Abstract
The basic leucine zipper (bZIP) transcription factor family plays crucial roles in various aspects of biological processes. Currently, no information is available regarding the bZIP family in the important tropical crop cassava. Herein, 77 bZIP genes were identified from cassava. Evolutionary analysis indicated that MebZIPs could be divided into 10 subfamilies, which was further supported by conserved motif and gene structure analyses. Global expression analysis suggested that MebZIPs showed similar or distinct expression patterns in different tissues between cultivated variety and wild subspecies. Transcriptome analysis of three cassava genotypes revealed that many MebZIP genes were activated by drought in the root of W14 subspecies, indicating the involvement of these genes in the strong resistance of cassava to drought. Expression analysis of selected MebZIP genes in response to osmotic, salt, cold, ABA, and H2O2 suggested that they might participate in distinct signaling pathways. Our systematic analysis of MebZIPs reveals constitutive, tissue-specific and abiotic stress-responsive candidate MebZIP genes for further functional characterization in planta, yields new insights into transcriptional regulation of MebZIP genes, and lays a foundation for understanding of bZIP-mediated abiotic stress response.
Due to their sessile nature, plants cannot move to avoid unfavorable conditions, thus they have to cope with various harsh environmental factors1. To survive in those stressors, plants developed various mechanisms for their protection, such as synthesis of functional proteins with diverse functions. Expression of functional proteins is largely controlled by specific transcription factors (TFs)2. Among TF families, the basic leucine zipper (bZIP) transcription factor family is one of the largest and most conserved families. bZIP TFs are named according to the conserved bZIP domain that is composed of 60–80 amino acids and contains two functional regions: a basic region and a leucine zipper. These two regions were linked by a hinge region3,4. The basic region is conserved, in which an invariant motif N-x7-R/K-x9 with about 18 amino acid residues is responsible for nuclear localization and DNA binding. The following leucine zipper motif that consists of several repeats of leucine or other hydrophobic amino acids is involved in recognition and dimerization of bZIPs3,4,5.
In plants, there is considerable evidence showing that bZIP TFs play crucial roles in various aspects of biological processes, including organ differentiation, embryogenesis, seed maturation, flower and vascular development3. Increasing evidences have also indicated that bZIP TFs take part in the regulation of plants’ response to biotic and abiotic stress3,5. Phytohormone abscisic acid (ABA) plays a central role in plants responding to abiotic stress through regulating the expression of numerous genes and related physiological process. In recent years, ABA perception and signal transduction have begun to clarify, which showed that RCAR/PYR/PYL ABA receptors, group A PP2Cs, and SnRK2s control the ABA signaling pathway in land plants6. SnRK2s can activate AREB/ABFs by phosphorylation in the ABA-dependent signaling network induced by abiotic stress5,6. AREB/ABFs, belonging to group A bZIPs, have been confirmed to modulate transcription of ABA-dependent genes that carry the ABRE cis-element, resulting in ABA response or environmental adaptation. In Arabidopsis, accumulated evidence has suggested the roles of group A bZIPs, such as, AtbZIP39, AtbZIP36, AtbZIP38, AtbZIP66, AtbZIP40, AtbZIP35 and AtbZIP37, in abiotic stress response or ABA signaling5,6. For example, ABF2/AREB1 is activated by phosphorylation of class 3 SnRK2s and overexpression of ABF2/AREB1 increased plants resistance to drought stress and sensitivity to ABA6,7,8. In rice, some bZIP family members, including OsbZIP23, OsbZIP46, OsbZIP71, and OsbZIP16, was also confirmed to positively regulate plants resistance to abiotic stress9. Together, these evidences indicate that bZIPs are crucial transcription factors involved in plants response to abiotic stress.
To date, numerous bZIP family members have been identified by genome-wide analyses in various species4,10,11,12. However, no evidence is available regarding the bZIP family in the important tropical crop cassava. Cassava is the third most important crop after rice and maize in Africa, Asia, and Latin America13. Due to its high starch production and limited input, cassava can provide source of dietary carbohydrate for over 600 million people worldwide, and is also considered as a producer of industrial starch and bioethanol14,15. Notably, cassava can effectively utilize light, heat and water resources; thus it shows resistance to drought and low-fertility environment16. However, the mechanisms by which cassava responds to abiotic stress are poorly understood. Thus, understanding of the molecular mechanisms underlying the resistance of cassava to abiotic stress may provide effective methods for genetic improvement of stress resistance for cassava and other crops. Previously, we get the high-quality sequencing data of cassava wild ancestor and cultivated varieties, thus providing an excellent opportunity for genome-wide analysis of cassava genes17. Due to the significance of bZIP TFs in various aspects of biological processes, especially for its crucial roles in abiotic stress response and ABA signaling, the bZIP family was chosen as a candidate for a systematic analysis in cassava.
Results
Identification and evolutionary analysis of bZIPs in cassava
To extensively identify cassava bZIP genes, we use BLAST and Hidden Markov Model searches to search the cassava genome database with bZIP sequences from Arabidopsis and rice as queries. After these programs, 77 putative bZIP members were characterized from cassava, and further conserved domain detection confirmed that all the identified bZIPs harbor the conserved bZIP domain that is the basic characteristics of bZIP family. The 77 predicted full-length cassava bZIP proteins varied from 129 (MebZIP69) to 766 (MebZIP1) amino acid residues and the relative molecular mass ranged from 15.0 (MebZIP69) to 83.2 (MebZIP1) kDa, with isoelectric points in the range of 5.23–10.18 (Supplementary Table S1).
