Abstract
Chemical inhibition of proteins involved in chromatin-mediated signaling is an emerging strategy to control chromatin compaction with the aim to reprogram expression networks to alter disease states. Protein methyltransferases constitute one of the protein families that participate in epigenetic control of gene expression, and represent a novel therapeutic target class. Recruitment of the protein lysine methyltransferase DOT1L at aberrant loci is a frequent mechanism driving acute lymphoid and myeloid leukemias, particularly in infants, and pharmacological inhibition of DOT1L extends survival in a mouse model of mixed lineage leukemia. A better understanding of the structural chemistry of DOT1L inhibition would accelerate the development of improved compounds. Here, we report that the addition of a single halogen atom at a critical position in the cofactor product S-adenosylhomocysteine (SAH, an inhibitor of SAM-dependent methyltransferases) results in an 8-fold increase in potency against DOT1L, and reduced activities against other protein and non-protein methyltransferases. We solved the crystal structure of DOT1L in complex with Bromo-deaza-SAH and rationalized the observed effects. This discovery reveals a simple strategy to engineer selectivity and potency towards DOT1L into the adenosine scaffold of the cofactor shared by all methyltransferases, and can be exploited towards the development of clinical candidates against mixed lineage leukemia.
Keywords: Protein methyltransferase, DOT1L, Leukemia, Inhibitor, Crystallography
1. Introduction
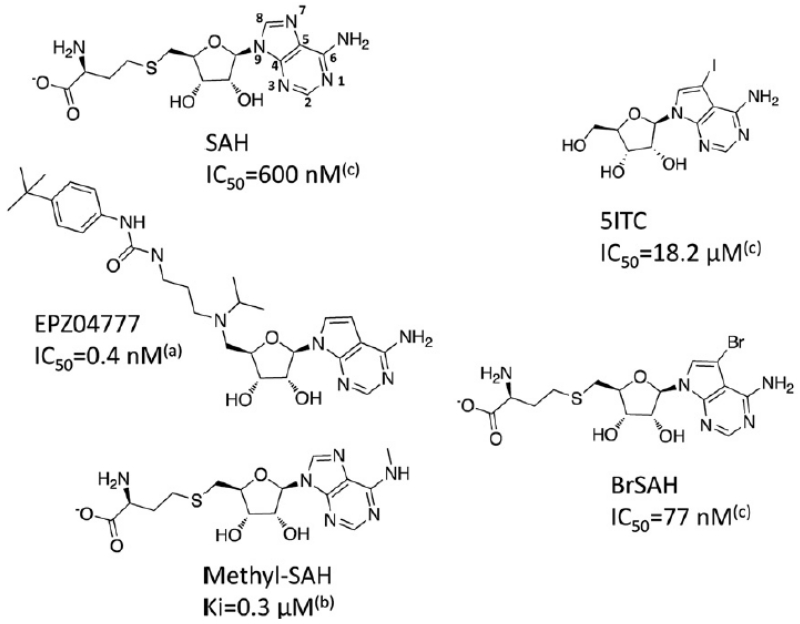
Chromosomal translocation at the mixed lineage leukemia (MLL) gene can result in oncogenic fusions between MLL and a number of proteins, including AF4, AF9 and AF10, and formation of translocation complexes that recruit the H3K79 methyltransferase DOT1L at aberrant loci in leukemia. The resulting illicit methylation of H3K79 activates expression of genes essential for leukemogenesis, and is a key driver in 70% of acute lymphoid and over 35% of acute myeloid leukemias in infants.1-3 Pharmacological inhibition of DOT1L as a therapeutic strategy was recently supported by the discovery of EPZ004777, a small molecule DOT1L inhibitor that selectively kills mixed lineage leukemia (MLL) cells and extends survival in a mouse MLL xenograft model.4 EPZ004777 competes with S-adenosylmethionine (SAM), the cofactor shared by SAM-dependent methyltransferases including lysine-, arginine-, DNA-, and small-molecule methyltransferases. EPZ004777 retains the adenosine scaffold of SAM, but is a much larger and more hydrophobic compound due to the substitution of the amino-acid end by a tertbutyl-phenyl-urea group (Fig. 1). This inhibitor, and