Abstract
Autophagy not only recycles intracellular components to compensate for nutrient deprivation but also selectively eliminates organelles to regulate their number and maintain quality control. Mitophagy, the specific autophagic elimination of mitochondria, has been identified in yeast, mediated by autophagy-related 32 (Atg32), and in mammals during red blood cell differentiation, mediated by NIP3-like protein X (NIX; also known as BNIP3L). Moreover, mitophagy is regulated in many metazoan cell types by parkin and PTEN-induced putative kinase protein 1 (PINK1), and mutations in the genes encoding these proteins have been linked to forms of Parkinson’s disease.
Autophagy is the process of catabolism of cellular components, such as the cytosol, organelles and protein aggregates, through their encapsulation by a double-membrane structure known as the autophagosome1,2. In yeast, the autophagosome is thought to develop from a single pre-autophagosomal structure (PAS), possibly through the coalescence of smaller vesicles3, whereas in mammalian cells its origin is multifocal. For example, in mammals the autophagosome can develop from a specialized double-membrane extension of the endoplasmic reticulum (ER) known as the omegasome4. Furthermore, recent reports indicate that, in some forms of autophagy, lipid for the autophagosomal membrane can be contributed by the ER (through contact with mitochondria, from which the autophagosomal membranes bud5) and the plasma membrane6. The newly formed double-membrane, which is known as an isolation membrane, surrounds cytosolic cargo, and its edges seal to sequester the contents into the autophagosome. This then fuses with a lysosome, allowing degradation of the cargo and the inner bilayer of the double autophagosomal membrane, which can be reused or catabolized for energy production (FIG. 1). The different steps of autophagy, including the expansion of the isolation membrane and autophagosome production, are mediated and/or regulated by autophagy-related (Atg) proteins, over 30 of which have been identified7.
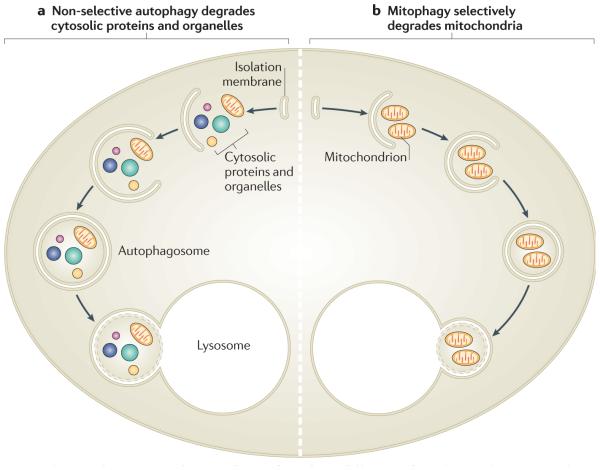
Figure 1. Non-selective autophagy and mitophagy have different roles.
a | Non-selective autophagy occurs when cells are deprived of nutrients. It degrades a range of cytosolic contents, including proteins and many types of organelles. After their recruitment into isolation membranes, cytosolic components are sealed into autophagosomes that fuse with lysosomes. The degradation of these components in the lysosome supplies building blocks for re-use and for metabolism to provide ATP. b | By contrast, mitophagy occurs to eliminate mitochondria, either to regulate their number or to specifically remove ones that are damaged. Mitochondria are selectively recruited into isolation membranes, which seal and then fuse with lysosomes to eliminate the trapped mitochondria.
Two types of macroautophagy have been identified to date (FIG. 1). Non-selective autophagy occurs on nutrient deprivation to supply cells with essential metabolic building blocks and energy until nutrients can once again be obtained from the extracellular environment. By contrast, cargo-specific autophagy occurs under nutrient-rich conditions to mediate the removal of superfluous or damaged organelles and protein aggregates that otherwise could be toxic. This can occur following changes in nutrient sources and during developmental processes. For example, a shift in the main carbon source of methylotrophic yeast species from glucose to methanol leads to increased proliferation of peroxisomes, which are removed by cargo-specific autophagy (in this case, termed pexophagy) when the carbon source is changed back to glucose (reviewed in ReF. 8). Other cargo-specific autophagic processes include ribophagy (ribosome elimination) and xenophagy (the capture of intracellular pathogens).
