Abstract
Of the three dominant marine microalgal groups, dinoflagellates and diatoms can undergo genetic transformation; however, no transformation method has been established for haptophytes to date. Here, we report the first stable genetic transformation of a coccolithophore, Pleurochrysis carterae, by means of polyethylene glycol (PEG)-mediated transfer of a bacterial hygromycin B-resistance gene. Together with the novel transient green fluorescent protein (GFP) expression system, this approach should facilitate further molecular-based research in this phylum.
The contemporary ocean is dominated by the three kinds of microalgae: diatoms (Heterokontophyta), dinoflagellates (Dinophyta), and coccolithophores (Haptophyta). Although successful transformation systems were developed for the former two groups in the late 1990s1,2, neither a transient nor stable transformation system has been established for Haptophyta until now. Coccolithophores, the main members of this phylum, are algae sharing the common characteristic of intracellular calcification, and cover themselves with fine oval structures of calcite crystals called coccoliths (Fig. 1a). Through coccolith synthesis and photosynthesis, these algae contribute to global carbon circulation3,4,5. A recent pan-genome analysis conducted on a representative coccolithophore species, Emiliania huxleyi, revealed extensive intraspecific genome variability, suggesting its potential for investigating the relationship between genomic plasticity and global/historical climate change6. In addition to their importance to ocean ecology, there is considerable interest in the commercial application of coccolithophores for bioenergy production. Pleurochrysis carterae is regarded as a promising candidate for biodiesel production for the following reasons: a high lipid content up to 33% of their dry weight, capability for long-term outdoor culture, and low potential for contamination of protozoans or other algae owing to the high pH of the growth medium during outdoor cultivation7. Given these advantages and the urgent need for alternative sources of biodiesel production, establishment of genetic transformation technologies for haptophytes, especially coccolithophores, is required. Thus, in the present study, we developed the methods of transient and stable transformation for P. carterae.
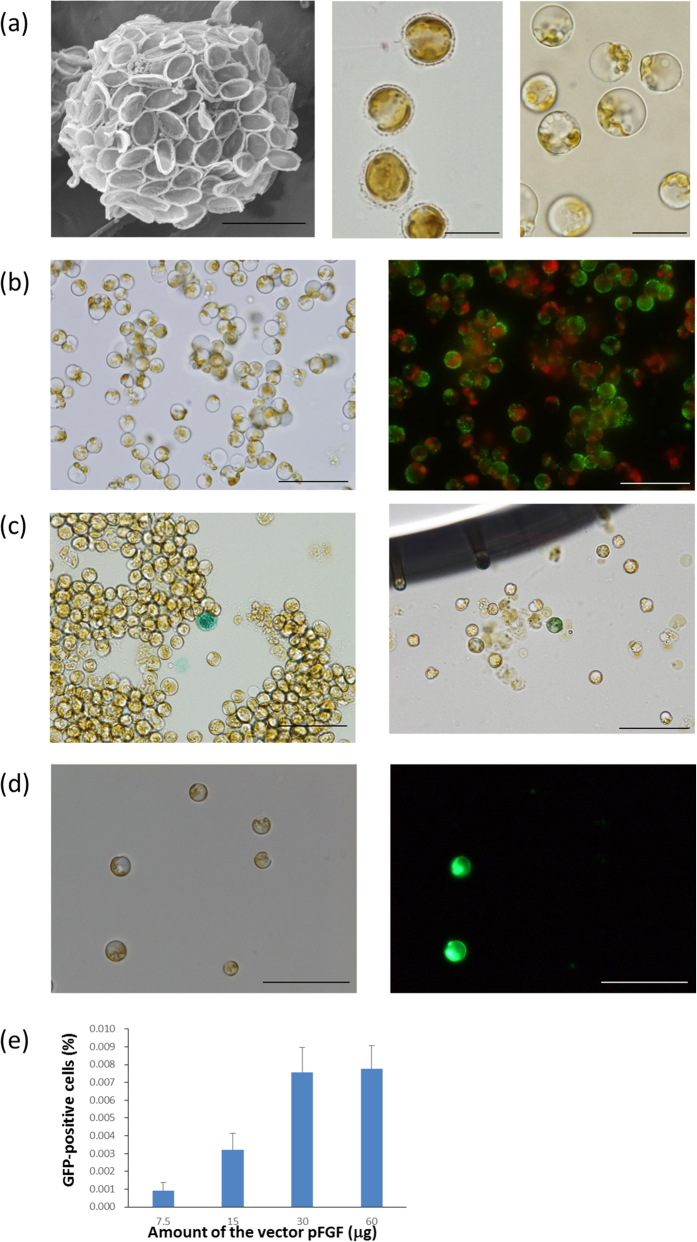
Figure 1. Microscopic views of the coccolithophore, Pleurochrysis carterae.
(a) Left: scanning electron microscopy of a calcified cell. Scale bar, 3 μm. Center: light microscopy of calcified cells; a thick coccosphere can be observed on the cell surface. Right: light microscopy of protoplasts; the coccosphere containing coccoliths is completely removed. Scale bars, 10 μm. (b) Fluorescein isothiocyanate (FITC)-labeled dextran-transferred protoplasts. Left: bright-field view. Right: fluorescent view. Green fluorescence can be observed in the cytosol with red fluorescence derived from the chloroplasts. Scale bars, 50 μm. (c) Representative transient GUS-expressing cells 3 days after pFGS introduction. Since P. carterae cells were sensitive to a detergent (TritonX-100), disrupted cells were frequently observed during the experiment (data not shown). Scale bars, 50 μm. (d) Representative transient GFP-expressing cells 24 h after pFGF introduction. Scale bars, 50 μm. (e) GFP transient expression analysis. Dose-response relationship between the vector pFGF and GFP-expressing cells. The positive cells were counted at 24 h after vector administration. n = 8, bars represents mean +S.E.