analogs, reveal that important chemical modifications are tolerated at the homocysteine end of SAM, which can be exploited to improve pharmacokinetics.4, 5 Follow-up structural analysis by Basavapathruni et al. and by us revealed that the presence of the tertbutyl-phenyl-urea group induced a dramatic remodeling of the DOT1L catalytic site. Introduction of a bromine on the adenosine scaffold improved IC50 from 0.5 to 0.3 nM, and the effect on specificity was not characterized6, 7. Yao et al. identified more direct SAM analogs as specific inhibitors of DOT1L: (1) while the reaction by-product S-adenosylhomocysteine (SAH) binds with high affinity to numerous protein methyltransferases8, and acts as SAM competitive inhibitor, increased selectivity was achieved by alkylating the primary amine at the adenine ring of SAH (see methyl-SAH in Fig. 1); (2) increased potency was obtained through substitution at the homocysteine end, which induced covalent binding to the substrate lysine.9 Unfortunately, no cellular activity has been reported for these compounds, presumably owing to their high hydrophilicity and poor cell penetrance.
Figure 1. Structure of DOT1L inhibitors.
All reported DOT1L inhibitors are mimetics of the cofactor by-product SAH. (a): Daigle et al 2011;4 (b): Yao et al 2011;9 (c): this work
Here we demonstrate that bromination of the adenosine ring is sufficient to engineer both potency and selectivity into SAH. The accompanying structural analysis rationalizes this effect, and provides a framework for the development of improved inhibitors.
2. Results
2.1. BrSAH is a potent DOT1L inhibitor
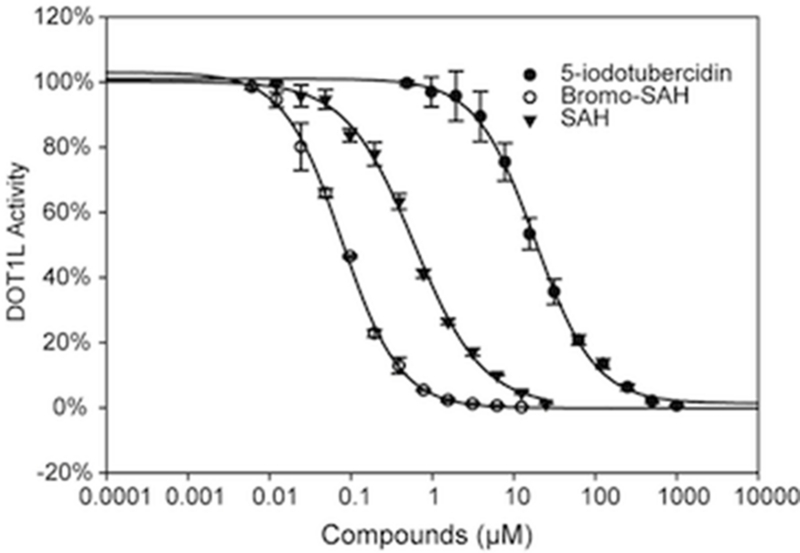
DOT1L is one of three known protein lysine methyltransferases (PKMT) that lack a catalytic SET domain, but are structurally related to protein arginine methyltransferases (PRMTs).10-12 Structural analysis indicates that the SAM binding site of DOT1L is more enclosed and therefore more likely to be druggable than that of other PKMTs.13 Since a common adenosine scaffold is shared by the cofactor of methyltransferases and kinases, we screened a library of 3120 kinase inhibitors, assembled at the Structural Genomics Consortium, by differential scanning fluorimetry (DSF) to find novel SAM-competitive inhibitors of DOT1L1. This screen, and a virtual screen conducted previously (24 compounds selected, 8 compounds in stock and tested experimentally, Supplementary Table 1), identified the kinase inhibitor 5-iodotubercidine (5ITC – Fig. 1) as a potential DOT1L inhibitor.14, 15 We found that 5-ITC stabilized DOT1L with a Tm shift of 2.5 °C at 50 μM. Follow-up methyltransferase assay confirmed that 5-ITC inhibits DOT1L methyltransferase activity with an IC50 value of 18.2 ± 2.1 μM (Fig. 2).