A well-studied type of cargo-specific autophagy is mitophagy, which mediates the selective removal of mitochondria (FIG. 1b). Mitophagy was first observed in mammalian cells by early electron microscopy studies, which identified increased mitochondrial sequestration in lysosomes following stimulation of hepatocyte catabolism with glucagon (reviewed in ReF. 9). Using this model, as well as starvation and photodamage-induced autophagy, Lemasters and colleagues10 coined the term mitophagy to describe the engulfment of mitochondria into vesicles that are coated with the autophagosome marker MAP1 light chain 3 (LC3; a homologue of yeast Atg8), a process that can occur within 5 minutes. LC3 (and Atg8) is an ubiquitin-like protein that is covalently attached to phosphatidylethanolamine during autophagosome biogenesis, which allows it to integrate into, and sculpt, the growing isolation membrane11 and participate in cargo recruitment12. Another study found that in neurons cultured with caspase inhibitors to prevent cell dissolution, mitochondria were completely removed by mitophagy following apoptosis induction13. As mitochondrial outer membrane permeabilization occurs upstream of caspase activation during apoptosis, these results suggest that mitochondrial damage can induce mitophagy. These early studies indicated that mitophagy regulates mitochondrial number to match metabolic demand and might also be a form of quality control to remove damaged mitochondria.
In yeast14 and mammalian cells15 mitophagy is preceded by mitochondrial fission16, which divides elongated mitochondria into pieces of manageable size for encapsulation and also quality control segregation of damaged mitochondrial material for selective removal by mitophagy. Beyond quality control, mitophagy has been shown to be required for steady-state turnover of mitochondria17, for the adjustment of mitochondrion numbers to changing metabolic requirements18 and during specialized developmental stages in mammalian cells, such as during red blood cell differentiation19,20.
In this Progress article we discuss recently identified pathways that mediate mitophagy in yeast and mammalian cells, including those involving the kinase PTEN-induced putative kinase protein 1 (PINK1) and the E3 ubiquitin ligase parkin (which have no yeast homologues). We also highlight how defects in mitophagy may contribute to human conditions such as Parkinson’s disease.
Mitophagy in yeast
Mitophagy has been observed in yeast17,18, which has facilitated the study of its molecular mechanisms. The product of uth1 is a SUN domain-containing protein that is located partially in the outer mitochondrial membrane (OMM), and this was found to be required for eliminating excess mitochondria that had been made redundant following nutrient changes18. Furthermore, ancient ubiquitous protein 1 (Aup1; also known as Ptc6), a member of the phosphatase 2C (PP2C) superfamily, was shown to facilitate mitophagy in stationary phase yeast grown under conditions in which respiration is favoured, perhaps reflecting the quality control elimination of overworked mitochondria17.
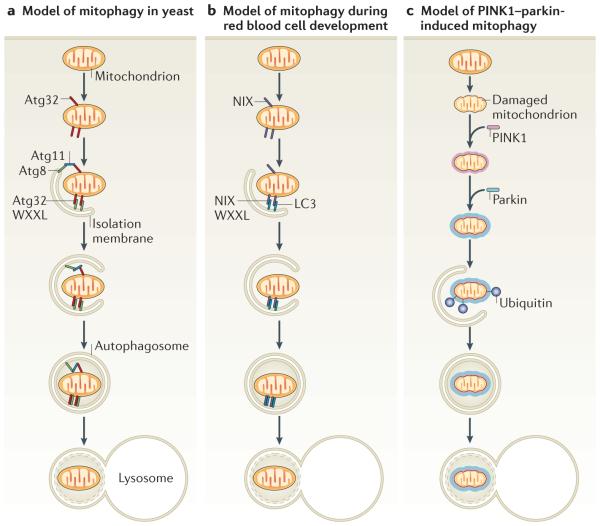
More recently, two groups independently used mitophagy screens to identify dozens of new gene products that are required for this process, some of unknown function and others with functions that were not previously linked to autophagy21,22. In these screens, mitophagy was identified by the accumulation of mitochondrial proteins in the vacuole of stationary-phase respiring cells17. Although Aup1 and Uth1 were not identified in these studies, perhaps owing to the use of different yeast strains, both groups found that the autophagy protein Atg32 is necessary for mitophagy21,23. Atg32 (which has no known metazoan homologue) is a 60-kDa protein spanning the OMM, with a carboxy-terminal domain in the intermembrane space and a ~40-kDa amino-terminal region in the cytosol. It is required for mitophagy that occurs in response to enforced respiration but not for non-selective