Abstract
No-flow ischemia occurs during cardiac arrest, hemorrhagic shock, liver resection and transplantation. Recovery of blood flow and normal physiological pH, however, irreversibly injures the liver and other tissues. Although the liver has the powerful machinery for mitochondrial quality control, a process called mitophagy, mitochondrial dysfunction and subsequent cell death occur after reperfusion. Growing evidence indicates that reperfusion impairs mitophagy, leading to mitochondrial dysfunction, defective oxidative phosphorylation, accumulation of toxic metabolites, energy loss and ultimately cell death. The importance of acetylation/deacetylation cycle in the mitochondria and mitophagy has recently gained attention. Emerging data suggest that sirtuins, enzymes deacetylating a variety of target proteins in cellular metabolism, survival and longevity, may also act as an autophagy modulator. This review highlights recent advances of our understanding of a mechanistic correlation between sirtuin 1, mitophagy and ischemic liver injury.
Keywords: Autophagy, Mitochondria, Liver, Ischemia/Reperfusion, Acetylation
INTRODUCTION
Timely removal of unnecessary cellular constituents and abnormal organelles is essential to sustain cell viability. Intracellular protein degradation and protein synthesis are tightly balanced to maintain cell survival. Two catabolic pathways account for protein degradation: 1) the ubiquitin-proteasome pathway for degradation of short-lived proteins, and 2) the autophagy or “self-eating” pathway for degradation of long-lived proteins and abnormal organelles (1). In the liver, long-lived proteins constitute more than 99% of cell proteins and thus, autophagic degradation is the primary catabolic process for proteins (2). Three major forms of autophagy have been described in mammalian cells: microautophagy, macroautophagy and chaperone-mediated autophagy (1). Among the three forms, macroautophagy is of particular importance in the liver as this form of autophagy not only clears unneeded intracellular proteins but also digests injured or dysfunctional organelles such as abnormal mitochondria. Although macroautophagy is generally considered a random process, growing evidence shows the existence of selective macroautophagy, especially for clearance of the mitochondria, termed mitophagy (3).
The liver is vulnerable to hypoxic and anoxic stresses. Although prolonged interruption of blood flow (ischemia) eventually causes hepatocyte death, extensive injury paradoxically occurs mostly after the restoration of blood flow and oxygen, a phenomenon called reperfusion injury (4). Ischemia/reperfusion (I/R) injury is a major pathological event in low flow disease states, including hemorrhagic shock, and cardiac arrest, and intentionally during surgical procedures such as liver resection and transplantation. Mitochondrial dysfunction is a causal mechanism attributing to I/R injury (4,5). Injured mitochondria hamper energy production and precipitate further injury to neighboring mitochondria through release of cytotoxic compounds (5). Therefore, elimination of damaged mitochondria in a timely manner is critical to sustain viability in ischemic hepatocytes. As mitophagy is the only known cellular mechanism to dispose abnormal mitochondria, active recruitment of mitophagy has a therapeutic potential for mitochondria-related diseases (4).
THE LIVER
The liver, the largest internal organ located in the upper right quadrant of the abdomen, performs multiple functions in the body, including synthesis of essential proteins and cofactors; regulation of glycogen synthesis and degradation; storage of vitamins and minerals; production and secretion of hormones and bile; and clearance of toxic metabolites. To accomplish these varied functions, distinct types of hepatic cells - hepatocytes, Kupffer cells, stellate cells, sinusoidal endothelial cells, cholangiocytes, lymphocytes and dendrite cells - require high amounts of cellular energy in order to synthesize and eliminate a plethora of complex molecules. Such a high demand for cellular energy in the liver can likewise be inferred from its unique dual-blood supply system: 1) the portal venous supply from the gut, pancreas, and spleen, and 2) arterial supply from the heart (6). Using these two sources of blood supply, the liver is continuously nourished with oxygen, energy substrates and nutrients. Hepatic blood vessels encompass about 22% of the liver mass/volume and the liver contains about 12% of the total blood volume (7). Blood flow through the liver amounts to 1,500~2,000 mL/min (8). The hepatic artery and the portal vein furnish approximately 25% and 75% of the resting cardiac output, respectively, and complete mixture of both portal and arterial blood occurs in the hepatic sinusoids (6). Hence, the liver is a highly aerobic organ and innately vulnerable to hypoxic and ischemic stresses. Sinusoidal blood flow to the hepatic lobules and subsequent addition of synthetic products and metabolic wastes to the blood create gradients of oxygen and metabolites between periportal and pericentral regions of the liver lobule (9). As a consequence, the liver exhibits a distinct zonal dependence on specific biochemical reactions. For instance, while ureagenesis, gluconeogenesis, beta-oxidation of fatty acids, and cholesterol synthesis are enriched in the periportal hepatocytes, lipogenesis, glycolysis and drug detoxification occurs mainly in the pericentral hepatocytes (9). Hepatic disease likewise demonstrates zonal dependency. Hypoxic injury is observed first in the pericentral region due to the intralobular oxygen gradient (10). Hemosiderin accumulation in hemochromatosis and drug-induced hepatotoxicity frequently exist in periportal and pericentral area, respectively (7).
LIVER SURGERY AND I/R INJURY
Chronic liver disease is the fifth leading cause of death worldwide (11). Total deaths worldwide from liver cirrhosis and cancer have increased from 676,000 in 1980 to over 1 million in 2010 (12). Liver resection refers to the surgical removal of portions of the liver that contain cancer, benign tumors or cystic disease. A common technique employed in liver resection in order to reduce intraoperative blood loss is the Pringle maneuver, which involves the clamping of the portal triad (hepatic artery, portal vein, and bile duct) (6). When this technique is employed, the blood flow to the liver is interrupted, resulting in oxygen and nutrient depletion and subsequently impeding mitochondrial oxidative phosphorylation and ATP generation (4). At the same time, stored glycogen hydrolyzes into glucose that prompts anaerobic glycolysis (13). Although this cytosolic glucose utilization pathway replenishes some ATP to the liver, the end product, lactic acid, accumulates in the cell resulting in tissue acidosis. Moreover, hydrogen ions released from ATP hydrolysis and acidic organelles during ischemia further enhance tissue acidosis (14,15). Restoration of blood flow upon release of vascular clamping results in a return to physiologic pH, an event that worsens ischemic tissue injury (4,16). Another setting from which I/R injury can occur to the liver is liver transplantation. Approximately 6,700 liver transplantations are performed annually in the United States (17). Donor livers exposed to ischemia during harvest, storage and transport undergo subsequent reperfusion injury once they are anastomosed to recipient vasculature, and blood flow is restored.
MECHANISMS OF I/R INJURY
The mechanisms underlying hepatic I/R injury are multifactorial and include Ca2+ deregulation, mitochondrial dysfunction, generation of reactive oxygen and nitrogen species, loss of cellular antioxidants, stimulation of catabolic enzymes, and loss of autophagy (4). Progression of I/R injury can be viewed as three different stages: Within the first few minutes of reperfusion, sequential events of calcium overloading and reactive oxygen species (ROS) accumulation in the mitochondria cause mitochondrial dysfunction and the onset of mitochondrial permeability transition (MPT), leading to ATP depletion and necrotic death of hepatocytes. (4,5) Increased ROS also activate Kupffer cells to promote even greater ROS production. During the next 6 hrs of reperfusion, activated Kupffer cells continue to release ROS, cytokines and chemokines to recruit neutrophils. Finally, at the late phase of reperfusion, neutrophil infiltration becomes uncontrollable and incites irreversible systemic inflammation. Although tissue inflammation is an important pathology in hepatic I/R injury, it should be noted that the damage to hepatocytes after reperfusion is the earliest event that conduces to permanent I/R injury to the liver (4,5).