Figure 2. BrSAH is a potent inhibitor of DOT1L.
IC50 values for 5-ITC, SAH and BrSAH were determined (18.4, 0.6 and 0.077 μM respectively) at Km of SAM, using a radioactivity based assay as described in material and methods.
Based on the chemical similarity between SAH and 5ITC, we hypothesized that 5ITC was occupying the portion of the cofactor site centered on the adenosine moiety of SAH. Superimposing 5ITC on the structure of SAH bound to DOT1L (PDB code 3qox) suggested that (1) the iodine atom of 5ITC was occupying a hydrophobic cleft juxtaposed to the adenine ring, and significantly contributed to the interaction, and (2) grafting the homocysteine moiety of SAH on 5ITC would further enhance binding affinity. To test this model, we synthesized N7-Bromo-deaza-S-adenosyl-L-homocysteine (BrSAH), where the iodine present in 5ITC was replaced by a bromine to simplify chemical accessibility (Fig. 1). BrSAH inhibited DOT1L methyltransferase activity with an IC50 value of 77 ± 4 nM (Fig. 2). Recently published SAH analogs were also designed to exploit the hydrophobic cleft juxtaposed to the cofactor adenine ring.9 We note that these compounds, substituted at the primary amine position of the adenine ring (e.g methyl-SAH Fig. 1), are less potent inhibitors than SAH with Ki values more than double that reported for SAH.9 On the other hand, BrSAH is about 8 times more potent than SAH (the IC50 of SAH is 600 ± 4 nM; Fig. 2). We therefore conclude that a bulky hydrophobic group such as an iodine or bromine substituent at the 7 position of the adenine ring better exploits the DOT1L cofactor site.
To verify the binding mode of BrSAH, we solved the DOT1L-BrSAH co-crystal structure at 2.28 Å resolution (Supplementary Table 2), and confirmed the overall similarity to the DOT1L-SAH complex (RMSD of 0.5 Å). The bromine atom is located in a hydrophobic cavity composed of residues F245, V249, L224 and P133 (Fig. 3A and 3B).
Figure 3. Structure of DOT1L-BrSAH complex.
BrSAH is buried in an enclosed pocket, and makes numerous hydrogen-bonds with surrounding polar residues, as previously observed in the DOT1L-SAM complex12 (A,B). The bromine atom (magenta) is occupying a cleft formed by a cluster of hydrophobic side-chains (A,B). Superimposing our structure of BrSAH with cofactor molecules co-crystallized with diverse methyltransferases shows that risks of cross-reactivity are low with SET domain PMTs, and moderate with PRMTs, DNMTs and small-molecule methyltrasferases (C-F. See text for details). PDB codes are SETD7: 1o9s, PRMT3: 2fyt, DNMT1: 3pta, NNMT: 3rod. Molecular surface coloring are green: hydrophobic, red: hydrogen-bond acceptors, blue: hydrogen-bond donor. Sticks color-code is blue: nitrogen, red: oxygen.