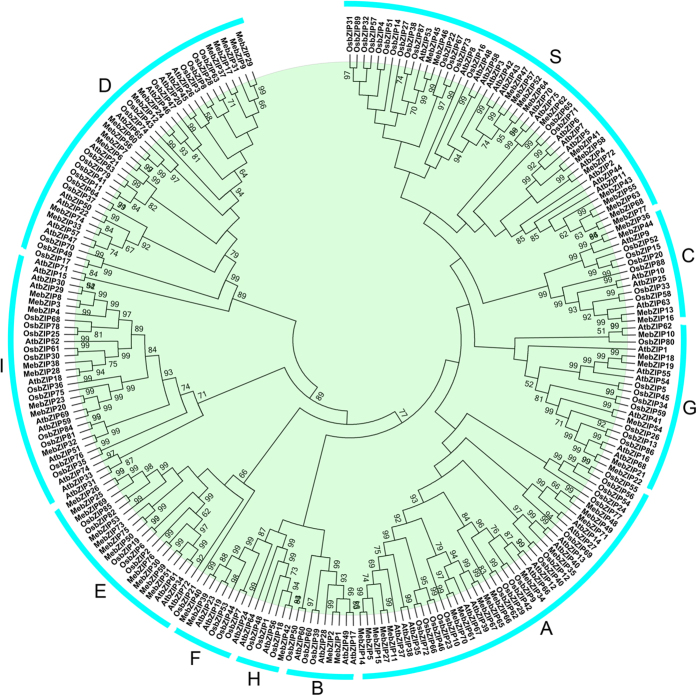
To characterize the evolutionary relationships between bZIPs from cassava and other known bZIPs from Arabidopsis and rice, an unrooted Neighbor-Joining tree was created (Fig. 1; Supplementary Table S2). The results showed that the 77 MebZIPs could be assigned to 10 subfamilies, together with their orthologous bZIPs from Arabidopsis and rice. Subfamily H only contains one MebZIP protein, whereas subfamilies A and S each have the maximum 15 MebZIP members, suggesting the existence of a diversified bZIP family in cassava with diverse functions. Generally, bZIPs from cassava have closer relationships with the bZIPs from Arabidopsis than that from rice, which is accord with the current understanding of plant evolutionary history. Evolutionary analysis also identified some closely related orthologous bZIPs between cassava and Arabidopsis, indicating that an ancestral set of bZIP genes existed prior to the divergence of cassava and Arabidopsis.
Figure 1. Evolutionary analysis of bZIP proteins from cassava, Arabidopsis, and rice.
A total of 77 bZIPs from cassava, 72 bZIPs from Arabidopsis and 89 bZIPs from rice were used to creat the NJ tree with 1000 bootstrap. The bZIP proteins are grouped into 10 subgroups (A,B,C,D,E,F,G,H,I,S).
Gene structure and conserved motifs of cassava bZIPs
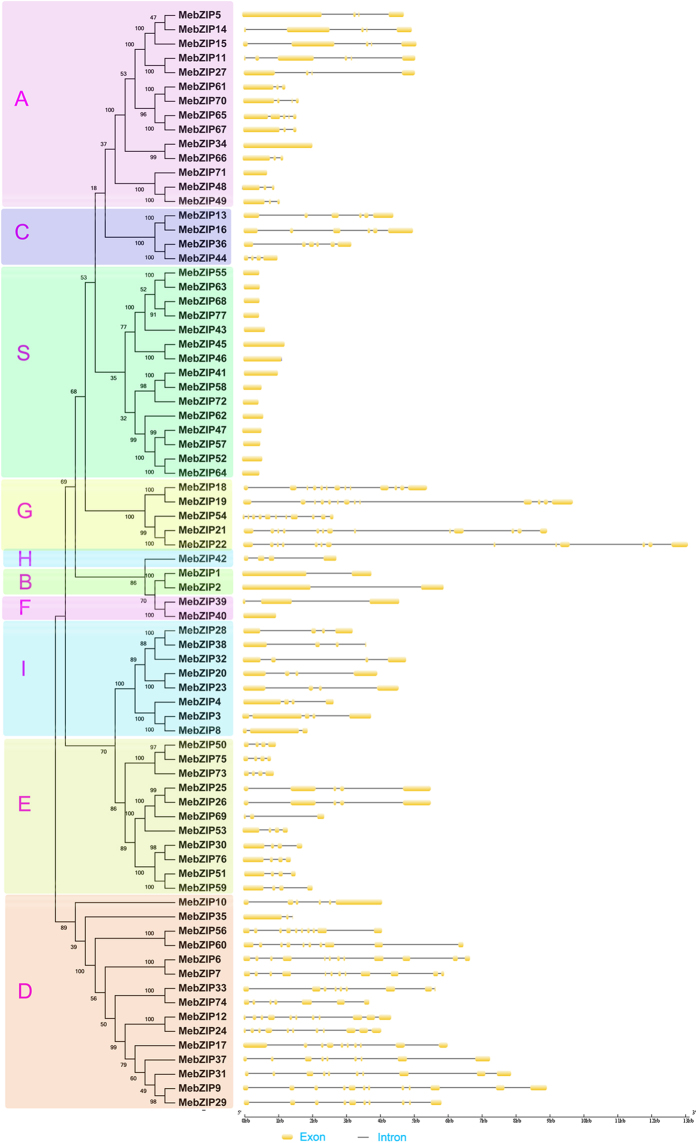
To better understand the structural features of MebZIP genes, intron/exon structure was detected based on their evolutionary relationships (Fig. 2). Gene structure analysis showed that the number of introns of MebZIP genes varied from 0 to 12. Subfamilies A, C, S, H, B, F, I, and E contained 0–5 introns, whereas subfamilies G and D had 5–12 introns, except for MebZIP35. Interestingly, no intron was detected in MebZIP genes of subfamily S. Generally, most of MebZIP genes in the same subfamilies showed similar exon-intron structure, which supports their close evolutionary relationship and the classification of subfamilies.
Figure 2. The exon-intron structure of MebZIP genes based on the evolutionary relationship.
The NJ evolutionary tree was created with 1000 bootstrap based on the full length sequences of MebZIPs. Exon-intron analyses of MebZIP genes were carried out with GSDS. Lengths of exons and introns of each MebZIP gene were exhibited proportionally.