Mitochondrial dysfunction is the main causative mechanism of hepatocyte death after I/R (4,16). The mitochondrion, a power plant in the cell, contains both inner and outer membranes. In contrast to the mitochondrial outer membrane, the mitochondrial inner membrane is virtually impermeable to all solutes except for those having specific carriers or exchangers. While acidic pH during ischemia prevents mitochondrial permeabilization, the opening of MPT pores upon reperfusion disrupts the inner membrane barrier in the mitochondria, which allows an unregulated influx of solutes up to 1,500 Da into the mitochondrial matrix (5,16). Free diffusion of solutes successively induces mitochondrial swelling, uncoupling of oxidative phosphorylation, and depolarization of the mitochondrial membrane potential. As the proton motive force in the mitochondria is governed equally by mitochondrial membrane potential and pH gradient, the onset of MPT after reperfusion collapses the proton motive force, causing ATP depletion and cellular necrosis. Thus, MPT onset and subsequent mitochondrial dysfunction are the key mechanisms contributing to hepatocyte death after I/R (5,16). Besides necrosis, the onset of MPT can also provoke apoptotic cell death. The loss of mitochondrial membrane integrity releases cytochrome c that is normally sequestered in the intermembrane space in the mitochondria. Once released, cytochrome c binds to the apoptosis-inducing factor-1 and pro-caspases to form a protein complex, the apoptosome, which, in turn, activates downstream effector caspases and develops apoptosis (16,18). Thus, MPT onset is a common pathway to both necrotic and apoptotic hepatocyte death after reperfusion. In contrast to necrosis, apoptosis requires ATP. The availability of glycolytic ATP is a critical determinant of cell death fate (18): When hepatocytes are depleted of ATP after MPT onset, necrosis is a predominant cell death fate. However, when ATP is available, hepatocytes undergo apoptotic death instead (16).
Molecular composition of the MPT pores remains incompletely understood; however, three major components, the adenine nucleotide translocator (ANT) on the inner mitochondrial membrane, voltage-dependent anion channel (VDAC) on the outer mitochondrial membrane, and cyclophilin D in the mitochondrial matrix have been identified (16,19). Multiple studies have proposed that the conformational change from trans to cis ANT by cyclophilin D results in MPT onset (16,19–21). Other proteins might also play a role, including hexokinase and Bcl-2 family members (16). Low pH and cyclosporine A (CsA), an immunosuppressive agent, suppress the MPT by inhibiting cyclophilin D (21). While the permanent opening of MPT pores inevitably accompanies cell death, transitory opening plays an integral role for sustaining normal cellular physiology. A transient cycle of mitochondrial pore opening and closing is necessary in Ca2+ and ROS-mediated signal transduction, while concomitantly serving protective functions against unexpected overloading of intramitochondrial Ca2+ and ROS (20).
Therapeutic strategies to prevent hepatic I/R injury have included antioxidant therapy, steroids, Ca2+ chelators, MPT blockers, and protease inhibitors but the clinical outcomes from these therapies to date remain disappointing (22–26). For instance, MPT blockers like CsA, N-methylvaline cyclosporin, and N-methyl-4-isolleucine cyclosporin (NIM811) have a narrow range of therapeutic efficacy (16,27,28) and CsA-insensitive MPT can also develop when MPT induction increases (21). It should also be noted that mitochondrial overloading of Ca2+ and overproduction of ROS, events upstream to the MPT, still prevail even after the MPT is blocked (10). Therefore, it is unlikely that significant and persistent levels of cytoprotection could be achieved with current therapeutic strategies. As mitophagy selectively targets and removes abnormal mitochondria, this endogenous mitochondrial quality control machinery could have therapeutic potentials for I/R injury and other mitochondrial diseases.
AUTOPHAGY
Autophagy is an evolutionarily conserved catabolic process that eliminates protein aggregates and surplus or damaged intracellular organelles. Autophagy was first described by Christian de Duve and is defined as a cell’s “self-eating” event (29). As macroautophagy clears both cellular constituents and dysfunctional mitochondria, this review focuses on macroautophagy and refers it to as autophagy hereafter.
Autophagy is a sequential process that begins with the initiation and formation of an autophagosome, a double membrane structure that sequesters and delivers cellular cargo to the lysosome. Canonical autophagy relies on the recruitment of multiple autophagy-related proteins (ATG) onto a cup-shaped double membrane complex termed a phagophore. Non-canonical autophagy is incompletely understood but can occur in the absence of some key ATG proteins where the expansion of phagophore membrane is dependent on vesicular transport vesicles originated from the Golgi and endosomes (30). Autophagy can selectively or non-selectively enclose cargo material. Selective autophagy includes the removal of specific cellular constituents and intracellular organelles: peroxisomes (pexophagy) (31), mitochondria (mitophagy) (3), ribosomes (ribophagy) (32), endoplasmic reticulum (reticulophagy) (33), lipids (lipophagy) (34), and iron (ferrintinophagy) (35).
Autophagy is slow under basal conditions but becomes stimulated by certain conditions such as nutrient depletion or starvation (1,29). Under nutrient and amino acid-rich conditions, UNC-51 like kinase 1 (ULK1), a mammalian ortholog of yeast ATG1, is phosphorylated at the residue of Ser757 by mammalian or mechanistic target of rapamycin complex 1 (mTORC1) and dissociated from adenosine monophosphate-activated protein kinase (AMPK). This process promotes cell growth and proliferation but prevents autophagy initiation (36). However, under nutrient insufficiency, AMPK senses the changes in energy contents and initiates autophagy by phosphorylating the residues of Ser317, 555, 777 of ULK1 (37,38). Activated ULK1 serves in recruiting and phosphorylating both ATG13 and focal adhesion kinase family interacting protein of 200 kDa (FIP200), generating the ULK1-ATG13-FIP200 complex on the surface of phagophores (39,40). Moreover, ULK1 subdues the kinase activity of mTORC1 through its binding with raptor (41). Thus, the orchestrated coordination between mTORC1, AMPK, and ULK1 is an integral part of autophagy initiation. Another important event in autophagy initiation is the phosphorylation and activation of Beclin -1 (BECN1). When nutrients are abundant, anti-apoptotic protein, B cell lymphoma-2 (Bcl-2), binds to BECN1 and inhibits autophagy. In contrast, under nutrient insufficiency, Bcl-2 is phosphorylated by stress-responsive c-Jun N-terminal protein kinase 1 (JNK1). The change in phosphorylation status of Bcl-2 dissociates BECN1 from Bcl-2, and liberated BECN1 then stimulates autophagy (42,43). BECN1 is also a key component in the BECN1-ATG14-VPS34-VPS15 class III PI3K core complexes (44). Vacuolar sorting protein 34 (VPS34), a class III lipid kinase, modulates vesicle trafficking and the formation of autophagosomal membranes (45,46). The initiation of autophagy is further regulated by other factors, including ultraviolet irradiation resistance-associated gene (UVRAG) (47), BIF-1 (48), ATG14L (49) or RUN domain Beclin-1-interacting cysteine-rich-containing protein (RUBICON) (50).