2.2. BrSAH is selective for DOT1L
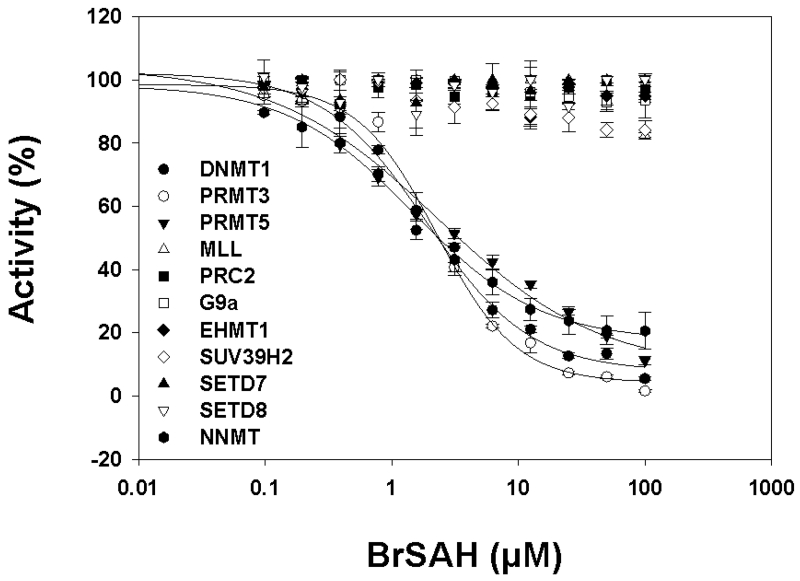
Structural analysis suggests that the bromide of BrSAH confers selectivity towards DOT1L. First, it is located at the N7 position of the adenine ring, a position that is facing the bottom of a binding groove and is engaged in a critical hydrogen-bond in all SET-domain PKMT structures.16 Introduction of a bulky group at this position, such as a bromide, would break this hydrogen-bond, and introduce unacceptable allosteric clashes in all SET domain PKMTs (Fig. 3C). We confirmed experimentally that BrSAH is inactive against the PKMTs MLL, EZH2 (present in the PRC2 complex), EHMT2/G9a, EHMT1/GLP, SUV39H2, SETD7 and SETD8 (Fig. 4 and Table 1).
Figure 4. Selectivity profile of BrSAH across a panel of methyltransferases.
Inhibitory effect of BrSAH on lysine methyltransferases MLL1, EZH2 (PRC2 complex), EHMT2/G9a, GLP/EHMT1, SUV39H2, SETD7, SETD8, and protein arginine methyltransferases PRMT3, PRMT5 as well as DNA-methyltransferase DNMT1 and the small molecule methyltransferase NNMT were assessed as described in materials and methods. BrSAH inhibited NNMT, PRMT3 and PRMT5 with IC50 values of 1.5, 2 and 2 μM respectively.
Table 1.
Selectivity profile of BrSAH across a panel of methyltransferases
| MTase | Ki (μM)* |
|---|---|
| DOT1L | 0.038 ± 0.002 |
| NNMT | 0.75 ± 0.05 |
| DNMT1 | 0.95 ± 0.1 |
| PRMT3 | 1.15 ± 0.1 |
| PRMT5 | 1.15 ± 0.15 |
| MLL | No inhibition |
| PRC2 | No inhibition |
| G9a | No inhibition |
| EHMT1 | No inhibition |
| SUV39H2 | No inhibition |
| SETD7 | No inhibition |
| SETD8 | No inhibition |
IC50 values were determined at cofactor concentration equal to Km value for SAM for each enzyme, and used to calculate Ki values as described by Cer et al.29
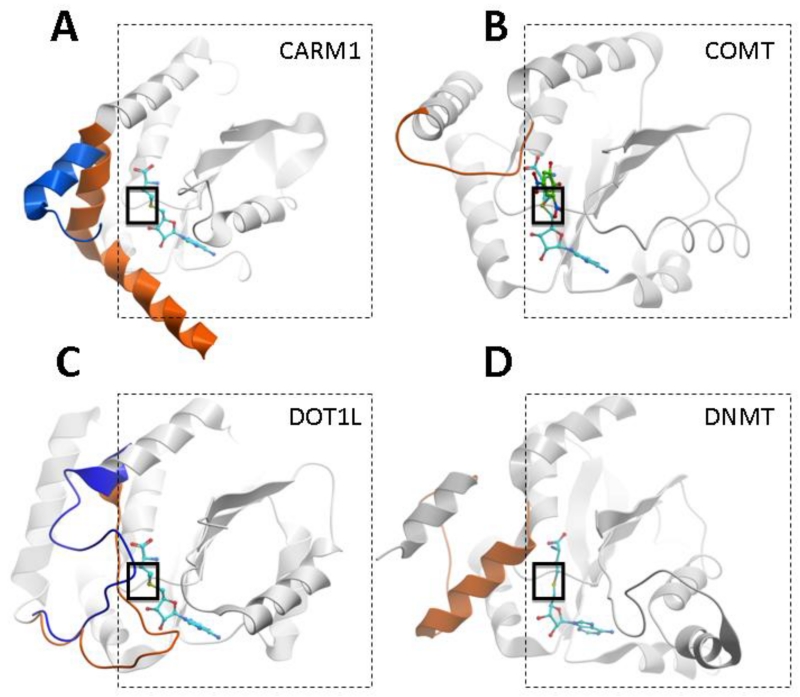
The observed specificity against PKMTs comes as no surprise, but the cofactor binding site of DOT1L, PRMTs, DNA methyltransferases (DNMTs) and small molecule methyltransferases is organized around the same SAM dependent MT-fold,17 and these enzymes share a structurally similar cofactor binding domain (Fig. 5). To further investigate possible cross-reactivity with these classes of enzymes we characterized the effect of BrSAH on the catalytic activities of PRMT3, PRMT5, DNMT1 and a small molecule methyltransferase, nicotinamide-N-methyltransferase (NNMT).
Figure 5. The SAM-dependant MT-fold is shared by non-SET domain methyltransferases.