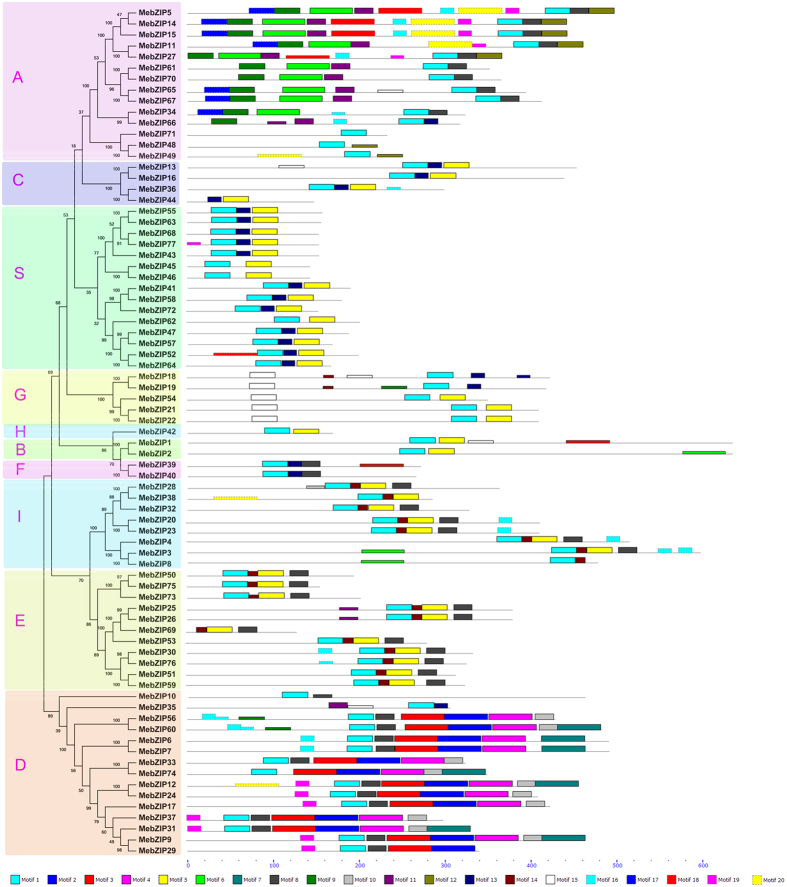
To investigate the structural diversity and functional prediction of MebZIPs, a total of 20 conserved motifs in the MebZIPs were captured by MEME software and subsequent annotation with InterPro (Supplementary Figure S1; Fig. 3; Supplementary Table S3). Motif 1 was annotated as basic-leucine zipper (bZIP) domain; motif 2 was annotated as transcription factor TGA like domain; and motif 15 was annotated as G-box binding, MFMR domain (Supplementary Table S3). Notably, all the MebZIPs, except for MebZIP44 and MebZIP69, had bZIP domain. MebZIP44 and MebZIP69 showed high identity with MebZIP36 and MebZIP26, respectively. It was observed that MebZIP44 and MebZIP69 exhibited C-terminal deficiency, which may result from the alternative splicing of MebZIP36 and MebZIP26, respectively. These two genes may not be functional bZIP transcription factors. Beside the bZIP domain, each subfamily of MebZIPs shares some common motifs. Most of bZIP members in subfamily A contain motifs 6, 8, 9, and 11; subfamily C shares motifs 5 and 13; subfamilies B, S, and H possess motif 5; subfamily G harbors motif 15; subfamily F shows motifs 8 and 13; motif 14 is present in subfamily I; motifs 5, 8, and 14 appeared in subfamily E; subfamily D have motifs 2 and 3. Additionally, some motifs represent the variable motifs across the bZIP gene family. For instance, motifs 9, 12, and 17 only appeared in subfamily A; and motifs 2, 4, 7, and 10 only appeared in subfamily D. These results indicated that bZIP members clustered in same subfamilies showed similar motif characteristic, suggesting functional similarities among members in the same subfamily.
Figure 3. Conserved motifs of cassava bZIP proteins according to the evolutionary relationship.
The conserved motifs in the MebZIP proteins were identified by MEME software. Grey lines represent the non-conserved sequences, and each motif is indicated by a colored box numbered at the bottom. The length of motifs in each protein was exhibited proportionally.
Expression profiles of MebZIP genes in different tissues of two cassava genotypes
To seek insights into the roles of MebZIP genes in cassava development, total RNA was isolated from stems, leaves, and storage roots of cultivated varieties (Arg7) and wild subspecies (W14) for transcriptome analysis. Transcriptome analysis showed a transcript abundance of 55 MebZIP genes in different tissues, while the rest of the 22 MebZIP genes were not covered in the RNA-seq libraries (Fig. 4; Supplementary Table S4).
Figure 4. Expression profiles of MebZIP genes in different tissues of two cassava genotypes.
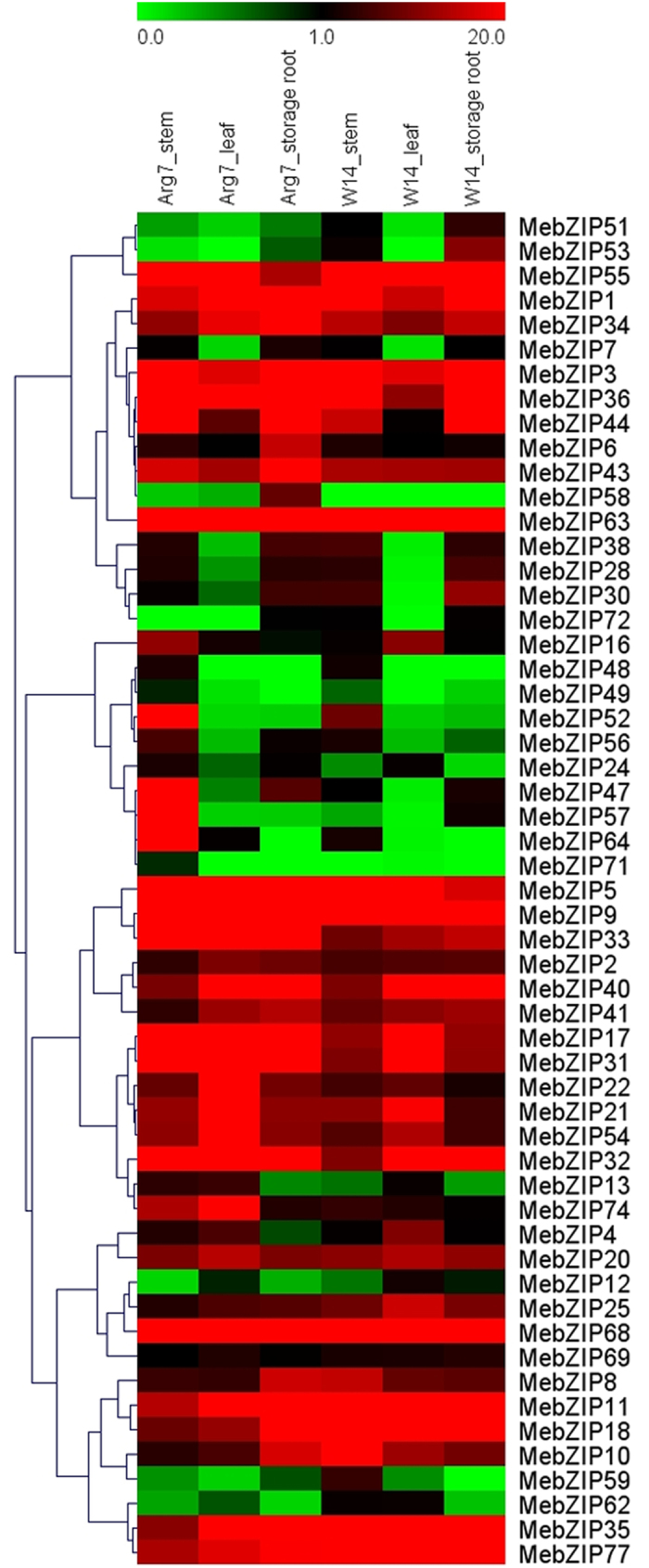
FPKM value was used to create the heat map with clustering. The scale represents the relative signal intensity of FPKM values.