Results
Algal cell walls contain several substances such as elastic or rigid polysaccharides, and mucilaginous, siliceous, cellulose theca, or sulfated polysaccharides8. In the case of P. carterae, the plasma membrane is coated with a thick layer consisting of as-yet-uncharacterized viscous polysaccharides containing uncalcified coccolith base plates and highly calcified coccoliths. This complicated cell wall structure (generally called the “coccosphere”, corresponding to the cell wall of other algae or higher plants) is the first barrier for effective gene transfer; therefore, removal of the coccosphere and protoplast preparation were the primary objectives for the development of a novel gene transfer system. Takayanagi et al.9 reported a simple protoplast preparation method for Pleurochrysis haptonemofera (a species closely related to P. carterae), in which the calcified cells “molt” their coccospheres in a K+-containing hypoosmotic buffer. In the present study, we found that pre-treatment with proteinase K was effective for degrading the coccosphere of P. carterae cells, and thus a sufficient number of protoplast cells could be easily obtained with our improved method (Fig. 1a). We also found that polyethylene glycol (PEG) treatment could effectively promote the transfer of exogenous macromolecules into the protoplast. Indeed, uptake of fluorescein isothiocyanate (FITC)-dextran was observed in the majority of the PEG-treated cells (Fig. 1b).
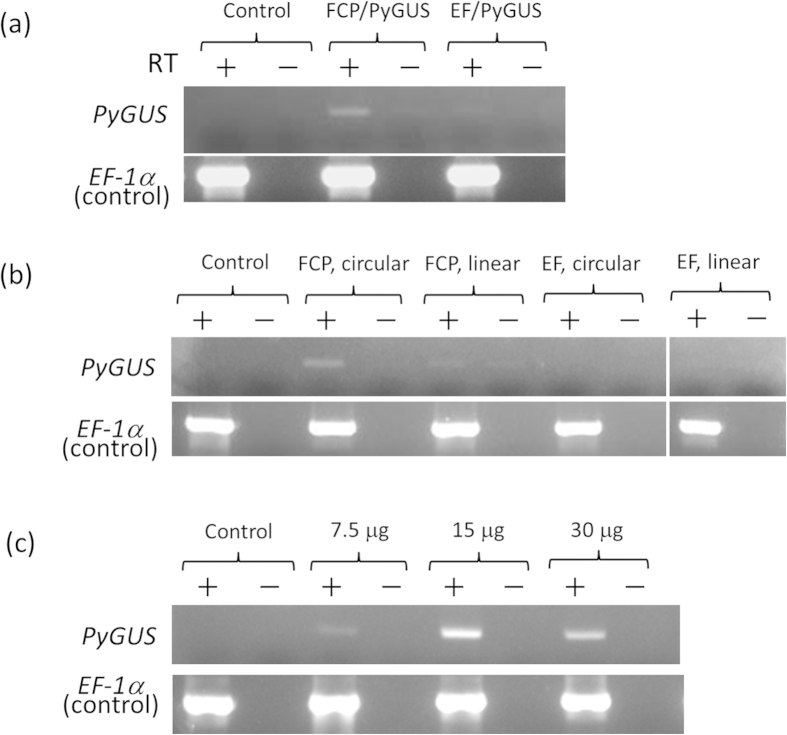
According to our preliminary results from expressed sequence tag (EST) and expression analyses of P. carterae (unpublished data), we selected the promoters of the fucoxanthin chlorophyll a/c-binding protein (FCP) and elongation factor-1α (EF1α) genes as candidates to drive the marker genes in a gene transformation system. The EST analysis also revealed that codon preference of P. carterae is highly GC-biased and closely resembles that of a macroalga, Pyropia yezoensis (Bangiales, Rhodophyta). Thus, P. yezoensis β-glucuronidase (PyGUS)10 and aminoglycoside phosphotransferase (PyAph7)11 were used as selectable marker genes without further modifications (Fig. 2). Three days after vector administration, the mRNA of PyGUS was clearly detected in the cells transferred with the FCP promoter-containing vector (pFGS), whereas the cells induced with the vector containing the EF-1α promoter (pEGS) showed very weak PyGUS expression (Figs 2a,b and 3a). This result prompted us to focus on the FCP promoter for the subsequent analyses. The topology of the introduced vector also affected the expression of marker genes. The supercoiled form (covalently closed and circular) of the vector resulted in relatively high expression compared to the linearized form (Fig. 3b). In general, the super-coiled plasmid form is desired for transfection since this can ensure efficient access to the nucleus of eukaryotic cells. For example, in the case of the diatom Phaeodactylum tricornutum, a substantially higher transformation frequency (approximately 30-fold greater) was confirmed with supercoiled DNA compared to linearized DNA1. The amount of the transferred plasmid DNA also affected the expression efficiency. In the present study, 15 and 30 μg of the plasmid DNA per 4.2 × 105 cells showed the best result (Fig. 3c). Consistent with the results of the reverse transcription-polymerase chain reaction (RT-PCR) analysis described above, GUS staining revealed that the infected cells clearly expressed GUS enzyme. Approximately 10–20 positive cells (~0.0024–0.0048%) were detected in one experimental group (4.2 × 105 cells) when 15 μg of the vector was introduced in the cells (Fig. 1c). Green fluorescent protein (GFP) expression was also successfully observed when another transient expression construct (pFGF) was introduced to the cells (Figs 1d and 2c). Furthermore, dose dependency was observed when the amount of the expression vector increased from 7.5 to 30 μg (Fig. 1e).