ATG12 and microtubule-associated protein 1 light chain 3 (LC3) play a major role in autophagosome maturation. Upon the initial activation by ATG7, an E1-ubiquitin like enzyme, ATG12 is covalently connected to ATG5 by ATG10, an E2-like ubiquitin carrier protein. Conjugated ATG12-ATG5 complexes then interact with Atg16L1 later. Other proteins including ATG4B, ATG7, ATG3, are also involved in the maturation of autophagosomes (51). The conjugation of LC3-I to phosphatidylethanolamine by ATG7 and ATG3 generates LC3-II that localizes to autophagosomal membranes. On the contrary, unconjugated LC3-I resides in the cytosol. Due to its distinct location and unique chemical structure, LC3-II is commonly used to monitor autophagy (52,53). The conversion of LC3-I to LC3-II requires ATG4B, a cysteine protease. Importantly, ATG4B can also act on LC3-II to release LC3 from phosphatidylethanolamine (54). The removal of phospholipid not only relocates LC3 from the autophagosomal membrane, but also facilitates its subsequent fusion with the endosome/lysosome.
The mature autophagosome fuses with the lysosome to produce the autolysosome. It has been shown that Ras-like GTPases are involved in this tethering process (55). Specifically, overexpression of Rab7 promotes autophagy, whereas its silencing prevents autophagy (56). The interaction of UVRAG-BECN1-PI3KIII complex with the class C vacuolar protein sorting complex further facilitates Rab7-mediated fusion (57). The formation of autolysosomes is also fine-tuned by soluble N-ethylmaleimide-sensitive fusion factors (SNARES) (57,58), showing the complexity of autophagy network. Once the formation of autolysosomes is completed, its luminal contents are rapidly degraded by acidic proteases, lipases, nucleases, and glycosidases. The final end products following this process are later recycled back to the cell for other metabolic purposes (37).
MITOPHAGY
Mitophagy mediates mitochondrial turnover, which occurs every 15 to 25 days (59). Therefore, functional mitophagy not only prevents the accumulation of abnormal or damaged mitochondria, but also is essential to maintain a stable number of healthy mitochondria. Lemasters’ group has proposed that three different types of mitophagy exist in the cell (3). The mechanisms of type I mitophagy are similar to those in canonical autophagy described above. This mitophagy requires PI3KIII signaling and can occur at the phagophore assembly. In contrast, mitochondrial depolarization instigates the onset of type II mitophagy. Type III mitophagy, termed “micromitophagy,” depends on the formation of mitochondria-derived vesicles enriched in oxidized mitochondrial proteins that bud off and transit into multivesicular bodies. Overall, both type I and II mitophagy engulf an entire mitochondrion for removal, while type III selectively eliminates damaged and oxidized mitochondrial components.
Several proteins have been proposed to induce mitophagy. Under normal conditions, the mitochondrial serine protease, presenilin-associated rhomboid like protein (PARL) cleaves tension homolog-induced putative kinase protein 1 (PINK1) in the mitochondria (60). When the mitochondria depolarize under stress conditions, a decrease in PARL activity and following inhibition of PINK cleavage translocate a full length PINK1 to the outer mitochondrial membrane. Soon after, PINK1 recruits PARKIN, an E3 ubiquitin ligase, to the mitochondria where PARKIN directs the ubiquitination of target proteins such as p62 and VDAC of damaged mitochondria (61–63). Transcription factor p62 is known to act as a linker protein between autophagic cargo and autophagosomes (64). The mitochondrial accumulation of PARKIN appears to be voltage-dependent, and does not require changes in pH or ATP levels (65). Mitochondrial receptors Bcl2/adenovirus EB 19-kDA interacting protein 3 (BNIP3) or FUN14 domain-containing protein-1 (FUNDC1) likely also plays a role in mitophagy (66). BNIP3, also called BNIP3L or NIX, shares homology with Bcl-2 in the BH3 domains. FUNDC1-mediated mitophagy requires ULK1, wherein activated ULK1 phosphorylates FUNDC1 upon mitochondrial depolarization (67,68). Multiple studies have posited that BNIP3 and FUNDC1 trigger mitophagy by binding to LC3 through a WXXL motif (66,69,70).
MITOPHAGY IN LIVER I/R INJURY
Autophagy is a highly energy-dependent process. Hence, ATP depletion during hepatic I/R adversely impacts the autophagic machinery. Anoxia during ischemia impedes the formation of autophagic vesicles, as evidenced by lack of LC3-II increase in the presence of lysosomal inhibitors such as bafilomycin or chloroquine (71,72). Although a transient repolarization of the mitochondrial membrane potential during the early stage of reperfusion can provide some ATP to cells and operate autophagy temporarily, the demand for mitophagy to remove swollen and injured mitochondria exceeds the autophagic capacity in reperfused hepatocytes. A few minutes after reperfusion, hepatocytes thus encounter accumulation of abnormal or dysfunctional mitochondria, uncontrolled Ca2+ and ROS overloading, activation of injurious enzymes, the onset of MPT and eventually cell death (5,71,72). To make things worse, key autophagy proteins such as ATG7 and BECN1 become hydrolyzed by calpains as a consequence of Ca2+ overloading (5,71,72). Hence, loss of key autophagy proteins and depletion of ATP synergistically impair mitophagy after I/R. The importance of mitophagy in ischemic livers is substantiated by findings that both pharmacological and genetic approaches that stimulate mitophagy confer cytoprotection against hepatic I/R injury (5,71,72). Of note, observations that initial MPT onset occurs in a subset of mitochondria prior to widespread MPT in the cell suggest that some mitochondria are more prone to I/R stress. Toxic metabolites and byproducts from injured mitochondria can propagate to neighboring healthy mitochondria, culminating in widespread mitochondrial dysfunction (73). Since mitophagy enhancement blocks the onset of MPT and cell death after reperfusion, timely clearance of these stress-prone mitochondria appears to be indispensable for sustaining functional bioenergetics and cell survival.
SIRTUINS IN THE LIVER
Acetylation is a post-translational modification of proteins by covalent addition of an acetyl group to lysine residues. In general, removal of positively-charged lysine neutralizes the total charge balance of target proteins and thus alters the steric environment of their active sites. Acetylation or deacetylation can impact a variety of cellular functions such as DNA binding affinity, catalytic activity, stability and localization of target proteins (74). Protein acetylation is a highly dynamic process that is governed by balanced action between lysine acetyltransferases (KATs, formerly known as histone acetyltransferases, HATs) and deacetylases (KDACs, formerly termed as histone deacetylases, HDACs). KATs are categorized into three major groups: 1) KAT2/GCN5-related N-acetyltransferases (GNAT family), 2) E1A binding protein p300 (EP300/CREBBP family), and 3) MYST family (75). KDACs are further subdivided into 4 classes, based on their sequence homology to the original yeast enzymes and domain organization. Designated as Class III KDACs, sirtuins have some distinctive features from other classes. They are mammalian ortholog of yeast silent information regulator 2 (Sir2) and utilize oxidized nicotinamide adenine dinucleotide (NAD+) as a cofactor for their enzyme activity (76–80). In mammals, seven different isoforms of sirtuins (SIRT1-7) have been identified. Although individual isoforms contain a uniquely conserved NAD+ deacetylase domain, deacetylation activity varies among the isoforms. The conserved catalytic domain of sirtuins contains up to 270 amino acid residues and forms a characteristic reverse Rossmann-fold, and zinc ribbon (81,82). Recent reports that acetyl-coenzyme A (AcCoA), a major component of the Krebs cycle and β-oxidation in the mitochondria as well as glycolysis and catabolism of branched amino acids in the cytosol, donates its acetyl moiety to the target lysine residues (83,84) demonstrate the involvement of acetylation reactions in cellular energy metabolism. Expectedly therefore, studies confirm that deacetylation reactions by sirtuins are distinctly coupled to transcription, mitochondrial biogenesis, oxidative phosphorylation, and autophagy (85,86).