autophagy in response to nutrient deprivation or for pexophagy23. Under mitophagy-inducing growth conditions, Atg32 binds to Atg11, which is known to recruit a range of cargo, including peroxisomes, into autophagosomes by interacting with Atg8. Interestingly, the cytosolic domain of Atg32 contains a WXXL-like Atg8-binding motif, which is required for binding to Atg8 and for mitophagy. Thus, Atg32 can interact with Atg8 indirectly through Atg11 and directly through the WXXL-like motif (FIG. 2a). The direct and indirect (when bridged by Atg11) association of mitochondrial membrane-anchored Atg32 with isolation membrane-bound Atg8 is thought to recruit mitochondria into autophagosomes. Although Atg32 expression increases under oxidizing conditions, which suggests that it might also participate in mitochondrial quality control21, no defects in mitochondria were identified in Atg32-null yeast. How the expression and activity of Atg32 are regulated to eliminate the appropriate number of mitochondria remains unknown and is an intriguing area for further study. The range of genes that have been identified as being necessary for mitophagy in yeast suggests that distinct pathways probably mediate mitophagy for different purposes.
Figure 2. The atg32, NiX and PiNK1–parkin pathways of mitophagy.
Mitophagy requires the specific labelling of mitochondria and their recruitment into isolation membranes. a | In yeast this occurs through the outer mitochondrial membrane protein autophagy-related 32 (Atg32), which can bind the isolation membrane protein Atg8 through its WXXL-like motif. How mitochondria are specified for removal to calibrate the mitochondrial population density according to the cell’s metabolic demand remains unknown but may occur through the adaptor protein Atg11, which binds both Atg32 and Atg8 and could physically link mitochondria and isolation membranes. Finally, the isolation membranes seal mitochondria in autophagosomes and fuse with lysosomes to allow degradation. b | During differentiation, red blood cells lose their mitochondria through mitophagy. Expression of the outer mitochondrial membrane protein NIP3-like protein X (NIX; also known as BNIP3L) increases during red blood cell differentiation and is required for mitochondrial removal. NIX has a WXXL-like motif, which binds to MAP1 light chain 3 (LC3; a homologue of yeast Atg8) on isolation membranes and is thought to mediate the binding and sequestration of mitochondria into autophagosomes. c | When mitochondria are damaged and lose membrane potential, the kinase PTEN-induced putative kinase protein 1 (PINK1) accumulates (shown as a halo around the mitochondrion) and recruits the E3 ubiquitin ligase parkin from the cytosol specifically to the damaged mitochondrion. Parkin ubiquitylates mitochondrial proteins and causes mitochondria to become engulfed by isolation membranes that then fuse with lysosomes. This may mediate mitochondrial quality control.
Mitophagy in red blood cells
Removal of undamaged mitochondria also occurs during key developmental processes. In most mammals, mature red blood cells lack mitochondria, and this is achieved by mitophagy during the maturation of immature red blood cells, which are known as reticulocytes20,24,25. The OMM protein NIP3-like protein X (NIX; also known as BNIP3L) could be involved in this process, as its expression greatly increases during the terminal differentiation stages of red blood cells26, and mature red blood cells from mice lacking NIX retain residual mitochondria19,27. NIX (which has no yeast homologue) was originally thought to be a BCL-2 homology domain 3 (BH3)-only protein involved in apoptosis, so it could potentially induce mitophagy through apoptotic signalling. However, phylogenetic studies indicated that NIX is not a BH3-only protein28, and NIX-mediated mitophagy in reticulocytes does not seem to involve apoptotic mechanisms as it does not require the pro-apoptotic B cell lymphoma 2 (BCL-2) family proteins BAX or BCL-2 antagonist/killer (BAK)19.