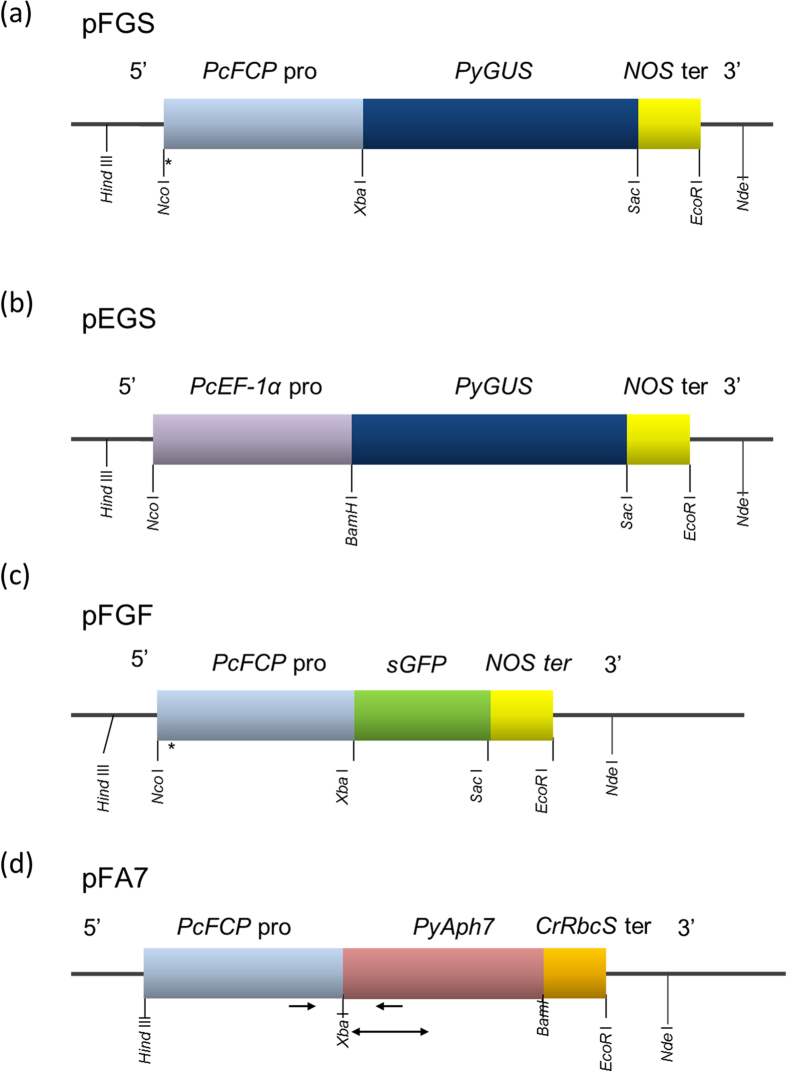
Figure 2. Schematic representations of the expression constructs used in the present study.
(a) pFGS, (b) pEGS, (c) pFGF, (d) pFA7. PcFCPpro, Pleurochrysis carterae FCP promoter; PyGUS, Pyropia yezoensis GUS gene; NOSter, nopaline synthase terminator derived from pBI221 vector; PcEF-1αpro, P. carterae EF-1α promoter; sGFP, a synthetic GFP gene; PyAph7, a hygromycin B-resistant gene optimized for P. yezoensis; CrRbcSter, Chlamydomonas reinhardtii ribulose 1,5-bisphosphate carboxylase/oxygenase small subunit terminator. *Twenty base pairs derived from the pGEM-T Easy vector were inserted. Arrows in (d) indicate a primer set used in the genomic PCR analysis. The double-headed arrow in (d) indicates the probe used in Southern hybridization analysis.
Figure 3. Analysis of transient PyGUS gene expression.
(a) Comparison of PyGUS expression with the FCP promoter and EF-1α promoter. (b) Comparison of PyGUS expression depending on vector topology: the super-coiled (circular) form and linearized form (linear). (c) Relationship between the amount of vector used and PyGUS expression.
Next, we tried to establish an antibiotic-resistant mutant strain using the pFA7 vector, which contains a codon-optimized hygromycin phosphotransferase gene (PyAph7) (Fig. 2d). Based on the results of the transient expression analysis described above, supercoiled DNA was used in the following experiments. Previously, Uji and colleagues succeeded in the stable nuclear transformation of P. yezoensis using PyAph7 and hygromycin B. According to their report, we chose hygromycin B as a suitable selectable marker. In the hygromycin B-resistant test, the minimum inhibitory concentration (MIC) of P. carterae was estimated to be 1.0–1.5 mg/mL (Supplementary Fig. 1); the concentrations of the selective media were set to be slightly higher than the MIC to avoid false-positive results. The cells were tested at two concentrations of antibiotics, 2.5 and 5.0 mg/mL, in SLEP medium (see Methods). In both conditions, the intact cells (control experiment) were killed within 4–5 weeks, whereas the pFA7-introduced cells survived for more than 12 weeks. In addition, some cells were observed to undergo mitosis during the antibiotic selection experiment (Fig. 4a). The surviving cells were then transferred onto solid selective plates containing 2.5 or 5.0 mg/mL hygromycin B. As shown in Fig. 4b, the transformed cells survived on the plate and formed visible resistant colonies within 4 weeks. The cells that survived after selection with 2.5 mg/mL hygromycin B were considered to be antibiotic-resistant, and were used in subsequent experiments.