Sirtuin 1 (SIRT1) localizes in the cytosol and nucleus (87) and is known to regulate circadian rhythms (88–90), autophagy (91–95), gluconeogenesis (96,97), fatty acid oxidation (96,98), mitochondrial biogenesis (96,99,100), cell proliferation (101,102) and antioxidant defense (92). Embryonic lethality in SIRT1-null transgenic mice implicates its essential role in tissue viability (103). In the liver, SIRT1 can deacetylate a myriad of non-histone targets including peroxisome proliferator-activated receptor gamma-coactivator-1 alpha (PGC-1α) (97), CREB regulated transcription coactivator 2 (104), Forkhead transcription factors (FOXO) (105), fibroblast growth factor 21 (FGF21) (106), and signal transducer and activator of transcription 3 (STAT3) (107), all of whch are closedly associated with hepatic energy homeostatsis and metabolism.
The roles of other sirtuin isoforms in physiology and pathophysiology are beginning to be elucidated as well. The mitochondria-localized SIRT3, for instance, modulates intramitochondrial metabolic activities and ROS formation by deacetylating the NDUFA9 subunit of Complex 1 in the mitochondrial electron transfer chain (108–112). Another connection of SIRT3 with mitochondrial energy homeostasis comes from experiments demonstrating that SIRT3 can directly deacetylate mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2, a late limiting enzyme in the synthesis of ketone body (112–114). SIRT5 is another mitochondrial isoform of sirtuins encompassing deacetylation (115), desuccinylation (116), demalonylation (116), and deglutarylation (117). This mitochondrial matrix enzyme has long been known to regulate carbamoyl phosphate synthase 1 (CPS1), a rate limiting enzyme for the urea cycle and ammonia clearance (115–117). Taken together, both mitochondrial and extramitochondrial sirtuins affect cellular energy metabolism and homeostasis.
ROLE OF ACETYLATION/DEACETYLATION IN AUTOPHAGY
Although sirtuins are not an essential component of the autophagic machinery, evidence is accumulating to indicate that autophagy is regulated by the cycle of acetylation/deacetylation. Table 1 summarizes autophagy-related protein targets that are regulated by acetylation status. Acetylation/deacetylation-dependent modulation of autophagy occurs transcriptionally and translationally. For example, spermidine, a natural inhibitor of KATs, enhances autophagy through hypoacetylation of ATG7 promoter (118). Studies have shown that changes in acetylation status significantly impact the activity of FOXO1 or FOXO3, transcription factors associated with autophagy induction (119). More direct evidences come from the studies which demonstrate that p300 can acetylate ATG5, ATG7, LC3 and ATG12 (120), whereas SIRT1 can deacetylate ATG5, ATG7 and LC3 under nutrient insufficiency (91). SIRT1-dependent deacetylation also determines intracellular distribution of autophagy components. Huang et al. recently reported that deacetylation of LC3 by SIRT1 redistributes LC3 from the nucleus to the cytoplasm, and that deacetylated cytosolic LC3 produces more stable autophagosomes (121). Thus, acetylation/deacetylation cycle not only regulates the activity of autophagy but also ensures an effective redistribution of autophagy elements between intracellular compartments. BECN1 appears to be another autophagy target that is regulated by acetylation/deacetylation since acetylated BECN1 inhibits autophagosome maturation and endocytic trafficking (122). Studies also suggest that acetylated ULK1 can enhance autophagy process by stimulating its kinase activity (123,124). Thus, changes in acetylation status highly affect both initiation and elongation stage of autophagy.
Table 1.
Summary of autophagy related proteins regulated by acetylation/deacetylation
| Protein | Description | References |
|---|---|---|
| ULK1 | Enhancing autophagy upon acetylation by TIP60 | (123,124) |
| ATG3, ATG5, ATG7 | Decreasing autophagy upon acetylation by p300 Enhancing autophagy upon deacetylation by SIRT1 |
(91,120) |
| ATG12 | Decreasing autophagy upon acetylation by p300 | (120) |
| LC3 | Decreasing autophagy upon acetylation by p300 Enhancing autophagy upon deacetylation by SIRT1 |
(91,120,121) |
| BECN1 | Decreasing autophagy upon acetylation by p300 Enhancing autophagy upon deacetylation by SIRT1 |
(122) |
| FOXO1, FOXO3 | Decreasing autophagy upon acetylation by p300 Enhancing autophagy upon deacetylation by sirtuins (SIRT1, 2 and 3) |
(119) |
| MFN2 | Enhancing autophagy upon deacetylation by SIRT1 | (125) |
| Tubulin | Enhancing autophagy upon acetylation by α-TAT1/MEC17 Decreasing autophagy upon deacetylation by HDAC6 |
(128,129) |
| Hsp70 | Decreasing autophagy upon acetylation by p300 Enhancing autophagy upon deacetylation by HDAC6 |
(130) |
SIRT1, AUTOPHAGY AND I/R INJURY
We recently demonstrated in human liver biopsies that hepatic inflow occlusion during liver resection decreases SIRT1 expression to 30% of basal levels (125). Such a reduction was also evident in the mouse livers and hepatocytes after in vivo and in vitro I/R, respectively. Calpain activation due to Ca2+ overloading during I/R appears to be, at least in part, responsible for SIRT1 loss. Although reperfusion after prolonged ischemia leads to near-complete depletion of SIRT1 in both the cytosol and nucleus, subcellular fractionation assays revealed that cytosolic SIRT1 loss precedes nuclear SIRT1 loss, implying that cytosolic SIRT1 is more susceptible to hepatocellular I/R injury. Adenoviral overexpression or pharmacological activation of SIRT1 by resveratrol or SRT1720 enhanced cytosolic levels of SIRT1 and mitophagy, and sustained mitochondrial structural integrity after reperfusion, substantiating the importance of cytosolic SIRT1 in mitochondrial quality. The mechanism by which SIRT1 elevates mitophagy is likely associated with ATG7 because SIRT1 overexpression significantly increases ATG7 expression, whereas levels of other autophagy-related proteins such as ATG3, ATG4B, ATG12-5, ATG14L, BECN1, RUBICON, LAMP2A, and Cathepsin D remain unaltered by this treatment. Importantly, both treatments conferred cytoprotection against global MPT onset and necrosis, which was not observed in hepatocytes from SIRT1 conditional knockout mice. In contrast to wild-type hepatocytes, the cells from SIRT1-deficient mice exhibited a rapid onset of the MPT and increased cell death even after short ischemia. Taken together, these results indicate that SIRT1 depletion contributes to I/R injury in hepatocytes and cytosolic SIRT1 is required for mitochondrial integrity and function (Fig. 1).