NIX is localized to the OMM and contains a WXXL-like motif facing the cytosol, which binds the mammalian Atg8 orthologue, LC3, and the LC3 homologue, GABA receptor-associated protein (GABARAP), in vitro and in vivo29,30. This motif may directly recruit isolation membranes to mitochondria. The induction of mitophagy by NIX is thought to be mediated, at least in part, by direct binding to LC3 and GABARAP through its WXXL-like motif (FIG. 2b). Consistent with this theory, mitophagy is not fully restored in NIX-null reticulocytes that are rescued with NIX lacking the WXXL-like motif. Although mitophagy can be rescued in NIX-deficient reticulocytes by depolarizing mitochondria with carbonylcyanide p-t rifluoromethoxyphenylhydrazone (FCCP) or the apoptosis-inducing BH3-mimetic ABT737 (ReF. 27), such depolarization seems to activate a NIX-independent mitophagy pathway25. The core autophagic proteins, UNC51-like kinase 1 (ULK1) and ATG7 are also required for mitophagy in reticulocytes20,24,25.
Mitophagy in disease
Recent work has linked defects in mitophagy to Parkinson’s disease. Two genes, which encode the OMM kinase PINK1 (ReF. 31) and the cytosolic E3 ubiquitin ligase parkin32, are mutated in autosomal recessive Parkinson’s disease. These proteins have been found to work in the same pathway to suppress mitochondrial damage in flies33–35, which can lead to loss of flight muscle and dopaminergic neurons, and to male sterility that is ameliorated by promotion of mitochondrial fission36–38. Below we describe the signalling pathway mediated by PINK1 and parkin that induces mitophagy in mammalian cells.
Parkin translocates to damaged mitochondria
Parkin is an E3 ubiquitin ligase, dysregulation of which is linked to Parkinson’s disease and loss of neurons of the substantia nigra. Parkin is robustly expressed in many tissues, including brain, skeletal muscle, heart and liver tissues, which suggests that it has a widespread physiological role39. One hour after stress induction by mitochondrial uncouplers (which mimic damage by decreasing the mitochondrial inner membrane electrochemical gradient that is used to produce ATP), parkin translocates from the cytosol to mitochondria in many cultured cell types, including neurons and HeLa cells40 (Supplementary information S1 (movie)). Administration of other mitochondrial poisons, such as Paraquat in many cell types40 and azide in cells lacking mitochondrial DNA (mtDNA)39, also induces parkin translocation to mitochondria. Interestingly, when mitochondrial fusion is blocked and a subset of mitochondria become impaired and lose membrane potential, parkin translocates to the subset of damaged mitochondria without accumulating on healthy mitochondria in the same cell40. Furthermore, when cells are incubated for up to 1 day with mitochondrial uncouplers, parkin mediates selective elimination through mitophagy of all mitochondria in a large proportion of the cells40. Based on the selective recruitment of parkin to damaged mitochondria and the induction of their removal by mitophagy, it was proposed that parkin mediates a mitochondrial quality control pathway to maintain organelle population fidelity by removing damaged mitochondria. Consistent with this proposal, overexpression of parkin in heteroplasmic cybrid osteosarcoma cells eliminates mitochondria that have deleterious mutations in cytochrome oxidase subunit I (COXI; encoded by mtDNA) and not wild-type mitochondria. This selectively eliminates the mitochondria with mutations in COXI, thereby enriching cells with wild-type mtDNA and rescuing cytochrome oxidase activity39.
Parkin is recruited by PINK1
The translocation of parkin to uncoupled mitochondria and the induction of parkin-mediated mitophagy require the activity of PINK1 (ReFs 41–44). Indeed, ectopic expression of PINK1 (ReFs 45,46) or experimentally induced accumulation of PINK1 on mitochondria41 are sufficient to induce parkin translocation and mitophagy in the absence of mitochondrial uncoupling. Genetic studies in flies also show that PINK1 and parkin function in the same pathway to prevent pathology in vivo34,35,47, which is consistent with the hypothesis that mitophagy induction may be a key physiological function of PINK1 and parkin. Interestingly, NIX has been recently reported to promote parkin translocation and parkin-mediated mitophagy in mouse embryonic fibroblasts48.