For Arg7 variety, all the 55 MebZIP genes (100%) showed transcripts in different tissues, in which 28 (50.9%), 26 (47.3%), and 27 (49.1%) genes exhibited high transcriptional abundance (value >10) in stem, leaf, and storage root tissues, respectively. Moreover, there were 21 MebZIP genes (MebZIP-9, -5, -63, -36, -68, -3, -1, -11, -40, -43, -33, -17, -34, -31, -32, -35, -77, -55, -21, -54, and -20) that had high expression levels (value >10) in all the tested tissues. Additionally, MebZIP72 did not show transcripts in stem; MebZIP-72, -53, -48, -71 did not show transcripts in leaf; and MebZIP-64, -49, -48, and -71 did not show transcripts in storage root.
For W14 subspecies, all the 55 MebZIP genes, except for MebZIP58, had transcripts in different tissues, in which 23 (42.6%), 27 (50%), 25 (46.3%) genes had high transcriptional abundance (value >10) in stem, leaf, and storage root tissues, respectively. Moreover, 19 MebZIP genes (MebZIP-68, -55, -63, -9, -3, -36, -1, -40, -18, -77, -35, -32, -11, -5, -34, -43, -17, -31, and -20) showed high transcriptional levels (value > 10) in all the tested tissues. Additionally, MebZIP-71 and -58 did not show transcripts in stem; MebZIP-53, -72, -49, -48, and -58 did not show transcripts in leaf; and MebZIP-59, -64, -71, -48, and -58 did not show transcripts in storage root.
For comparing the expression profiles of MebZIP genes in different tissues between Arg7 and W14, 49 genes (89.1%) had transcripts accumulation in all tissues of Arg7, while 47 genes (85.5%) in W14, suggesting the constitutive expression patterns for these genes. On the contrary, the absence of detection might represent the distinct temporal or spatial expression patterns for the remaining MebZIP genes. Notably, 18 MebZIP genes (MebZIP-9, -5, -63, -36, -68, -3, -1, -11, -40, -43, -17, -34, -31, -32, -35, -77, -55, and -20) showed high transcript abundance (value >10) in all tested tissues of Arg7 and W14, indicating key roles for these genes in tissue development. Overall, MebZIP genes had similar expression profiles in different tissues of Arg7 and W14, indicating that most of bZIP genes played similar roles in tissue development of the two genotypes. However, there were some genes that exhibited differential expression profiles. MebZIP-18, -10, and -8 had high transcript abundance (value >10) in stem of W14, whereas low in stem of Arg7. On the contrary, MebZIP-33, -52, -54, -74, -64, -16, -47, and -57 had high transcriptional abundance (value >10) in stem of Arg7, while low in stem of W14. This phenomenon was also observed in leaf and storage root of Arg7 and W14. These findings indicated differential roles for these genes in tissue development in different genotypes.
Expression profiles of MebZIP genes responding to drought stress in different cassava genotypes
To get some clues on the roles of MebZIP genes responding to drought stress, cassava seedlings of three genotypes were subjected to water withholding and then the leaves and roots tissues were sampled to extract RNA for subsequent RNA-seq analysis (Fig. 5; Supplementary Table S5). According to the transcriptome data, all the cassava bZIP genes, except for MebZIP50, showed the corresponding expression data. In Arg7 variety, 32 (42.1%) and 23 (30.3%) MebZIP genes were induced by drought in leaves and roots, respectively. In South China 124 (SC124) variety, 28 (36.8%) and 26 (34.2%) MebZIP genes showed upregulation after drought in leaves and roots, respectively. In W14 subspecies, 21 (27.6%) and 40 (52.6%) MebZIP genes were upregulated by drought in leaves and roots, respectively. These results suggest that the number of MebZIP genes upregulated by drought was greater in W14 than those in Arg7 and SC124, suggesting the comprehensive response of bZIP genes to drought at transcriptional levels in W14 subspecies. Based on the transcriptome data, we also found that the number of bZIP genes up-regulated by drought was significantly more in roots than those in leaves in W14, whereas less in Arg7 and SC124.
Figure 5. Expression profiles of MebZIP genes in leaves and roots of three cassava genotypes after drought treatment.
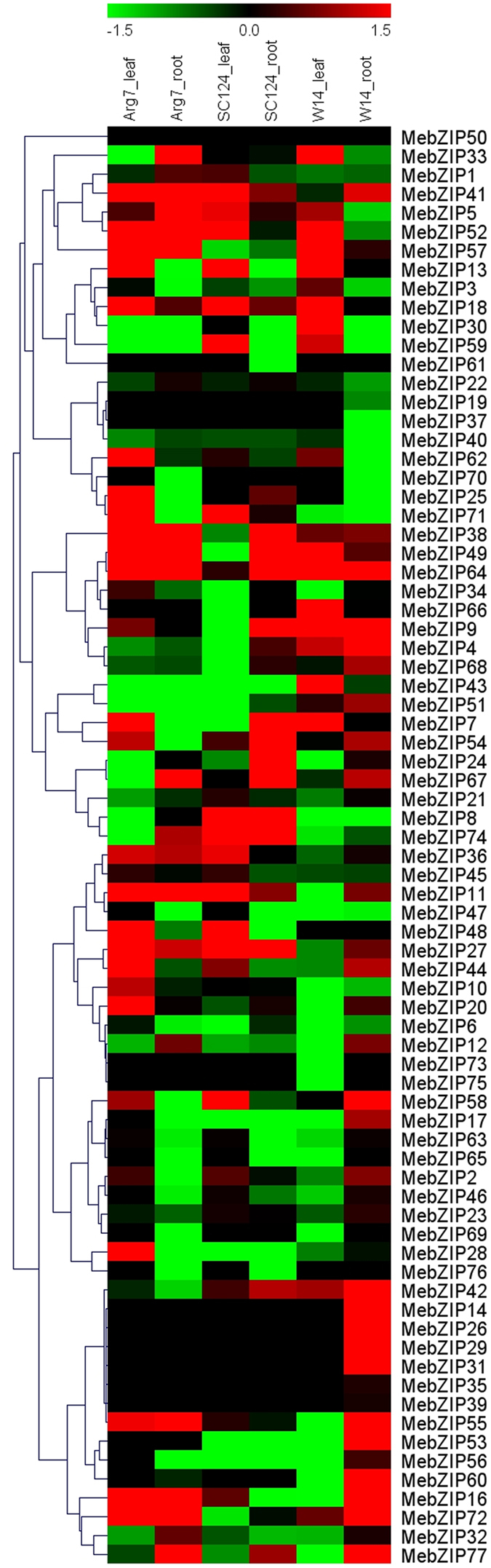
Log2 based FPKM value was used to create the heat map with clustering. The scale represents the relative signal intensity of FPKM values.