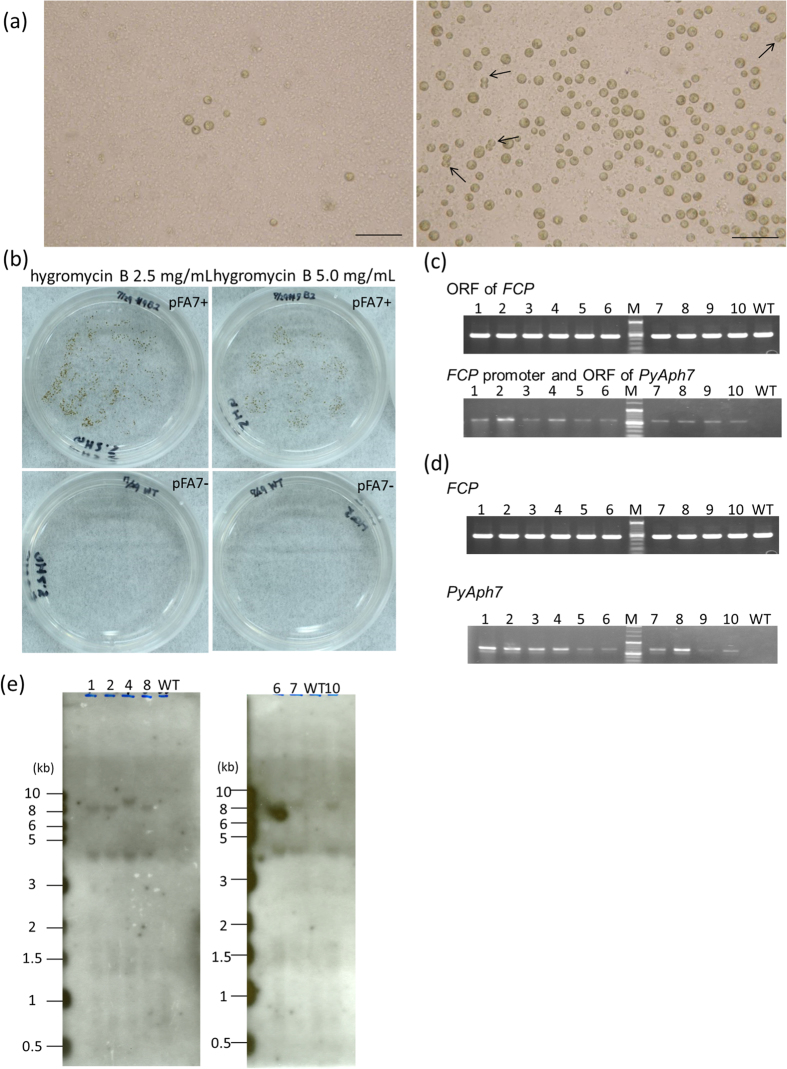
Figure 4. Antibiotic selection and gene expression of mutant strains.
(a) Intact cells (left) and pFA7-introduced cells (right) after 3 weeks of selection on hygromycin B (2.5 mg/mL)-containing SLEP (representative image of n = 4). Arrows indicate the cells undergoing mitosis. Scale bars, 50 μm. (b) pFA7-introduced cells (upper panels) and intact cells (lower panels) on the hygromycin B-containing selective plates after 4 weeks of selection. Brown colonies were observed only in the transformed groups (representative image of n = 4). (c) Genomic PCR for ORF of endogenous FCP (upper), and for a region including the 3′-end of the FCP promoter and the 5′-end of the PyAph7 ORF derived from pFA7 (lower) in the ten transformed mutant strains (1–10) and wild-type strain (WT); M, molecular marker. (d) Expression of FCP (upper) and PyAph7 (lower) in the ten transformed mutant strains (1–10) and wild-type (WT) strain; M, molecular marker. (e) Southern hybridization for seven mutant strains and a WT strain; molecular marker bands are on the left.