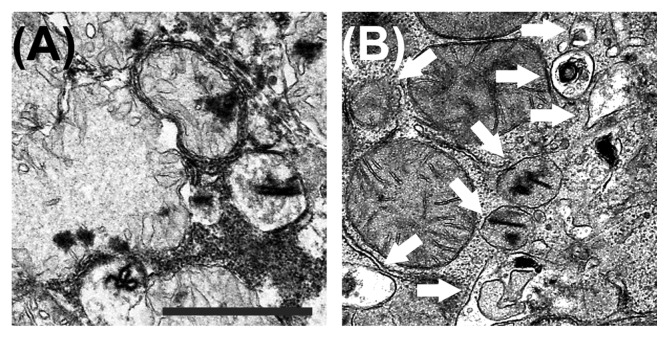
Fig. 1.
The effects of sirtuin 1 on the mitochondrial integrity and autophagy in reperfused hepatocytes. (A) Scanning electron microgram shows that in the control cell, reperfusion of ischemic hepatocytes causes a marked swelling and structural disruption of the mitochondria. Note lack of autophagy induction under this condition. (B) However, overexpression of sirtuin 1 prior to I/R sustains the integrity of mitochondrial structure. Under this condition, numerous autophagic vesicles are visible (arrows). Scale bar: 2 μm.
Many mitochondrial proteins exist in an acetylated form. About 35% of all mitochondrial proteins are endogenously acetylated, 24% of which are mechanistically linked to energy homeostasis (126). Hyperacetylation or defective deacetylation of the mitochondrial proteins has been shown to account for liver steatosis and obesity (114,127). The mechanisms underlying SIRT1-mediated cytoprotection against I/R injury are likely linked to deacetylation of mitofusin 2 (MFN2), a mitochondrial outer membrane protein, by SIRT1 and subsequent augmentation of mitophagy. Immunoprecipitation and immunoblotting approaches unveiled that while cytosolic SIRT1 physically interacts with both MFN1 and MFN2, it deacetylates only MFN2. The importance of SIRT1/MFN2 interaction in hepatic I/R injury was further supported by the result that knock-down of MFN2 with a small hairpin RNA abolishes a series of beneficial effects by SIRT1, including SIRT1-mediated mitophagy induction, cytoprotection against mitochondrial dysfunction, and cell death after reperfusion. Though the acetylated residues of mouse MFN2 are currently unknown, bioinformatic analysis conforming to either X6-K-[Y,W,F]-X5 or X6-KX5-[Y,W,F] motif predicts at least five different SIRT1 target sites of MFN2: K37, K215, K357, K655, and K662 (125). Noticeably, K215 localizes at the GTPase domain, a critical site for catalytic activity of MFN2. Both K655 and K662 reside in the C-terminal flanking domain that directs mitochondrial localization of MFN2. Consistent with this prediction, deletion of N-terminal regions of MFN2 blunted SIRT1-mediated autophagy induction. One interesting observation is that while hepatocytes are relatively tolerant of a large loss in SIRT1, a similar reduction of MFN2 causes greater cell death after reperfusion, implying a central role of MFN2 in I/R injury to the liver. It has been reported that MFN2-null animals are embryonically lethal, whereas SIRT1 knockout mice are born alive (125). Hence, it is speculated that MFN2-deficient cells may be more prone to I/R injury than SIRT1-null counterpart. Although the minimal levels of MFN2 needed for adequate responses to I/R and other stresses remain to be determined, the interaction of MFN2 and its subsequent deacetylation by SIRT1 are likely pivotal events in autophagy regulation and cell survival after I/R (Fig. 2).
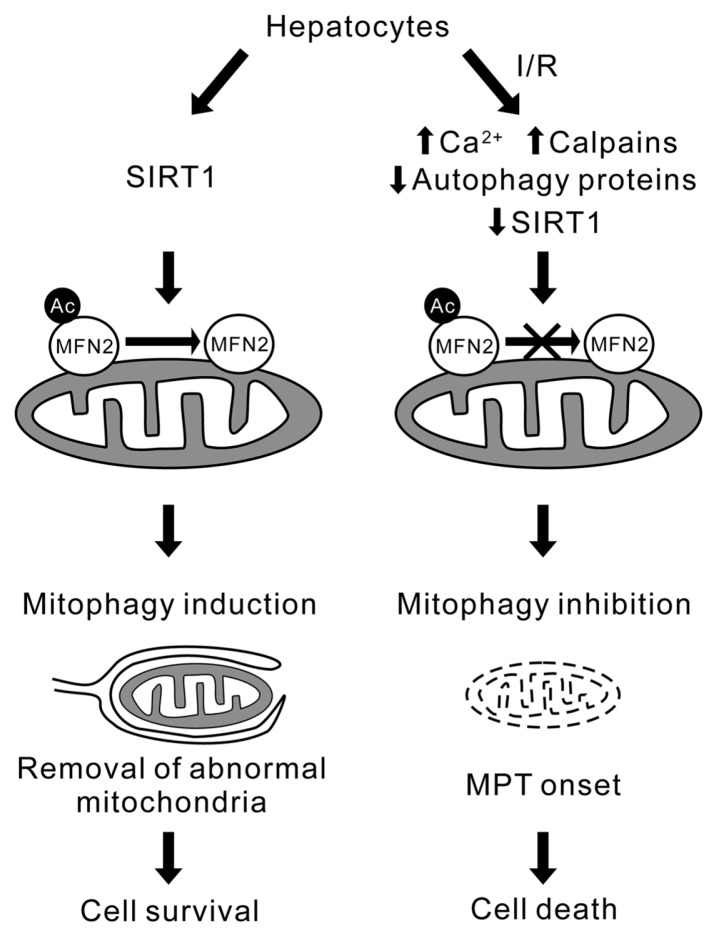
Fig. 2.
Proposed mechanism of sirtuin 1-mediated cytoprotection against ischemia/reperfusion injury in the liver. In normal hepatocytes, cytosolic sirtuin 1 (SIRT1) deacetylates mitofusin 2 (MFN2), a mitochondrial outer membrane protein, which, in turn, induces mitophagy and maintains a stable number of heathy mitochondria and cell viability thereafter. In sharp contrast, ischemia/reperfusion (I/R) to hepatocytes causes Ca2+ overloading and calpain activation that subsequently hydrolyzes both key autophagy proteins and SIRT1. As a consequence, MFN2 remains acetylated (Ac-MFN2) and the onset of mitophagy fails. Hepatocytes further accumulate intramitochondrial Ca2+ and reactive oxygen species, resulting in the mitochondrial permeability transition (MPT) and cell death.