Generally, the expression patterns of MebZIP genes in response to drought were similar between Arg7 and SC124, which differs from W14. Seven genes (MebZIP-33, -30, -43, -66, -4, -3, and -51) showed induction in leaves of W14, whereas downregulation or no response in leaves of Arg7 and SC124 after drought treatment. Seventeen genes (MebZIP-14, -26, -29, -60, -58, -53, -31, -44, -17, -51, -2, -56, -35, -46, -39, -63, and -21) were upregulated in roots of W14, whereas downregulated or no response in roots of Arg7 and SC124 after drought treatment. This differential expression profile suggested that bZIP-mediated response underlying cassava tolerant to drought stress might be different between cultivated varieties and wild subspecies. In addition, some bZIP genes had close evolutionary relationships; however, they displayed different responses to drought stress, such as, MebZIP-5/-14, MebZIP-65/-67, MebZIP-66/-34, MebZIP-48/-49, MebZIP-13/-16, MebZIP-36/-44, MebZIP-55/-63, MebZIP-68/-77, MebZIP-18/-19, MebZIP-1/-2, and MebZIP-39/-40. Together, the expression profiles of bZIP genes responding to drought in cultivated varieties and wild subspecies will be benefit for further investigation of the mechanisms underlying strong drought resistance in cassava.
Expression profiles of MebZIP genes upon exposure to various stress and related signaling
To examine the response of MebZIP genes to various environmental stresses and related signaling at transcriptional levels, the transcripts of MebZIP genes under these treatments were determined by quantitative real-time PCR. Eight MebZIP genes (MebZIP-4, -11, -27, -41, -52, -55, -64, and -72) induced by drought based on RNA-seq data in different cassava genotypes were chosen for further examination of their expression patterns after osmotic, salt, cold, ABA and H2O2 treatments (Fig. 6; Supplementary Table S6, S7). Under salt treatment, MebZIP-11, -27, and -52 were upregulated, whereas MebZIP-41 and -64 were downregulated during all the treated time points. MebZIP-4, -55, and -72 were repressed at several time points (Supplementary Figure S2). Under osmotic treatment, MebZIP11 showed induction during 2 h–18d treatment. MebZIP27 was induced after 6 h–24d treatment. MebZIP-52, -55, -64, and -72 were upregulated at several time points. MebZIP-4 and -41 were repressed after 2–6 h treatment (Supplementary Figure S3). Under cold treatment following recovery, MebZIP-4, -55, -64, and -72 transcripts increased during all the treated time points. MebZIP-41 and -52 showed upregulation at several time points. On the contrary, cold stress caused a seriously decrease in transcriptional levels of MebZIP11. MebZIP27 did not show obvious trends after cold treatment (Supplementary Figure S4). Under ABA treatment, MebZIP-11, -27, -52, and -64 were induced during all the treated time points. MebZIP-55 and -72 were upregulated at several time points. MebZIP-4 and -41 showed downregulation at several time points (Supplementary Figure S5). Under H2O2 treatment, MebZIP-4 and -52 were upregulated at several time points. MebZIP55 transcripts decreased during 2–48 h treatment. MebZIP-64 and -72 did not show obvious trends after H2O2 treatment. Notably, MebZIP-11, -27, and -41 were significantly repressed during all the treated time points (Supplementary Figure S6).
Figure 6. Expression profiles of MebZIP genes in leaves under various stimuli.
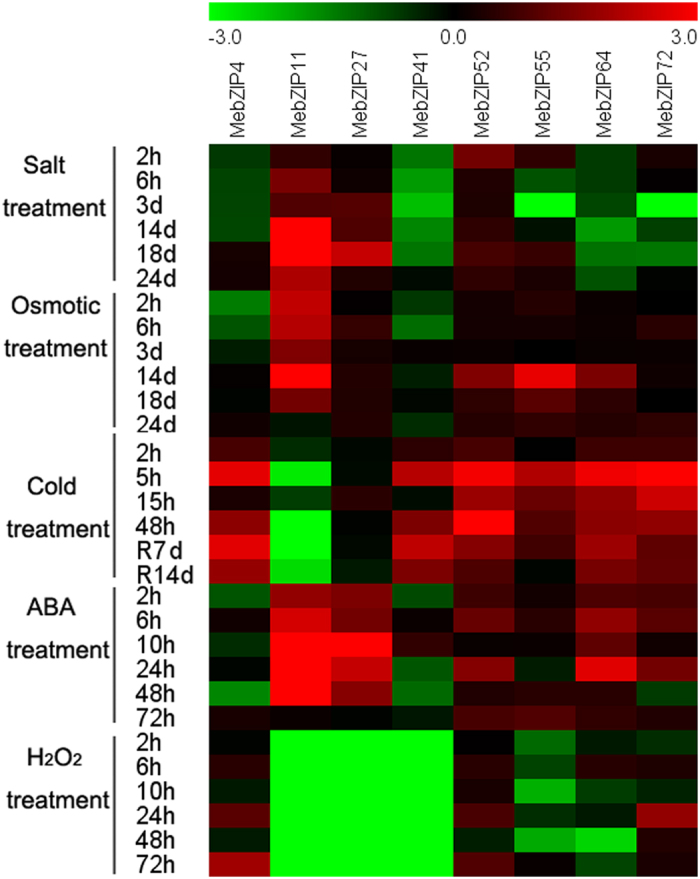
Log2 based values from three replicates of quantitative real-time PCR data were used to create the heatmap. The scale represents the relative signal intensity values.