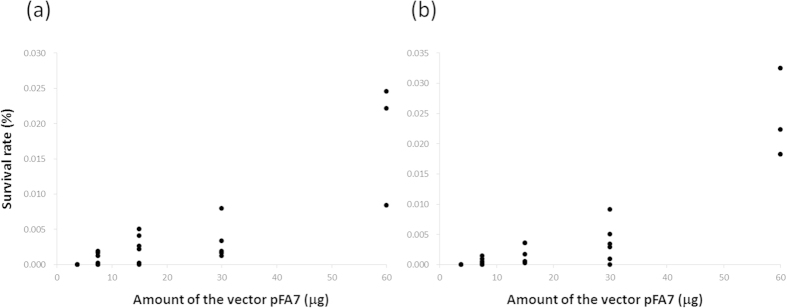
Ten surviving colonies were randomly selected from the selection plate to establish 10 mutant strains, inoculated in the conditioned SWEP medium (see Methods), and grown at a larger scale in non-selective SWEP medium for 8 weeks. To confirm that the transformed cell lines harbored the exogenous PyAph7 gene, genomic PCR was performed with gene-specific primers. In all 10 strains, as well as the wild-type strain, the fragment of the open reading frame region derived from the endogenous FCP gene was detected at the expected size. However, the region including the FCP promoter and PyAph7 derived from pFA7 was amplified only from the 10 transformed strains and not from the wild-type (Fig. 4c). This result indicates that the mutant strains maintained the exogenous PyAph7 gene. RT-PCR analysis revealed that all of the mutant strains also expressed PyAph7 under the non-selecting condition, whereas endogenous FCP expression was detected in all of the strains, including the wild-type (Fig. 4d). To verify the stable integration of PyAph7 into the genome of the antibiotic-resistant strains, we conducted a Southern blot analysis using the seven fastest-growing mutants out of the ten strains described above. Hybridization with a PyAph7-derived probe revealed one or two hybridizing bands in all of the mutant strains (Fig. 4e). The larger signals were approximately 8 kb (strain Nos. 1, 2, 8, and 10), 9 kb (strain No. 4), and 7 kb (strain No. 6) in size. In addition, a band of approximately 4.3 kb was commonly observed in all of the mutant strains. Although the total length of the pFA7 construct (5.5 kb) is slightly larger than the position of the observed band, appearance of the band at 4.3 kb raised the possibility that the resistant construct might have remained as a circular plasmid in the mutant cells. Therefore, we conducted PCR analysis to amplify the flanking region of the PyAph7 gene cassette using a primer set designed based on the pBI221 plasmid sequence. However, we could not detect any of the expected bands in the mutant strains, indicating that the strains did not carry the intact plasmids (Supplementary Fig. 2). These results indicated that at least 4 different mutant cell lines with distinct genotypes were obtained in this study. No hybridizing signals were observed in the wild type. Based on this result, the transformation efficiency is calculated to be 9.5 cells/106 cells (4 mutant cell lines/4.2 × 105 cells). To evaluate whether the transformation rate could be further increased by adding more plasmid DNA, the cells infected with the pFA7 vector were incubated in a non-selective medium for two or three days and then directly subjected to solid-plate selection. The number of surviving colonies increased remarkably in a dose-dependent manner within the range of the amount of the vector (Fig. 5).
Figure 5. Survival rate analysis.
The left and right panels represent colony numbers at 2 days (a) and 3 days (b) of preincubation before hygromycin B selection, respectively.
Discussion
Various methods for effective gene transfer in many algal species have been reported, including electroporation, biolistic DNA delivery (particle gun), and agitation with glass beads8,12. Among these techniques, the particle gun approach has recently been accepted as the most reliable, as it directly delivers DNA into the cells. In general, use of a gene gun results in higher transformation efficiency than PEG; however, this was not the case in our study. We detected transient GUS gene expression in the cells bombarded with pFGS vector, but failed to isolate hygromycin B-resistant strains using the pFA7 vector (data not shown). In the case of a gene gun, the expression vector is coated on the gold particle prior to bombardment. Thus, it is possible that the supercoiled plasmid DNA can be converted to a relaxed form during this step, resulting in low or unstable transformation efficiency. PEG-mediated transient or stable transformation is not as common as the other methods described above. However, effective gene transfer mediated by PEG has been reported in the green microalgae Chlamydomonas reinhardtii13 and Chlorella ellipsoidea14, and both of these studies were conducted with protoplasts or cell wall-deficient cells in logarithmic phase. We presume that the main obstacles that have hindered development of transgenic techniques for haptophytes thus far are their rigid calcified coccospheres and difficulties in controlling the proliferation rate. In this study, we have overcome these problems while developing a novel preparation method of protoplasts, which could easily enter the log phase of growth immediately after transfer to an appropriate medium.
The method presented here unambiguously meets the criteria for successful stable transformation, including successful integration of a foreign gene into the genome and expression of mRNA and functional translated products. Our next aim is to establish a more robust gene-manipulation tool such as a genome-editing technique. We believe that our newly developed method will provide valuable insight for achieving this next goal.
Methods
Cultivation of the coccolithophorid alga
Pleurochrysis carterae was grown in seawater-based Eppley’s medium15 (SWEP) for usual cultivation or in artificial seawater-based Eppley’s medium (SLEP) for the transformation experiments (Table 1). The culture conditions were 20 °C, with a photoperiod of 16-h light:8-h dark and no agitation or aeration. In usual cultivation, the medium was renewed every 4 weeks.
Table 1. Media and solutions.