CONCLUSION AND FUTURE PERSPECTIVES
I/R injury has a profound impact on the burden of liver diseases. Efforts to improve liver function after I/R, however, have not been successful largely due to an incomplete understanding of I/R injury. The mechanisms behind hepatic I/R injury are multifactorial, including defective mitophagy, the onset of MPT and mitochondrial dysfunction. Such a complexity of reperfusion injury is further underscored by the recent study which shows that the SIRT1/MFN2 axis controls mitophagy and mitochondrial function in ischemic livers. Although enhancing mitophagy has emerged as a new potential strategy against reperfusion injury, there exist a few unanswered questions, such as the effects of mitophagy on non-parenchymal cells after I/R, roles of different types of mitophagy in ischemic livers, similarities and differences in mitophagy signaling pathways between normal and ischemic livers, and impact of other mitochondrial sirtuins and their potential interactions with SIRT1 before and after ischemia. It should also be noted that current strategies including treatment with autophagy inducers prior to ischemia and the viral delivery of specific autophagy genes and lack of specific autophagy enhancers without compromising the immune system all limit their clinical applications. Better understanding of the pathological complexity of reperfusion injury and its mechanistic insights into mitophagy could lead to the development of promising treatment strategies for hepatic I/R injury and mitochondrial diseases.
ACKNOWLEDGEMENTS
This work was supported in part by US National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases grant DK079879 and DK090115 (J-S Kim) and National Institute on Aging AG028740 (J-S Kim).
REFERENCES
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 3.Lemasters JJ. Variants of mitochondrial autophagy: Types 1 and 2 mitophagy and micromitophagy (Type 3) Redox Biol. 2014;2:749–754. doi: 10.1016/j.redox.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cursio R, Colosetti P, Gugenheim J. Autophagy and liver ischemia-reperfusion injury. Biomed Res Int. 2015;2015:417590. doi: 10.1155/2015/417590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JS, Nitta T, Mohuczy D, O’Malley KA, Moldawer LL, Dunn WA, Jr, Behrns KE. Impaired autophagy: A mechanism of mitochondrial dysfunction in anoxic rat hepatocytes. Hepatology. 2008;47:1725–1736. doi: 10.1002/hep.22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumgart LH, Hann LE. Surgical and Radiological Anatomy of the Liver, Biliary Tract and Pancreas, Blumgart’s Surgery of the Liver, Biliary tract and Pancreas. 5th Edition. Elsevier; Philadelphia: 2012. p. 31. [Google Scholar]
- 7.Arias IM, Alter HJ, Boyer JL, Cohen DE, Fausto N, Shafritz DA, Wolkoff AW. The liver: Biology and pathobiology. 5th Edition. Wiley-Blackwell; New Jersey: 2009. [DOI] [Google Scholar]
- 8.Bradley SE, Ingelfinger FJ, Bradley GP, Curry JJ. The estimation of hepatic blood flow in man. J Clin Invest. 1945;24:890–897. doi: 10.1172/JCI101676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford BU, Marotto M, Lemasters JJ, Thurman RG. New, simple models to evaluate zone-specific damage due to hypoxia in the perfused rat liver: time course and effect of nutritional state. J Pharmacol Exp Ther. 1986;236:263–268. [PubMed] [Google Scholar]
- 10.Malhi H, Gores GJ, Lemasters JJ. Apoptosis and necrosis in the liver: a tale of two deaths? Hepatology. 2006;43:S31–44. doi: 10.1002/hep.21062. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization (WHO) Study of liver disease mortality. 2013. [Google Scholar]
- 12.Mokdad AA, Lopez AD, Shahraz S, Lozano R, Mokdad AH, Stanaway J, Murray CJ, Naghavi M. Liver cirrhosis mortality in 187 countries between 1980 and 2010: a systematic analysis. BMC Med. 2014;12:145. doi: 10.1186/s12916-014-0145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang L, Tian F, Tao W, Cui J. Hepatocellular glycogen in alleviation of liver ischemia-reperfusion injury during partial hepatectomy. World J Surg. 2007;31:2039–2043. doi: 10.1007/s00268-007-9186-0. [DOI] [PubMed] [Google Scholar]
- 14.Lemasters JJ, Caldwell-Kenkel JC, Gao W, Nieminen AL, Herman B, Thurman RG. In: Hypoxic, ischemic and reperfusion injury in the liver in Pathophysiology of Reperfusion Injury. Das DK, editor. CRC; Florida: 1992. pp. 101–135. [Google Scholar]
- 15.Bronk SF, Gores GJ. Efflux of protons from acidic vesicles contributes to cytosolic acidification of hepatocytes during ATP depletion. Hepatology. 1991;14:626–633. doi: 10.1002/hep.1840140409. [DOI] [PubMed] [Google Scholar]
- 16.Kim JS, He L, Lemasters JJ. Mitochondrial permeability transition: a common pathway to necrosis and apoptosis. Biochem Biophys Res Commun. 2003;304:463–470. doi: 10.1016/S0006-291X(03)00618-1. [DOI] [PubMed] [Google Scholar]
- 17.United Network for Organ Sharing (UNOS) 2014. [Google Scholar]
- 18.Kim JS, Qian T, Lemasters JJ. Mitochondrial permeability transition in the switch from necrotic to apoptotic cell death in ischemic rat hepatocytes. Gastroenterology. 2003;124:494–503. doi: 10.1053/gast.2003.50059. [DOI] [PubMed] [Google Scholar]
- 19.Leung AW, Halestrap AP. Recent progress in elucidating the molecular mechanism of the mitochondrial permeability transition pore. Biochim Biophys Acta. 2008;1777:946–952. doi: 10.1016/j.bbabio.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Hausenloy D, Wynne A, Duchen M, Yellon D. Transient mitochondrial permeability transition pore opening mediates preconditioning-induced protection. Circulation. 2004;109:1714–1717. doi: 10.1161/01.CIR.0000126294.81407.7D. [DOI] [PubMed] [Google Scholar]
- 21.He L, Lemasters JJ. Regulated and unregulated mitochondrial permeability transition pores: a new paradigm of pore structure and function? FEBS Lett. 2002;512:1–7. doi: 10.1016/S0014-5793(01)03314-2. [DOI] [PubMed] [Google Scholar]
- 22.Akhtar MZ, Henderson T, Sutherland A, Vogel T, Friend PJ. Novel approaches to preventing ischemia-reperfusion injury during liver transplantation. Transplant Proc. 2013;45:2083–2092. doi: 10.1016/j.transproceed.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Yamashita Y, Shimada M, Hamatsu T, Rikimaru T, Tanaka S, Shirabe K, Sugimachi K. Effects of preoperative steroid administration on surgical stress in hepatic resection: prospective randomized trial. Arch Surg. 2001;136:328–333. doi: 10.1001/archsurg.136.3.328. [DOI] [PubMed] [Google Scholar]
- 24.Pan LJ, Zhang ZC, Zhang ZY, Wang WJ, Xu Y, Zhang ZM. Effects and mechanisms of store-operated calcium channel blockade on hepatic ischemia-reperfusion injury in rats. World J Gastroenterol. 2012;18:356–367. doi: 10.3748/wjg.v18.i4.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gurusamy KS, Gonzalez HD, Davidson BR. Current protective strategies in liver surgery. World J Gastroenterol. 2010;16:6098–6103. doi: 10.3748/wjg.v16.i48.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uchida M, Takemoto Y, Nagasue N, Dhar DK, Kohno H, Nakamura T. Effect of verapamil on hepatic reperfusion injury after prolonged ischemia in pigs. J Hepatol. 1994;21:217–223. doi: 10.1016/S0168-8278(05)80398-8. [DOI] [PubMed] [Google Scholar]
- 27.Kon K, Kim JS, Jaeschke H, Lemasters JJ. Mitochondrial permeability transition in acetaminophen-induced necrosis and apoptosis of cultured mouse hepatocytes. Hepatology. 2004;40:1170–1179. doi: 10.1002/hep.20437. [DOI] [PubMed] [Google Scholar]