Discussion
Cassava is the third most important crop after rice and maize in Africa, Asia, and Latin America. Research progress on cassava proceeded extremely slow compared to other important crops, such as rice, maize, wheat, and cotton. Cassava has the characteristic of strong resistances to drought and low-fertility environment. However, the mechanisms by which cassava responds to abiotic stress are poorly understood. The ABA signal pathway has been documented to play crucial roles in plants responding to abiotic stress, in which bZIP is important transcription factors for the ABA signaling pathway. Numerous studies have confirmed the roles of group A bZIPs (AREB/ABFs) in ABA response or environmental adaptation through regulating expression of ABA-dependent genes. However, the identity and function of cassava bZIPs have remained unknown.
Herein, we identified 77 bZIP family members from cassava. Previous studies have identified 75 bZIPs in Arabidopsis, 89 in rice, 92 in Sorghum, 64 in cucumber, 125 in maize, 55 in grapevine, 131 in soybean, and 96 in Brachypodium distachyon4,10,11. These data suggested that bZIP in cassava had expanded compared to that in Arabidopsis, cucumber, and grapevine, whereas had shrunk compared to that in rice, Sorghum, maize, soybean, and Brachypodium distachyon. Evolutionary analysis indicated that the cassava bZIPs could be clustered into 10 subfamilies, which is in line with previous evolutionary classification of bZIPs in Arabidopsis, grapevine and Sorghum5,10,12 (Fig. 1). This classification was further supported by gene structure analysis and conserved motif analysis. Gene structure analysis showed that the number of introns of MebZIPs varied from 0 to 12 (Fig. 2). In grapevine and Brachypodium distachyon, the number of introns varied from 0 to 10 and 0 to 13, respectively4,10. This suggested that there were similar gene structure diversity of bZIP genes in different species. We also observed that the number of introns was significantly more in subfamilies G and D than in subfamilies A, C, S, H, B, F, I, and E. According to a previous report18, the rate of intron loss is faster than the rate of intron gain after segmental duplication in rice. Thus, it can be concluded that subfamilies G and D might contain the original genes, from which those in other clusters were derived. Additionally, all the 15 bZIP members in subfamily S were intronless. This phenomenon was also found in other species, such as grapevine and Brachypodium distachyon4,10. Conserved motif analysis indicated that almost all the MebZIPs contained typical bZIP domain. Additioanlly, each subfamily had some common motifs and some subfamilies also contained the special motifs (Fig. 3). These features in conserved motifs of bZIPs were also observed in grapevine10. Generally, most of MebZIP genes in the same subfamilies showed similar gene structure and conserved motifs, which supports their close evolutionary relationship and the classification of subfamilies.
Although cassava has robust resistance to drought stress, the mechanisms by which cassava responds to abiotic stress are poorly understood. Genome-wide expression profiles have indicated that numerous bZIP genes showed transcriptional response to drought/osmotic stress4,10,11,12. Further biochemical and genetic evidences revealed that group A bZIPs played central roles in plants responding to abiotic stress and ABA signal in various species5,19,20,21,22. In this study, we found that many MebZIP genes could transcriptionally respond to drought stress in different genotypes, suggesting possible function of these genes in responsive to drought stress in cassava (Fig. 5). Further, we observed that the number of MebZIP genes upregulated by drought was greater in W14 than those in Arg7 and SC124, suggesting the comprehensive response of bZIP genes to drought at transcriptional levels in W14 subspecies (Fig. 5). W14 was reported to have strong resistance to drought stress17. After withholding water for 12 days, W14 showed less cured and wilted leaves relative to SC124 and Arg7, indicating that W14 was more tolerant to drought than SC124 and Arg7 (Supplementary Figure S7). Numerous evidences have confirmed that bZIPs play a positive role in response to drought or osmotic stress in plants9,21. Consequently, we speculated that the high ratio of MebZIP genes upregulated by drought in W14 might contribute to its robust drought resistance. Finally, we noted that the number of bZIP genes up-regulated by drought was significantly more in roots than those in leaves in W14, whereas less in Arg7 and SC124 (Fig. 5). Cassava can penetrate into deep soil layers and absorb water stored in soil because of its deep root system23. Therefore, the high ratio of drought induced MebZIP genes in roots indicates that MebZIP genes might function on water uptake from soil by roots, thus maintaining robust drought resistance of W14. Collectively, these findings suggested that MebZIP genes might contribute to the robust resistance of cassava to drought stress and provided a foundation for further research on the bZIP-mediated drought response in cassava.
To date, accumulated evidences have revealed the involvement of bZIP genes in abiotic stresses and related signal transduction pathways. However, no information was available for the response of bZIP genes to these stimuli in cassava. In this study, eight MebZIP genes (MebZIP-4, -11, -27, -41, -52, -55, -64, and -72) upregulated by drought based on RNA-seq data in different cassava genotypes were chosen for further examination of their transcriptional response various treatments (Fig. 6; Supplementary Table S6, S7).
Under salt treatment, MebZIP-11, -27, and -52 showed upregulation, whereas MebZIP-41, -64, -4, -55, and -72 exhibited downregulation at several time points (Supplementary Figure S2). There was some evidences showing that some bZIP genes positively regulate salt resistance, such as, ABF2, AtbZIP17, OsABI5, OsbZIP23, OsbZIP71, GmbZIP44, GmbZIP62, and GmbZIP789,19,24,25,26. Conversely, ABF3 and ABF4 overexpressing Arabidopsis plants showed salt-hypersensitive phenotype, indicating their negative effect on salt response27. These data suggested that bZIP family genes might be positively or negatively involved in salt stress response.
Biochemical and genetic evidences have suggested that bZIP genes, such as, ABF2, ABF3, ABF4, OsbZIP23, OsbZIP46, OsbZIP71, and OsbZIP16 could positively regulate plants resistance to osmotic/drought stress, indicating the crucial role of bZIP genes in osmotic/drought stress response9,21. We found that most of the selected MebZIP genes were upregulated after osmotic treatment in cassava (Supplementary Figure S3). In Brachypodium distachyon, 16 of 96 bZIP genes were upregulated; conversely, 28 of 96 genes were downregulated after 6 h osmotic treatment4. In sorghum, 6 of 16 bZIP genes showed induction, whereas only one exhibited downregulation after osmotic treatment12. Together, these results suggest the important roles of bZIP genes responding to osmotic/drought stress.