| Eppley’s medium | ||
|---|---|---|
| 1000x metal stock | FeCl3 · 6H2O | 1000 mg |
| CuSO4 · 5H2O | 4 mg | |
| Na2MoO4 · 4H2O | 130 mg | |
| ZnSO4 · 7H2O | 250 mg | |
| CoCl2 · 6H2O | 4 mg | |
| MnSO4 · H2O | 620 mg | |
| Na2EDTA | 6 g | |
| fill up to | 1000 mL | |
| 1000x NKP stock | KNO3 | 5.05 g |
| K2HPO4 | 0.87 g | |
| fill up to | 100 mL | |
| 1000x vitamine | thiamine | 200 mg |
| biotin | 1 mg | |
| cyanocobalamin | 10 mg | |
| fill up to | 1000 mL | |
| hypo-osmotic buffer | HEPES | 10 mM |
| KCl | 100 mM | |
| NaOH | 350 μM | |
| MaMg buffer | mannitol | 400 mM |
| (pH was adjusted to 5.8 with KOH) | MgCl2 | 15 mM |
| MES | 0.1% (w/v) | |
| 40% PEG in CMS solution | PEG | 4 g |
| CMS solution | 6 mL | |
| CMS solution | mannnitol | 400 mM |
| (pH was adjusted to 7.0 with KOH) | Ca(NO3)2 | 100 mM |
Expression constructs
The sequences of the primers and the PCR conditions are summarized in Table 2. The upstream regions of FCP and EF1α were obtained by inverse PCR as follows. Genomic DNA was extracted from the wild-type strain of P. carterae using the Wizard Genomic DNA Purification Kit (Promega). The DNA was digested with PstI for FCP or with XbaI for EF1α, and ligated using DNA Ligation Kit (TaKaRa). The first PCRs were carried out using the primer pairs FCPinvF4 and FCPinvR1 or EF1alINVF1 and EF1alINVR1. The nested PCR was carried out with the primer pairs FCPinvF5 and FCPinvR2 or EF1alINVF2 and EF1alINVR2. LA-Taq (TaKaRa) and KOD plus Neo (Toyobo) polymerases were used for FCP and EF1α, respectively. The PCR products of approximately 1,300 bp (FCP) and 2,700 bp (EF1α) were subcloned into the pGEM-T easy vector (Promega), and the sequences were determined. To prepare the expression constructs for transient expression, we used pBI221 as the basic vector. For transient expression of the GUS gene, the promoter region of pBI221 was replaced by the upstream region of FCP or EF1α(DDBJ accession nos. LC075595 and LC075596). Using the obtained sequences as the templates, approximately 1,200 bp of the promoter regions were amplified by PCR using FPCproLF1NcoI and FCPproLR2XbaI or EF1aproF1NcoI and EF1aproR1BamHI, and subcloned into the corresponding sites of the vector. Then, the GUS gene was replaced by an artificial PyGUS gene, in which codon usage was adapted to that of the macroalga Pyropia yezoensis (Bangiales, Rhodophyta). For transient expression of the GFP gene, the vector was prepared by replacing the GUS gene with the sGFP gene amplified from pPyAct1-sGFP16 using the primers PcGFP1F1XbaI and PcGFP1R1SacI. For stable expression, the expression construct was prepared using pEA711 as the basic vector. The promoter region of FCP was obtained by PCR using FPCproLF1HindIII and PCPproLR2XbaI and subcloned into the HindIII-XbaI site of the pEA7 vector. These four expression constructs were designated as pFGS, pEGS, pFGF, and pFA7, respectively (Fig. 2). The vectors were propagated in the Escherichia coli strains XL-1blue or DH5α, and extracted using NucleoBond (Macherey-Nagel) immediately before analysis.
Table 2. Sequences of the primers and PCR programs.
| Gene | Reaction | Primer(sense, 5′→3′) | Primer(antisense 5′→3′) | PCR condition |
|---|---|---|---|---|
| FCP | 1st inverse PCR | FCPinvF4 | FCPinvR1 | 94 °C 30 s, 50 °C 30 s, 72 °C 4 min |
| CAT GTT GAT GAA GCC GAC GAG | TGC GCC GAA CTG TTC CGA T | 25 cycles | ||
| Nested inverse PCR | FCPinvF5 | FCPinvR2 | 94 °C 30 s, 50 °C 30 s, 72 °C 4 min | |
| CGT TTT CGC CGG CAA TCG A | CCG AGA TCA CGT GGC ACA AGA T | 25 cycles | ||
| Subcloning of the promoter region | FCPproLF1NcoI | FCPproLR2XbaI | 98 °C 10 s, 57 °C 30 s, 68 °C 90 s | |
| CCA TGG CTG CAT GCA GTA TCA ACA GGC A | TCT AGA CTC GCG CAT GGC TTC ACG AGT GTG TGT G | 30 cycles | ||
| FCPproLF1HindIII | FCPproLR2XbaI | 98 °C 10 s, 55 °C 30 s, 68 °C 90 s | ||
| AAG CTT CTG CAT GCA GTA TCA ACA GGC A | see above | 25 cycles | ||
| Expression analysis | FCPExAF2 | FCPExAR2 | 94 °C 30 s, 58 °C 30 s, 72 °C 30 s | |
| ATG GCT CTC TCC CTG TCC G | CGA CCG TTG TTC AGC TCG AT | 30 cycles | ||
| EF1α | 1st inverse PCR | EF1alNVF1 | EF1alNVR1 | 94 °C 10 s, 59 °C 30 s, 68 °C 2.5 min |
| CCT CGA CAA GCA GAA CAT GCC | ATG GCG GTG GTG AAG TTA CC | 25 cycles | ||
| Nested inverse PCR | EF1alNVF2 | EF1alNVR2 | 94 °C 10 s, 59 °C 30 s, 68 °C 2.5 min | |
| TCG ACA TTC CGG GCG AGA TC | GCG CCG GAG ATC ATG TTC TTG | 25 cycles | ||
| Subcloning of the promoter region | EF1aproF1NcoI | EF1aproR1BamHI | 98°C 10 s, 57°C 30 s, 68°C 1 min | |
| CCA TGG CTG GGG CTG TTG CTG AGA TAC | GGA TCC CTT CTC CAT CTC ACG CTC CGG | 27 cycles | ||
| 94°C 30 s, 52°C 30 s, 72°C for 40 s | ||||
| 40 cycles | ||||
| Expression analysis | EF1aExAF1 | EF1aExAR2 | 94 °C 30 s, 52 °C 30 s, 72 °C for 40 s | |