- 28.Kim JS, Jin Y, Lemasters JJ. Reactive oxygen species, but not Ca2+ overloading, trigger pH- and mitochondrial permeability transition-dependent death of adult rat myocytes after ischemia-reperfusion. Am J Physiol Heart Circ Physiol. 2006;290:H2024–H2034. doi: 10.1152/ajpheart.00683.2005. [DOI] [PubMed] [Google Scholar]
- 29.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 30.Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2011;13:7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- 31.Dunn WA, Jr, Cregg JM, Kiel JA, van der Klei IJ, Oku M, Sakai Y, Sibirny AA, Stasyk OV, Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- 32.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 33.Hamasaki M, Noda T, Baba M, Ohsumi Y. Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic. 2005;6:56–65. doi: 10.1111/j.1600-0854.2004.00245.x. [DOI] [PubMed] [Google Scholar]
- 34.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung CH, Seo M, Otto NM, Kim DH. ULK1 inhibits the kinase activity of mTORC1 and cell proliferation. Autophagy. 2011;7:1212–1221. doi: 10.4161/auto.7.10.16660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai DF, Johnson SC, Villarin JJ, Chin MT, Nieves-Cintrón M, Chen T, Marcinek DJ, Dorn GW, Kang YJ, Prolla TA, Santana LF, Rabinovitch PS. Mitochondrial oxidative stress mediates angiotensin II-induced cardiac hypertrophy and Galphaq overexpression-induced heart failure. Circ Res. 2011;108:837–846. doi: 10.1161/CIRCRESAHA.110.232306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 46.Obara K, Ohsumi Y. Atg14: a key player in orchestrating autophagy. Int J Cell Biol. 2011;2011:713435. doi: 10.1155/2011/713435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mulé JJ, Pledger WJ, Wang HG. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, Akira S, Noda T, Yoshimori T. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 51.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 52.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 53.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 55.Chua CE, Gan BQ, Tang BL. Involvement of members of the Rab family and related small GTPases in autophagosome formation and maturation. Cell Mol Life Sci. 2011;68:3349–3358. doi: 10.1007/s00018-011-0748-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014;21:348–358. doi: 10.1038/cdd.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moreau K, Renna M, Rubinsztein DC. Connections between SNAREs and autophagy. Trends Biochem Sci. 2013;38:57–63. doi: 10.1016/j.tibs.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 58.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Söllner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/S0092-8674(00)81404-X. [DOI] [PubMed] [Google Scholar]
- 59.Menzies RA, Gold PH. The turnover of mitochondria in a variety of tissues of young adult and aged rats. J Biol Chem. 1971;246:2425–2429. [PubMed] [Google Scholar]
- 60.Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SH, Renton AE, Harvey RJ, Whitworth AJ, Martins LM, Abramov AY, Wood NW. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 62.Michiorri S, Gelmetti V, Giarda E, Lombardi F, Romano F, Marongiu R, Nerini-Molteni S, Sale P, Vago R, Arena G, Torosantucci L, Cassina L, Russo MA, Dallapiccola B, Valente EM, Casari G. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ. 2010;17:962–974. doi: 10.1038/cdd.2009.200. [DOI] [PubMed] [Google Scholar]
- 63.Okatsu K, Oka T, Iguchi M, Imamura K, Kosako H, Tani N, Kimura M, Go E, Koyano F, Funayama M, Shiba-Fukushima K, Sato S, Shimizu H, Fukunaga Y, Taniguchi H, Komatsu M, Hattori N, Mihara K, Tanaka K, Matsuda N. PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat Commun. 2012;3:1016. doi: 10.1038/ncomms2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 65.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin-induced mitophagy in the pathogenesis of Parkinson disease. Autophagy. 2009;5:706–708. doi: 10.4161/auto.5.5.8505. [DOI] [PubMed] [Google Scholar]
- 66.Feng D, Liu L, Zhu Y, Chen Q. Molecular signaling toward mitophagy and its physiological significance. Exp Cell Res. 2013;319:1697–1705. doi: 10.1016/j.yexcr.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 67.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, Huang L, Xue P, Li B, Wang X, Jin H, Wang J, Yang F, Liu P, Zhu Y, Sui S, Chen Q. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 68.Wu W, Tian W, Hu Z, Chen G, Huang L, Li W, Zhang X, Xue P, Zhou C, Liu L, Zhu Y, Zhang X, Li L, Zhang L, Sui S, Zhao B, Feng D. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014;15:566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen M, Sandoval H, Wang J. Selective mitochondrial autophagy during erythroid maturation. Autophagy. 2008;4:926–928. doi: 10.4161/auto.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang JH, Ahn IS, Fischer TD, Byeon JI, Dunn WA, Jr, Behrns KE, Leeuwenburgh C, Kim JS. Autophagy suppresses age-dependent ischemia and reperfusion injury in livers of mice. Gastroenterology. 2011;141:2188–2199. doi: 10.1053/j.gastro.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim JS, Wang JH, Biel TG, Kim DS, Flores-Toro JA, Vijayvargiya R, Zendejas I, Behrns KE. Carbamazepine suppresses calpain-mediated autophagy impairment after ischemia/reperfusion in mouse livers. Toxicol Appl Pharmacol. 2013;273:600–610. doi: 10.1016/j.taap.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pacher P, Hajnoczky G. Propagation of the apoptotic signal by mitochondrial waves. EMBO J. 2001;20:4107–4121. doi: 10.1093/emboj/20.15.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 75.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 76.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 77.North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Michan S, Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anderson KA, Green MF, Huynh FK, Wagner GR, Hirschey MD. SnapShot: Mammalian Sirtuins. Cell. 2014;159:956. doi: 10.1016/j.cell.2014.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nat Struct Biol. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- 82.Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/S0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- 83.Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015;21:805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 84.Mariño G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, Zamzami N, Scoazec M, Durand S, Enot DP, Fernández ÁF, Martins I, Kepp O, Senovilla L, Bauvy C, Morselli E, Vacchelli E, Bennetzen M, Magnes C, Sinner F, Pieber T, López-Otin C, Maiuri MC, Codogno P, Andersen JS, Hill JA, Madeo F, Kroemer G. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell. 2014;53:710–725. doi: 10.1016/j.molcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 85.Lombard DB, Tishkoff DX, Bao J. In: Mitochondrial sirtuins in the regulation of mitochondrial activity and metabolic adaptation in Histone Deacetylases: the Biology and Clinical Implication. Yao TP, Seto E, editors. Springer; Heidelberg: 2011. pp. 163–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Satoh A, Stein L, Imai S. In: The role of mammalian sirtuins in the regulation of metabolism, aging, and longevity in Histone Deacetylases: the Biology and Clinical Implication. Yao TP, Seto E, editors. Springer; Heidelberg: 2011. pp. 126–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 88.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 89.Belden WJ, Dunlap JC. SIRT1 is a circadian deacetylase for core clock components. Cell. 2008;134:212–214. doi: 10.1016/j.cell.