Cold stress does great harm to the growth and development of cassava, thus leading to decrease of cassava production. However, little is known for the mechanism underlying cassava response to cold stress. In the present study, we found that all the tested bZIP genes, except for MebZIP11, showed upregulation following cold treatment (Supplementary Figure S4). In Brachypodium distachyon, 12 of 96 bZIP genes were upgulated; conversely, 28 of 96 genes were downregulated after 6 h cold treatment4. In chinese cabbage, transcripts of 36 of 136 bZIP genes increased, while 17 of 136 genes decreased after cold treatment28. These results will be benefit for future researches on the function of bZIP genes and mechanisms underlying cold response.
The phytohormone ABA plays a crucial role in plants responding to abiotic stress, including drought, osmotic, salt and cold stresses. Accumulated evidence has suggested that some bZIP members are crucial components of ABA signaling pathway. In Arabidopsis, ABF2, ABF3, and ABF4 were significantly upregulated by ABA and abiotic stress treatments. Further functional analyses showed that these genes were involved in the regulation of ABA-mediated processes, such as root elongation and seed germination21. To study the response of bZIP genes to ABA signaling, we examined the transcriptional levels of 8 MebZIP genes following ABA treatment. The results found that 6 genes were upregulated, and 2 genes were downregulated after ABA treatment, indicating the comprehensive response of bZIP genes to ABA in cassava (Supplementary Figure S5).
H2O2, as an important signal molecule, is induced by environmental and developmental stimuli29. To determine whether cassava bZIP genes are involved in H2O2 signaling pathway, we examined the expression changes of 8 MebZIP genes after H2O2 treatment. The results showed that MebZIP-4, and -52 were upregulated at several time points, whereas MebZIP-55, -11, -27, and -41 were significantly repressed after H2O2 treatment (Supplementary Figure S6). This is consistent with the expression patterns of bZIPs in response to H2O2 in Brachypodium distachyon, in which 6 genes were induced, and 28 genes were repressed4. These results indicated that H2O2 might mainly cause downregulation of bZIP genes.
In conclusion, we identified 77 bZIP genes from cassava and built their basic classification and evolutionary relationship using evolutionary, conserved protein motif, and gene structure analyses, which will supply abundant information for functional characterization of bZIP genes. The expression profiles of MebZIPs in distinct tissues of two cassava genotypes indicated that bZIP members showed similar or differential expression patterns between Arg7 and W14, thus assisting in understanding the molecular basis for tissue development and function. Transcriptome analysis of three cassava genotypes responding to drought stress revealed that more MebZIP genes were activated in the root of W14 subspecies compared with Arg and SC124, which might contribute to its robust resistance to drought. Expression analysis of MebZIP genes under various treatments showed the comprehensive response of cassava bZIP genes to salt, osmotic, cold, ABA, and H2O2, indicating that they might represent convergence points of different signaling pathways. These data will lay a solid foundation for future research on the functional characterization of bZIP genes and bZIP-mediated signal transduction pathways, thereby advancing our understanding of the molecular basis of genetic enhancements to the cassava.
Methods
Plant materials and treatments
The characteristics of cassava genotypes W14, SC124 and Arg7 have been described in previous studies30,31. Segments cut from cassava stems were inserted into pots filled with soil and vermiculite (1:1) where they were regularly watered. The plants were grown from April to July 2013 during which time the temperature in the glass house ranged from 20 to 35 °C. To analyze transcripts of cassava bZIP genes in different tissues, stems (90-days-old), leaves (90-days-old) and storage roots (150-days-old) were collected from Arg7 and W14 under standard conditions. To examine the transcriptional changes of cassava bZIP genes to drought stress, leaf samples (90-days-old) were collected from and W14, Arg7 and SC124 under standard conditions or drought treatments for 12 days. For various stimuli, 60-days-old seedlings of Arg7 were subjected to 300 mM NaCl for 14 d, 200 mM mannitol for 14 d, 100 μM abscisic acid (ABA) for 24 h, 10% H2O2 for 24 h, and low temperature (4 °C) for 48 h, respectively.
Identification and evolutionary analyses
The whole bZIP protein sequence of cassava, Arabidopsis and rice was obtained from the Phytozome (http://www.phytozome.net/cassava.php), UniPort (http://www.uniprot.org/) and RGAP (http://rice.plantbiology.msu.edu/) databases, respectively32,33,34. To identify the cassava bZIP family genes, the local Hidden Markov Model-based searches (http://hmmer.wustl.edu/) was firstly built from known bZIPs to search the cassava genome database35; subsequently, BLAST searches were performed to check the predicted bZIPs in cassava database with all the Arabidopsis and rice bZIPs as queries. Finally, all candidate protein sequences were further examined by the CDD (http://www.ncbi.nlm.nih.gov/cdd/) and PFAM (http://pfam.sanger.ac.uk/) databases36,37. Then, multiple sequence alignments were applied to confirm the conserved domains of predicted MebZIP proteins. Additionally, sequence alignments of the full-length bZIP proteins from cassava, Arabidopsis and rice were performed by Clustal X 2.0. The bootstrap neighbor-joining evolutionary tree was created by MEGA 5.0 software with 1000 bootstrap replicates based on the sequence alignments38,39.
Protein properties and sequence analyses
The ExPASy proteomics server (http://expasy.org/) was employed to detect the molecular weight and isoelectric points of predicted MebZIP proteins40. Using the MEME program (http://meme.nbcr.net/meme/cgi-bin/meme.cgi), the conserved motifs in full-length cassava bZIP proteins were identified with the following parameters: maximum number of motifs was 20 and the optimum width of motifs was set between 10 and 5041. Subsequently, all identified motifs were annotated with the help of InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan/)42. The gene structure display server program (http://gsds.cbi.pku.edu.cn/) was employed to identify gene structures of cassava bZIPs43.
Transcriptome analysis
Leaves and storage roots of Arg7 and W14 under standard conditions, and leaves and roots of Arg7, SC124 and W14 under normal conditions and 12-days drought treatment were collected to extract total RNA using plant RNeasy extraction kit (TIANGEN, Beijing, China) for transcriptome analysis. Three μg total RNA of each sample were used to construct the cDNA libraries following the Illumina instructions, and subsequently sequenced by Illumina GAII. Data preprocessing and analyses were performed according to our previous study30. The transcriptiomic data was submitted to NCBI and the accession number was listed in Supplementary Table S8.