| CGA CAA CCT CAA CAA GAA GTC GAC | ATT GGC GAG TAG CCG AGC TT | 40 cycles | ||
| Genomic PCR | EF1aExAF1 | EF1aExAR2 | 94 °C 30 s, 60 °C 30 s, 72 °C 30 s | |
| see above | see above | 25 cycles | ||
| GFP | Subcloning of the ORF region | PcGFP1F1XbaI | PcGFP1R1SacI | 98 °C 10 s, 57 °C 30 s, 68 °C 1 min |
| TCT AGA ATG GTG AGC AAG GGC GAG | GAG CTC TTA CTT GTA CAG CTC GTC CAT G | 27 cycles | ||
| PyGUS | Expression analysis | PyGUSExAF2 | PyGUSExAR2 | 94 °C 30 s, 52 °C 30 s, 72 °C for 40 s |
| GCA GTT CCT GAT CAA CCA CA | AGA ACA TCA CGT TCA CGC AC | 40 cycles | ||
| PyAph7 | Expression analysis | AphExAF1 | AphExAR2 | 94 °C 30 s, 58 °C 30 s, 72 °C 45 s |
| GAC GCA GGA GTC CCT GCT | ACG AAG ATG TTG GTC CCG T | 30 cycles | ||
| FCP promoter -PyAph7 | Genomic PCR | checkFCPproF1232 | AphExAR1 | 94 °C 30 s, 60 °C 30 s, 72 °C 30 s |
| ACA CTG CAC CGT CCA GGT T | TCC GGG AAG ACC TCG GAG T | 30 cycles |
PEG-mediated transfer
The cells were harvested from 50 mL of a 4–5-week-old culture by centrifugation at 110 × g for 5 min, and treated with proteinase K in SLEP (250 μg/mL) at 30 °C for 2 h and then at 20 °C for 2.0–2.5 h. To promote “molting” of the cell wall (coccosphere), the cells were treated with a hypo-osmotic buffer (Table 1)9 for 5–10 min in a 100-mm-diameter glass Petri dish, and removal of the coccosphere was carefully checked under the microscope. The protoplasts were then filtrated with three ply of Miracloth (EMD Millipore), collected by centrifugation at 60 × g for 3 min, and incubated in a 0.4 M mannitol solution at 4 °C for 20 min. After filtration through a tetron filter (180-mesh), approximately 4.2 × 105 cells were resuspended in 320 μL of MaMg buffer (Table 1) and mixed with 7.5–60 μg of the expression vector or 15 μg of 500-kDa FITC-labeled dextran (SIGMA) diluted in 30 μL of double-distilled water. The same volume (350 μL) of a 40% PEG solution (Table 1) was added to the cell suspension and mixed by gentle shaking. PEG with a molecular weight of 6,000 (WAKO) was used throughout the present study. After 15 min of PEG reaction, the cells were washed with 5 mL of SLEP, collected by centrifugation at 60 × g for 5 min, and incubated in SLEP at 20 °C for 2 to 3 days.
Transient expression of GUS and GFP
After 3 days of incubation, transient GUS expression was visualized with a 50 mM phosphate buffer (pH 7.0)-based GUS staining solution, containing 0.5 mM of X-Gluc, 0.5 mM of K3Fe(CN)6 and K4Fe(CN)6, 0.005% of TritonX-100, and 0.4 M mannitol. The cells were incubated at 25 °C for 16 h and stained cells were observed under the microscope. Expression of GFP was observed under a fluorescent microscope at 24 h after the PEG treatment.
Isolation of antibiotic-resistant cells
To select for antibiotic-resistant cells, the cells were first cultured in SLEP containing 2.5 or 5.0 mg/mL of hygromycin B for 4 weeks. As a second selection step, the surviving cells were spread on a 0.2% gellan gum/45% SLEP plate containing 2.5 or 5.0 mg/mL of hygromycin B, and incubated until the colonies became visible and could be isolated (approximately 4–5 weeks). The colonies were picked and cultured in modified SLEP (SLEP with a 5-times concentration of vitamin mixture, 2.5 ppm of glycolic acid, and 0.625 mg/mL of ampicillin) for 3 days in a 96-well plate, and then transferred to a larger volume of SLEP in Petri dishes or glass flasks.
For the survival rate analysis, the cells infected with pFA7, at various amounts ranging from 0.375 to 60 μg, were directly restreaked onto the selective medium plate with 2.5 mg/mL of hygromycin B after 2 or 3 days of incubation. The number of colonies was counted after 3 weeks of selection.
RT-PCR and genomic PCR
The sequences of the primers and the conditions used for PCR are summarized in Table 2. Transient expression of PyGUS was examined by RT-PCR. After 3 days of incubation, total RNA was extracted from the cells using RNeasy Plant Mini Kit (Qiagen), and treated with TurboDNase Kit (Ambion) to avoid contamination of genomic DNA. Using 50 ng of total RNA as a template, cDNA was synthesized with the poly-T17 primer and SuperScriptIII reverse transcriptase (Invitrogen). To quantify the expression level of PyGUS and EF1α, PCR was performed using LA-Taq polymerase with the primer pairs PyGUSExAF2 and PyGUSExAR2, and EF1aExF1 and EF1aExAR2. To examine the expression of PyAph7, total RNAs were extracted from the ten mutant strains, and cDNAs were prepared from 500 ng of total RNA as described above. The Aph7 mRNA was amplified by PCR with the primers AphExAF1 and AphExAR2 using LA-Taq polymerase with GC buffer 2 (TaKaRa). Expression of endogenous FCP was also examined as a positive control using the primer pair FCPExAF2 and FCPExAR2.