So how does PINK1 detect mitochondrial damage and respond to uncoupling to recruit parkin? The significant increase in PINK1 following mitochondrial uncoupling41,44,49 does not explain how PINK1 distinguishes damaged and healthy mitochondria in individual cells to mediate parkin recruitment only to uncoupled organelles (FIG. 2c). However, the finding that PINK1 is rapidly turned over by proteolysis in healthy mitochondria suggests a novel mechanism for the detection of damaged mitochondria41,44. PINK1 seems to be expressed and imported into all mitochondria and then rapidly degraded by proteolysis and maintained at very low levels. When a subset of mitochondria becomes damaged, PINK1 proteolysis is inhibited, allowing the accumulation of PINK1 only in damaged mitochondria and the subsequent recruitment of parkin specifically to organelles that are accumulating PINK1 (ReF. 41). In fly cells, the protease Rhomboid 7 has been reported to cleave and activate PINK1 (ReF. 50). Which protease normally degrades and inhibits mammalian PINK1 remains unknown, and it is unclear how PINK1 cleavage is regulated by membrane potential. Furthermore, although it is reported that PINK1 binds43,51,52 and phosphorylates parkin45,53, the molecular mechanism by which PINK1 recruits parkin to mitochondria remains unclear.
Induction of mitophagy by parkin
The induction of mitophagy following parkin recruitment is thought to involve the parkin-mediated ubiquitylation of mitochondrial substrates; indeed, the E3 ubiquitin ligase activity of parkin increases following its translocation to mitochondria44. Furthermore, parkin ubiquitylates mitochondrial substrates following CCCP-induced uncoupling43,44,54–56, and mitochondrial substrates prominently display Lys63-linked polyubiquitin chains55,56, which are usually associated with signalling and not proteosomal degradation. Furthermore, the ubiquitin-binding adaptor p62 (also known as sequestosome 1), which can both aggregate ubiquitylated proteins by polymerizing with other p62 molecules and recruit ubiquitylated cargo into autophagosomes by binding to LC3 (ReF. 57), accumulates on mitochondria following parkin-mediated ubiquitylation43,48,54–56. Although it is clear that p62 binds to parkin-ubiquitylated mitochondrial substrates and mediates clumping of mitochondria, there have been contradicting reports showing that p62 is43,48,54 or is not55,56 essential for parkin to promote mitophagy. The histone deacetylase HDAC6, which also binds ubiquitylated substrates, accumulates on mitochondria following parkin translocation and is required for parkin-mediated mitophagy, indicating a possible link between mitochondrial transport and mitophagy54.
Many parkin substrates from several cellular compartments have been reported58. In mitochondria, four substrates of parkin have been identified: the mitofusin mitochondrial assembly regulatory factor (MARF) in flies59,60, and mitofusin 1, mitofusin 2 (ReFs 61,62) and voltage-dependent anion-selective channel protein 1 (vDAC1) in mammalian cells43, all of which are embedded in the OMM.
In addition to activating signalling pathways through Lys63-linked ubiquitylation of mitochondrial substrates, parkin triggers substrate degradation. Specifically, the ubiquitylation of mitofusins seems to mediate proteosomal degradation and to thereby promote mitochondrial fragmentation and possibly mitophagy62. As mitofusins mediate mitochondrial fusion, it is thought that, by inducing their degradation62, parkin actively inhibits the re-fusion of damaged and healthy mitochondria and thereby segregates impaired mitochondria for mitophagy. Ubiquitylation of vDAC1, the most abundant OMM protein, may43 or may not56 be required for parkin-mediated mitophagy.