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 Plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010;107:1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, Mitchell JR, Croteau DL, Bohr VA. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD+/SIRT1 reduction. Cell. 2014;157:882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jang SY, Kang HT, Hwang ES. Nicotinamide-induced mitophagy: event mediated by high NAD+/ NADH ratio and SIRT1 protein activation. J Biol Chem. 2012;287:19304–19314. doi: 10.1074/jbc.M112.363747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rickenbacher A, Jang JH, Limani P, Ungethum U, Lehmann K, Oberkofler CE, Weber A, Graf R, Humar B, Clavien PA. Fasting protects liver from ischemic injury through Sirt1-mediated downregulation of circulating HMGB1 in mice. J Hepatol. 2014;61:301–308. doi: 10.1016/j.jhep.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 96.Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 98.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aquilano K, Vigilanza P, Baldelli S, Pagliei B, Rotilio G, Ciriolo MR. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J Biol Chem. 2010;285:21590–21599. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 101.García-Rodríguez JL, Barbier-Torres L, Fernández-Álvarez S, Gutierré-de Juan V, Monte MJ, Halilbasic E, Herranz D, Alvarez L, Áspichueta P, Marin JJ, Trauner M, Mato JM, Serrano M, Beraza N, Martinez-Chantar ML. SIRT1 controls liver regeneration by regulating bile acid metabolism through farnesoid X receptor and mammalian target of rapamycin signaling. Hepatology. 2014;59:1972–1983. doi: 10.1002/hep.26971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kabra N, Li Z, Chen L, Li B, Zhang X, Wang C, Yeatman T, Coppola D, Chen J. SirT1 is an inhibitor of proliferation and tumor formation in colon cancer. J Biol Chem. 2009;284:18210–18217. doi: 10.1074/jbc.M109.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, Olefsky J, Guarente L, Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/ coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park JM, Kim TH, Bae JS, Kim MY, Kim KS, Ahn YH. Role of resveratrol in FOXO1-mediated gluconeogenic gene expression in the liver. Biochem Biophys Res Commun. 2010;403:329–334. doi: 10.1016/j.bbrc.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 106.Li Y, Wong K, Giles A, Jiang J, Lee JW, Adams AC, Kharitonenkov A, Yang Q, Gao B, Guarente L, Zang M. Hepatic SIRT1 attenuates hepatic steatosis and controls energy balance in mice by inducing fibroblast growth factor 21. Gastroenterology. 2014;146:539–549. doi: 10.1053/j.gastro.2013.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nie Y, Erion DM, Yuan Z, Dietrich M, Shulman GI, Horvath TL, Gao Q. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol. 2009;11:492–500. doi: 10.1038/ncb1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schwer B, Eckersdorff M, Li Y, Silva JC, Fermin D, Kurtev MV, Giallourakis C, Comb MJ, Alt FW, Lombard DB. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–606. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–928. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Peek CB, Affinati AH, Ramsey KM, Kuo HY, Yu W, Sena LA, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, Levine DC, Bacsik DJ, Gius D, Newgard CB, Goetzman E, Chandel NS, Denu JM, Mrksich M, Bass J. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science. 2013;342:1243417. doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci USA. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Van Hove JL, Watson PA, Birdsey N, Bao J, Gius D, Sack MN, Jing E, Kahn CR, Friedman JE, Jonscher KR. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J. 2011;433:505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu G, Ilkayeva OR, Muehlbauer MJ, Braulke T, Mühlhausen C, Backos DS, Olsen CA, McGuire PJ, Pletcher SD, Lombard DB, Hirschey MD, Zhao Y. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Eisenberg T, Knauer H, Schauer A, Büttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, Fussi H, Deszcz L, Hartl R, Schraml E, Criollo A, Megalou E, Weiskopf D, Laun P, Heeren G, Breitenbach M, Grubeck-Loebenstein B, Herker E, Fahrenkrog B, Fröhlich KU, Sinner F, Tavernarakis N, Minois N, Kroemer G, Madeo F. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 119.Daitoku H, Sakamaki J, Fukamizu A. Regulation of FoxO transcription factors by acetylation and protein-protein interactions. Biochim Biophys Acta. 2011;1813:1954–1960. doi: 10.1016/j.bbamcr.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 120.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009;284:6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang R, Xu Y, Wan W, Shou X, Qian J, You Z, Liu B, Chang C, Zhou T, Lippincott-Schwartz J, Liu W. Deacetylation of Nuclear LC3 Drives Autophagy Initiation under Starvation. Mol Cell. 2015;57:456–466. doi: 10.1016/j.molcel.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 122.Sun T, Li X, Zhang P, Chen WD, Zhang HL, Li DD, Deng R, Qian XJ, Jiao L, Ji J, Li YT, Wu RY, Yu Y, Feng GK, Zhu XF. Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat Commun. 2015;6:7215. doi: 10.1038/ncomms8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J, Ma C, Sun Y, Zhang S, Feng W, Zhu L, Le Y, Gong X, Yan X, Hong B, Jiang FJ, Xie Z, Miao D, Deng H, Yu L. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336:474–477. doi: 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- 124.Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y, Zhang SM, Lian G, Liu Q, Ruan K, Wang Z, Zhang CS, Chien KY, Wu J, Li Q, Han J, Lin SC. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012;336:477–481. doi: 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- 125.Biel TG, Lee S, Flores-Toro JA, Dean JW, Go KL, Lee MH, Law BK, Law ME, Dunn WA, Jr, Zendejas I, Behrns KE, Kim JS. Sirtuin 1 suppresses mitochondrial dysfunction of ischemic mouse livers in a mitofusin 2-dependent manner. Cell Death Differ. 2016;23:279–290. doi: 10.1038/cdd.2015.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Anderson KA, Hirschey MD. Mitochondrial protein acetylation regulates metabolism. Essays Biochem. 2012;52:23–35. doi: 10.1042/bse0520023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stančáková A, Goetzman E, Lam MM, Schwer B, Stevens RD, Muehlbauer MJ, Kakar S, Bass NM, Kuusisto J, Laakso M, Alt FW, Newgard CB, Farese RV, Jr, Kahn CR, Verdin E. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mackeh R, Lorin S, Ratier A, Mejdoubi-Charef N, Baillet A, Bruneel A, Hamaï A, Codogno P, Poüs C, Perdiz D. Reactive oxygen species, AMP-activated protein kinase, and the transcription cofactor p300 regulate α-tubulin acetyltransferase-1 (αTAT-1/MEC-17)-dependent microtubule hyperacetylation during cell stress. J Biol Chem. 2014;289:11816–11828. doi: 10.1074/jbc.M113.507400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McLendon PM, Ferguson BS, Osinska H, Bhuiyan MS, James J, McKinsey TA, Robbins J. Tubulin hyperacetylation is adaptive in cardiac proteotoxicity by promoting autophagy. Proc Natl Acad Sci USA. 2014;111:E5178–E5186. doi: 10.1073/pnas.1415589111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang Y, Fiskus W, Yong B, Atadja P, Takahashi Y, Pandita TK, Wang HG, Bhalla KN. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc Natl Acad Sci USA. 2013;110:6841–6846. doi: 10.1073/pnas.1217692110. [DOI] [PMC free article] [PubMed] [Google Scholar]