Quantitative real-time PCR analysis
Transcriptional changes of MebZIP genes responding to various stimuli, including osmotic, salt, cold, ABA and H2O2 were determined by quantitative real-time PCR analysis on Stratagene Mx3000P Real-Time PCR system using SYBR® Premix Ex Taq™ (TaKaRa, Japan). The PCR amplification conditions used for all reactions were implemented as follows: 10 min at 95 °C, and followed by 40 cycles of 10 s at 95 °C, 15 s at 50 °C and 30 s at 72 °C. The relative expression levels of the target genes were calculated by the 2–ΔΔCt method44. Reaction specificities for each primer pairs was tested using quantitative real-time PCR melting curve analysis, agarose gel electrophoresis and sequencing PCR products (Supplementary Table S9). β-tubulin gene (TUB) and elongation factors 1α gene (EF1) were employed as internal references to normalize the transcriptional levels of target genes45. Each treated sample contained a corresponding regularly-watered control and each sample had three independent biological replications. The treated and control plants at each time point were sampled for expression analysis. The relative expression levels of MebZIP genes in each treated time point were compared with that in each time point at normal conditions.
Additional Information
How to cite this article: Hu, W. et al. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 6, 22783; doi: 10.1038/srep22783 (2016).
Supplementary Material
Acknowledgments
This study was funded by the National Natural Science Foundation of China (31500981), the Natural Science Foundation of Hainan Province (314122, 20153048), the National Nonprofit Institute Research Grant of CATAS-ITBB (ITBB2015ZD04, ITBB140204, 1630052014003), the Major Technology Project of Hainan (ZDZX2013023-1).
Footnotes
Author Contributions M.P. and K.M.L. conceived the study. W.H., H.B.Y., Y.Y., Y.X.W., Z.H.D., W.W.T. and J.Z. performed the experiments and carried out the analysis. W.H. and H.B.Y. designed the experiments and wrote the manuscript.
References
- Llorca C. M., Potschin M. & Zentgraf U. bZIPs and WRKYs: two large transcription factor families executing two different functional strategies. Front Plant Sci. 5, 169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. et al. TaASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant Cell Environ. 36, 1449–1464 (2013). [DOI] [PubMed] [Google Scholar]
- Wei K. et al. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 19, 463–476 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. & Chu Z. Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genomics 16, 227 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M. et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111 (2002). [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yamaguchi-Shinozaki K. & Shinozaki K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci. 5, 170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y. et al. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17, 3470–3488 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julkowska M. M. & Testerink C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 20, 586–594 (2015). [DOI] [PubMed] [Google Scholar]
- Todaka D., Shinozaki K. & Yamaguchi-Shinozaki K. Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front Plant Sci. 6, 84 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. et al. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genomics 15, 281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baloglu M. C., Eldem V., Hajyzadeh M. & Unver T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS One 9, e96014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J Integr Plant Biol. 53, 212–231 (2011). [DOI] [PubMed] [Google Scholar]
- Oliveira E. J., Santana F. A., Oliveira L. A. & Santos V. S. Genetic parameters and prediction of genotypic values for root quality traits in cassava using REML/BLUP. Genet Mol Res. 13, 6683–6700 (2014). [DOI] [PubMed] [Google Scholar]
- Zidenga T., Leyva-Guerrero E., Moon H., Siritunga D. & Sayre R. Extending cassava root shelf life via reduction of reactive oxygen species production. Plant Physiol. 159, 1396–1407 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera P. I., Ordoñez C. A., Dedicova B. & Ortega P. E. Reprogramming of cassava (Manihot esculenta) microspores towards sporophytic development. AoB Plants 6, plu022 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C. et al. Chilling acclimation provides immunity to stress by altering regulatory networks and inducing genes with protective functions in cassava. BMC Plant Biol. 14, 207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. et al. Cassava genome from a wild ancestor to cultivated varieties. Nat Commun. 5, 5110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuruzzaman M. et al. Genome-wide analysis of NAC transcription factor family in rice. Gene 465, 30–44 (2010). [DOI] [PubMed] [Google Scholar]
- Liao Y. et al. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228, 225–240 (2008). [DOI] [PubMed] [Google Scholar]
- Ji X. et al. The bZIP protein from Tamarix hispida, ThbZIP1, is ACGT elements binding factor that enhances abiotic stresssignaling in transgenic Arabidopsis. BMC Plant Biol. 13, 1–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T. et al. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. et al. A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiol Plantarum. 153, 538–554 (2015). [DOI] [PubMed] [Google Scholar]
- Okogbenin E. et al. Phenotypic approaches to drought in cassava: review. Front Physiol. 4, 93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kang J. Y., Cho D. I., Park J. H. & Kim S. Y. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 40, 75–87 (2004). [DOI] [PubMed] [Google Scholar]
- Liu J. X., Srivastava R. & Howell S. H. Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis. Plant Cell Environ. 31, 1735–1743 (2008). [DOI] [PubMed] [Google Scholar]
- Zou M., Guan Y., Ren H., Zhang F. & Chen F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol. 66, 675–683 (2008). [DOI] [PubMed] [Google Scholar]
- Kang J. Y., Choi H. I., Im M. Y. & Kim S. Y. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Jung H. J., Park J. I., Yang T. J. & Nou I. S. Transcriptome analysis of newly classified bZIP transcription factors of Brassica rapa in cold stress response. Genomics 104, 194–202 (2014). [DOI] [PubMed] [Google Scholar]
- Costa A. et al. H2O2 in plant peroxisomes: an in vivo analysis uncovers a Ca2+-dependent scavenging system. Plant J. 62, 760–772 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. et al. Genome-wide gene phylogeny of CIPK family in cassava and expression analysis of partial drought-induced genes. Front Plant Sci. 6, 914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. et al. Genome-wide identification and expression analysis of the NAC transcription factor family in cassava. PLoS One 10, e0136993 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik S. et al. The Cassava genome: current progress, future directions. Trop Plant Biol. 5, 88–94 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. UniProt: A hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y. et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Clements J. & Eddy S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, 29–37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D. et al. The Pfam protein families database. Nucleic Acids Res. 42, D222–D230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E. et al. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P. et al. MEME-LaB: Motif analysis in clusters. Bioinformatics 29, 1696–1697 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder N. & Apweiler R. InterPro and InterProScan: Tools for protein sequence classification and comparison. Methods Mol. Biol. 396, 59–70 (2007). [DOI] [PubMed] [Google Scholar]
- Hu B. et al. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31, 1296–1297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time Quantitative PCR and the 2–ΔΔCt method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Salcedo A., Zambrana C. & Siritunga D. Comparative expression analysis of reference genes in field-grown cassava. Trop Plant Biol. 7, 53–64 (2014). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.