Link to Parkinson’s disease
Early onset hereditary forms of Parkinson’s disease can be caused by loss of function mutations in PINK1 or PARK2 (which incodes parkin). Several mutations in parkin found in patients with Parkinson’s disease caused defects in mitophagy, whereas parkin translocation was variably affected, indicating that discreet steps are involved in mitophagy upstream and downstream of parkin translocation41,43,44,54. In addition, PINK1 mutations in patients with Parkinson’s disease also caused defective parkin recruitment and parkin-induced mitophagy41,63. Moreover, mutations and deletions in mtDNA have been linked to Parkinson’s disease, which is consistent with the model that a defect in mitochondrial quality control may contribute to the aetiology of certain forms of the disease (BOX 1).
Box 1. Mitochondrial damage associated with Parkinson’s disease.
Parkinson’s disease is caused by the loss of dopaminergic neurons in the substantia nigra, a region in the midbrain that is important for modulating motor control. The disease is largely sporadic, with little known genetic component. However, numerous monogenic causes of Parkinson’s disease have been identified, with both autosomal dominant and recessive forms66. Two of the autosomal recessive genes, PTEN-induced putative kinase protein 1 (PINK1) and PARK2 (which encodes parkin), have roles in mitophagy of damaged mitochondria39–44,46. Consistent with this, excessive mitochondrial damage has been linked to Parkinson’s disease67. Furthermore, deletions in mitochondrial DNA (mtDNA) have been found to accumulate with age and appear most commonly in the substantia nigra68,69. Such deletions are found in patients with Parkinson’s disease more frequently than in age-matched controls69, and patients with a higher rate of mtDNA deletions owing to defective mtDNA replication develop Parkinson’s disease as a result of mtDNA deletions in their substantia nigra neurons70–72. Thus, Parkinson’s disease can be caused by the accumulation of mitochondrial damage. We suggest this can occur two ways: by excessive mitochondrial stress (as may occur following exposure to environmental toxins, such as MPTP or Paraquat, or defective mtDNA replication) or by the failure to clear normally occurring damaged mitochondria by mitophagy (as is suggested to occur in patients with mutations in PINK1 and PARK2).
Conclusion and perspectives
Selective elimination of mitochondria by autophagy, in conjunction with mitochondrial biogenesis, regulates the changes in steady-state mitochondrial number that are required to meet metabolic demand. Damaged mitochondria are also selectively removed by mitophagy to maintain quality control. Mitophagy involves distinct steps to recognize defective or superfluous organelles and to target them to autophagosomes. In yeast, Atg32 seems to recruit redundant and/or damaged mitochondria to autophagosomes. In mammals, NIX mediates developmental removal of mitochondria during erythropoiesis, and parkin and PINK1, the genes of which are mutated in autosomal recessive forms of Parkinson’s disease, can regulate the selective autophagy of damaged mitochondria in many cell types.
More precise and quantitative assays of mitophagy are needed in all these systems. For example, one weakness of the model suggesting that parkin induces mitophagy has been the requirement for parkin overexpression to show clearance of all mitochondria from a large percentage of cells. Although endogenous PINK1 has been shown to be required for mitophagy in cells expressing only endogenous parkin41, it is still not known whether knocking out parkin decreases the rate of mitophagy. This has been difficult to address because of the crude assays that are currently available to quantify mitophagy. A more quantitative assessment of mitophagy is needed to determine whether endogenous parkin mediates an increase in the mitochondrial turnover rate. Dopaminergic neurons that are derived from induced pluripotent stem cells of patients with mutations in PARK2 and PINK1 should soon be available and will be helpful for further studies of the role of mitophagy in Parkinson’s disease. The study of mitophagy in animal models in vivo is also needed to rigorously test the model that parkin and PINK1 mediate mitochondrial quality control. Beyond its role in Parkinson’s disease, parkin has also been found to be a tumour suppressor, suggesting it will be worth exploring the role of mitophagy in cancer biology64,65.
The rapid pace of current investigation indicates that we will soon better understand important missing steps such as how mitophagy is regulated to titrate mitochondrial numbers and how mitochondria are tagged for autophagosomal recognition. The proposal that defects in mitophagy may be linked to Parkinson’s disease suggests that this nascent research topic will also be important to understand and, hopefully, to treat human disease.
