol 33: 341–353, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reddy MA, Thimmalapura PR, Lanting L, Nadler JL, Fatima S, and Natarajan R. The oxidized lipid and lipoxygenase product 12(S)-hydroxyeicosatetraenoic acid induces hypertrophy and fibronectin transcription in vascular smooth muscle cells via p38 MAPK and cAMP response element-binding protein activation. Mediation of angiotensin II effects. J Biol Chem 277: 9920–9928, 2002 [DOI] [PubMed] [Google Scholar]
- 46.Reidy K, Kang HM, Hostetter T, and Susztak K. Molecular mechanisms of diabetic kidney disease. J Clin Invest 124: 2333–2340, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reilly KB, Srinivasan S, Hatley ME, Patricia MK, Lannigan J, Bolick DT, Vandenhoff G, Pei H, Natarajan R, Nadler JL, and Hedrick CC. 12/15-Lipoxygenase activity mediates inflammatory monocyte/endothelial interactions and atherosclerosis in vivo. J Biol Chem 279: 9440–9450, 2004 [DOI] [PubMed] [Google Scholar]
- 48.Ruggenenti P, Cravedi P, and Remuzzi G. The RAAS in the pathogenesis and treatment of diabetic nephropathy. Nat Rev Nephrol 6: 319–330, 2010 [DOI] [PubMed] [Google Scholar]
- 49.Sanchez AP. and Sharma K. Transcription factors in the pathogenesis of diabetic nephropathy. Expert Rev Mol Med 11: e13, 2009 [DOI] [PubMed] [Google Scholar]
- 50.Sharma K, Jin Y, Guo J, and Ziyadeh FN. Neutralization of TGF-beta by anti-TGF-beta antibody attenuates kidney hypertrophy and the enhanced extracellular matrix gene expression in STZ-induced diabetic mice. Diabetes 45: 522–530, 1996 [DOI] [PubMed] [Google Scholar]
- 51.Shevalye H, Lupachyk S, Watcho P, Stavniichuk R, Khazim K, Abboud HE, and Obrosova IG. Prediabetic nephropathy as an early consequence of the high-calorie/high-fat diet: relation to oxidative stress. Endocrinology 153: 1152–1161, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun G, Reddy MA, Yuan H, Lanting L, Kato M, and Natarajan R. Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol 21: 2069–2080, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Susztak K. Understanding the epigenetic syntax for the genetic alphabet in the kidney. J Am Soc Nephrol 25: 10–17, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki H, Kayama Y, Sakamoto M, Iuchi H, Shimizu I, Yoshino T, Katoh D, Nagoshi T, Tojo K, Minamino T, Yoshimura M, and Utsunomiya K. Arachidonate 12/15-lipoxygenase-induced inflammation and oxidative stress are involved in the development of diabetic cardiomyopathy. Diabetes 64: 618–630, 2015 [DOI] [PubMed] [Google Scholar]
- 55.Wang D, Zhou J, Liu X, Lu D, Shen C, Du Y, Wei FZ, Song B, Lu X, Yu Y, Wang L, Zhao Y, Wang H, Yang Y, Akiyama Y, Zhang H, and Zhu WG. Methylation of SUV39H1 by SET7/9 results in heterochromatin relaxation and genome instability. Proc Natl Acad Sci U S A 110: 5516–5521, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, and Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell 8: 1207–1217, 2001 [DOI] [PubMed] [Google Scholar]
- 57.Xu ZG, Li SL, Lanting L, Kim YS, Shanmugam N, Reddy MA, and Natarajan R. Relationship between 12/15-lipoxygenase and COX-2 in mesangial cells: potential role in diabetic nephropathy. Kidney Int 69: 512–519, 2006 [DOI] [PubMed] [Google Scholar]
- 58.Xu ZG, Miao LN, Cui YC, Jia Y, Yuan H, and Wu M. Angiotensin II type 1 receptor expression is increased via 12-lipoxygenase in high glucose-stimulated glomerular cells and type 2 diabetic glomeruli. Nephrol Dial Transplant 24: 1744–1752, 2009 [DOI] [PubMed] [Google Scholar]
- 59.Xu ZG, Yuan H, Lanting L, Li SL, Wang M, Shanmugam N, Kato M, Adler SG, Reddy MA, and Natarajan R. Products of 12/15-lipoxygenase upregulate the angiotensin II receptor. J Am Soc Nephrol 19: 559–569, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto S. Mammalian lipoxygenases: molecular structures and functions. Biochim Biophys Acta 1128: 117–131, 1992 [DOI] [PubMed] [Google Scholar]
- 61.Yuan H, Lanting L, Xu ZG, Li SL, Swiderski P, Putta S, Jonnalagadda M, Kato M, and Natarajan R. Effects of cholesterol-tagged small interfering RNAs targeting 12/15-lipoxygenase on parameters of diabetic nephropathy in a mouse model of type 1 diabetes. Am J Physiol Renal Physiol 295: F605–F617, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan H, Reddy MA, Sun G, Lanting L, Wang M, Kato M, and Natarajan R. Involvement of p300/CBP and epigenetic histone acetylation in TGF-beta1-mediated gene transcription in mesangial cells. Am J Physiol Renal Physiol 304: F601–F613, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ziyadeh FN. and Sharma K. Overview: combating diabetic nephropathy. J Am Soc Nephrol 14: 1355–1357, 2003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.