Genomic DNA was extracted from the wild-type strain or hygromycin B-resistant strains with Chelex 100 resin (BioRad). The primers, checkFCPproF1232 and AphExAR1, for genomic DNA were designed to amplify the FCP promoter and Aph7 gene, respectively (Fig. 2). As a control experiment, the open reading frame (ORF) of FCP was also amplified with the primers FCPExAF2 and FCPExAR2. Using 5 ng of genomic DNA as a template, PCR was carried out with LA-Taq polymerase and GC buffer 2.
Southern blot analysis
Genomic DNAs were extracted from the eight fastest-growing strains, strains Nos. 1, 2, 4–8, and 10. The DNA samples (2.5 μg) were digested with BssHII and transferred to a nylon membrane. The 32P-labeled DNA probe of PyAph7 (G3 to T692, Fig. 2d) was prepared by a random-prime method and hybridized with the membrane. Signals were visualized using autoradiography.
Additional Information
How to cite this article: Endo, H. et al. Stable Nuclear Transformation System for the Coccolithophorid Alga Pleurochrysis carterae. Sci. Rep. 6, 22252; doi: 10.1038/srep22252 (2016).
Supplementary Material
Acknowledgments
This work was supported in part by the Precursory Research for Embryonic Science and Technology program of the Japanese Science and Technology Agency, and Grant-in-Aid for Scientific Research No. 22228006 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Author Contributions H.E., N.S., K.I. and H.N. conceived of the study. H.E. designed all the experiments. H.E., M.Y. and T.U. conducted all the experiments. H.E. wrote the manuscript with support from all authors.
References
- Apt K. E., Kroth-Pancic P. G. & Grossman A. R. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol. Gen. Genet. 252, 572–279 (1996). [DOI] [PubMed] [Google Scholar]
- ten Lohuis M. R. & Miller D. J. Genetic transformation of dinoflagellates (Amphidinium and Symbiodinium): expression of GUS in microalgae using heterologous promoter constructs. Plant J. 13, 427–435 (1998). [Google Scholar]
- Poulton A. J., Adey T. R., Balch W. M. & Holligan P. M. Relating coccolithophore calcification rates to phytoplankton community dynamics: Regional differences and implications for carbon export. Deep-Sea Res. 54, 538–557 (2007). [Google Scholar]
- Shiraiwa Y. Physiological regulation of carbon fixation in the photosynthesis and calcification of coccolithophorids. Comp. Biochem. Physiol. B 136, 775–783 (2003). [DOI] [PubMed] [Google Scholar]
- Rost B. & Riebesell U. In Coccolithophores from molecular processes to global impact (eds Thierstein H. R. & Young J. R.) 99–125 (Springer, 2004). [Google Scholar]
- Read B. A. et al. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499, 209–213 (2013). [DOI] [PubMed] [Google Scholar]
- Moheimani N. R. & Borowitzka M. A. The long-term culture of the coccolithophore Pleurochrysis carterae (Haptophyta) in outdoor raceway ponds. J. Appl. Phycol. 18, 703–712 (2006). [Google Scholar]
- Coll J. M. Review. Methodologies for transferring DNA into eukaryotic microalgae. Spanish J. Agric. Res. 4, 316–330 (2006). [Google Scholar]
- Takayanagi T. et al. Protoplast formation of the coccolithophorid Pleurochrysis haptonemofera in hypoosmotic K+ solution: Shedding of the coccosphere and regrowth of the protoplast in normal medium. Mar. Biotechnol. 9, 56–65 (2007). [DOI] [PubMed] [Google Scholar]
- Fukuda S. et al. Factors influencing efficiency of transient gene expression in the red macrophyte Porphyra yezoensis. Plant Sci. 174, 329–339 (2008). [Google Scholar]
- Uji T., Hirata R., Fukuda S., Mizuta H. & Saga N. A codon-optimized bacterial antibiotic gene used as selection marker for stable nuclear transformation in the marine red alga Pyropia yezoensis. Mar. Biotechnol. 16, 251–255 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann A. Algal transgenics and biotechnology. Transgenic Plant J. 1, 81–98 (2007). [Google Scholar]
- Kindle K. L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. 87, 1228–1232 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-H. et al. Stable integration and functional expression of flounder growth hormone gene in transformed microalga, Chlorella ellipsoidea. Mar. Biotechnol. 4, 63–73 (2002). [DOI] [PubMed] [Google Scholar]
- Eppley R. W., Holmes R. W. & Strickland J. D. H. Sinking rates of marine phytoplankton measured with a fluorometer. J. Exp. Mar. Biol. Ecol. 1, 191–208 (1967). [Google Scholar]
- Uji T., Takahashi M., Mikami K. & Saga N. Visualization of nuclear localization of transcription factors with cyan and green fluorescent proteins in the red alga Porphyra yezoensis. Mar. Biotechnol. 12, 150–159 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.