Supplementary Material
Acknowledgements
We thank H. Abeliovich for valuable discussions and D. Suen for the movie that is used in Supplementary information 1. D.P.N. is a member of the US National Institutes of Health–Oxford–Cambridge Scholars Program.
Footnotes
Competing interests statement The authors declare no competing financial interests.
SUPPLEMENTARY INFORMATION See online article: S1 (movie)
Contributor Information
Richard J. Youle, Biochemistry Section, Surgical Neurology Branch, National Institute of Neurological Disorders and Stroke, National Institutes of Health, 35 Convent Drive, 2C-917, Bethesda, Maryland 20892, USA
Derek P. Narendra, Biochemistry Section, Surgical Neurology Branch, National Institute of Neurological Disorders and Stroke, National Institutes of Health, 35 Convent Drive, 2C-917, Bethesda, Maryland 20892, USA, Medical Research Council Mitochondrial Biology Unit, Hills Road, Cambridge, CB2 2XY, UK
References
- 1.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nature Rev. Mol. Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 2.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nature Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mari M, et al. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hailey DW, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nature Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klionsky DJ, et al. A comprehensive glossary of autophagy-related molecules and processes. Autophagy. 2010;6:438–448. doi: 10.4161/auto.6.4.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oku M, Sakai Y. Pexophagy in. Pichia pastoris. Methods Enzymol. 2008;451:217–228. doi: 10.1016/S0076-6879(08)03215-1. [DOI] [PubMed] [Google Scholar]
- 9.De Duve C, Wattiaux R. Functions of lysosomes. Annu. Rev. Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 10.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 12.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 13.Tolkovsky AM, Xue L, Fletcher GC, Borutaite V. Mitochondrial disappearance from cells: a clue to the role of autophagy in programmed cell death and disease? Biochimie. 2002;84:233–240. doi: 10.1016/s0300-9084(02)01371-8. [DOI] [PubMed] [Google Scholar]
- 14.Nowikovsky K, Reipert S, Devenish RJ, Schweyen RJ. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 2007;14:1647–1656. doi: 10.1038/sj.cdd.4402167. [DOI] [PubMed] [Google Scholar]
- 15.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westermann D. Mitochondrial fusion and fission in cell life and death. Nature Rev. Mol. Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 17.Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J. Biol. Chem. 2007;282:5617–5624. doi: 10.1074/jbc.M605940200. [DOI] [PubMed] [Google Scholar]
- 18.Kissova I, Deffieu M, Manon S, Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J. Biol. Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- 19.Schweers RL, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl Acad. Sci. USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kundu M, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamoto K, Kondo-Okamoto N, Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Kanki T, Wang K, Klionsky DJ. A genomic screen for yeast mutants defective in mitophagy. Autophagy. 2010;6:278–280. doi: 10.4161/auto.6.2.10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortensen M, et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc. Natl Acad. Sci. USA. 2010;107:832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, et al. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009;114:157–164. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aerbajinai W, Giattina M, Lee YT, Raffeld M, Miller JL. The proapoptotic factor Nix is coexpressed with Bcl-xL during terminal erythroid differentiation. Blood. 2003;102:712–717. doi: 10.1182/blood-2002-11-3324. [DOI] [PubMed] [Google Scholar]
- 27.Sandoval H, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aouacheria A, Brunet F, Gouy M. Phylogenomics of life-or-death switches in multicellular animals: Bcl-2, BH3-only, and BNip families of apoptotic regulators. Mol. Biol. Evol. 2005;22:2395–2416. doi: 10.1093/molbev/msi234. [DOI] [PubMed] [Google Scholar]
- 29.Schwarten M, et al. Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy. 2009;5:690–698. doi: 10.4161/auto.5.5.8494. [DOI] [PubMed] [Google Scholar]
- 30.Novak I, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valente EM, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 32.Kitada T, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 33.Greene JC, et al. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl Acad. Sci. USA. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park J, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 35.Clark IE, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 36.Poole AC, et al. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc. Natl Acad. Sci. USA. 2008;105:1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, et al. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl Acad. Sci. USA. 2008;105:7070–7075. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng H, Dodson MW, Huang H, Guo M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl Acad. Sci. USA. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suen DF, Narendra DP, Tanaka A, Manfredi G, Youle RJ. Parkin overexpression selects against a deleterious mtDNA mutation in heteroplasmic cybrid cells. Proc. Natl Acad. Sci. USA. 2010;107:11835–11840. doi: 10.1073/pnas.0914569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narendra DP, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 44.Matsuda N, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim Y, et al. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. BBRC. 2008;377:975–980. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 46.Kawajiri S, et al. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett. 2010;584:1073–1079. doi: 10.1016/j.febslet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 47.Yang Y, et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl Acad. Sci. USA. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding WX, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin–ubiquitin–p62-mediated mitochondrial priming. J. Biol. Chem. 2010;285:27879–27890. doi: 10.1074/jbc.M110.119537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin W, Kang UJ. Characterization of PINK1 processing, stability, and subcellular localization. J. Neurochem. 2008;106:464–474. doi: 10.1111/j.1471-4159.2008.05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitworth AJ, et al. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson’s disease factors Pink1 and Parkin. Dis. Model Mech. 2008;1:168–174. doi: 10.1242/dmm.000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shiba K, et al. Parkin stabilizes PINK1 through direct interaction. Biochem. Biophys. Res. Commun. 2009;383:331–335. doi: 10.1016/j.bbrc.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 52.Um JW, Stichel-Gunkel C, Lübbert H, Lee G, Chung KC. Molecular interaction between Parkin and Pink1 in mammalian neuronal cells. Mol. Cell Neurosci. 2009;40:421–432. doi: 10.1016/j.mcn.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 53.Sha D, Chin LS, Li L. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-κB signaling. Hum. Mol. Genet. 2010;19:352–363. doi: 10.1093/hmg/ddp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. Disease-causing mutations in Parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J. Cell Biol. 2010;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okatsu K, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narendra D, Kane L, Hauser D, Fearnley I, Youle R. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensible for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/ LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 58.Dawson TM, Dawson VL. The role of parkin in familial and sporadic Parkinson’s disease. Mov. Disord. 2010;25(Suppl. 1):S32–S39. doi: 10.1002/mds.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl Acad. Sci. USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poole AC, Thomas RE, Yu S, Vincow ES, Pallanck L. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS ONE. 2010;5:e10054. doi: 10.1371/journal.pone.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gegg ME, et al. Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 2010;19:4861–4870. doi: 10.1093/hmg/ddq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka A, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. doi: 10.1083/jcb.201007013. in the press; doi:10.1083/ jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Geisler S, et al. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010;6:871–878. doi: 10.4161/auto.6.7.13286. [DOI] [PubMed] [Google Scholar]
- 64.Cesari R, et al. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc. Natl Acad. Sci. USA. 2003;100:5956–5961. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poulogiannis G, et al. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc. Natl Acad. Sci. USA. 2010;107:15145–15150. doi: 10.1073/pnas.1009941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klein C, Schlossmacher MG. The genetics of Parkinson disease: implications for neurological care. Nature Clin. Pract. Neurol. 2006;2:136–146. doi: 10.1038/ncpneuro0126. [DOI] [PubMed] [Google Scholar]
- 67.Schapira AH. Mitochondria in the aetiology and pathogenesis of Parkinson’s disease. Lancet Neurol. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- 68.Kraytsberg Y, et al. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nature Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 69.Bender A, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nature Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 70.Luoma P, et al. Parkinsonism, premature menopause, and mitochondrial DNA polymerase γ mutations: clinical and molecular genetic study. Lancet. 2004;364:875–882. doi: 10.1016/S0140-6736(04)16983-3. [DOI] [PubMed] [Google Scholar]
- 71.Reeve AK, et al. Nature of mitochondrial DNA deletions in substantia nigra neurons. Am. J. Hum. Genet. 2008;82:228–235. doi: 10.1016/j.ajhg.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baloh RH, Salavaggione E, Milbrandt J, Pestronk A. Familial parkinsonism and ophthalmoplegia from a mutation in the mitochondrial DNA helicase twinkle. Arch. Neurol. 2007;64:998–1000. doi: 10.1001/archneur.64.7.998. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.