PRMTs (A), small-molecule methyltransferases (B), DOT1L (C), and DNMTs (D) share a common SAM binding fold (dashed-line square). A structural element variable in length and geometry (orange), immediately upstream of this conserved fold, covers the bound cofactor (cyan), contributes to formation of the site of methyl transfer (solid-line square), and to the recruitment of the methyl-accepting substrate (indicated in green by the co-crystallized substrate in B). In the absence of cofactor (A) or in the absence of the cofactor’s homocysteine end (C), this element can adopt different conformations (blue) that alter the binding pocket of SAM. The specificity of substrate recruitment and accuracy of substrate presentation relies on the nature of this critical and diverse structural element. PDB codes: CARM1: 3b3f (orange), 3b3j (blue), COMT: 1vid, DOT1L; 1nw3 (orange), 3uwp (blue); DNMT: 3pta. (structurally diverse domains not participating in cofactor binding were removed, and DNA substrate is not shown in D, for clarity).
Available structures suggest that BrSAH should have reduced activity against PRMTs. The 7 position of the adenine ring is located at the putative entrance of the cofactor pocket, next to a mobile alpha-X helix that can adopt diverse conformations depending on the presence of cofactor and peptide substrate (Fig. 5A).18 It is therefore likely that small structural rearrangements would be necessary to accommodate the presence of a hydrophobic substituent at this position. However, none of the available PRMT structures present a cluster of hydrophobic residues as observed in DOT1L (Fig. 3D), and the presence of a bromide should come at an energetic cost. We confirmed experimentally that BrSAH was about 25 times less active against PRMT3 and PRMT5 (IC50 = 2.3 μM) than DOT1L (Fig. 4 and Table 1).
Superimposing BrSAH on the structure of SAH in complex with DNMT1 places the bromide across the surface of the cofactor binding pocket, into the core of the protein structure, at a position partially occupied by L1247, P1225, and next to F1145 (Fig. 3E). It is unclear from the structure whether conformational rearrangements of these hydrophobic residues that would be necessary to accommodate the bromide are energetically acceptable. To explore this possibility, we tested the ability of BrSAH to inhibit DNMT1, and determined an IC50 of 1.9 μM (Fig. 4 and Table 1). This suggests that binding of BrSAH to DNMT1 is accompanied by unfavorable conformational strain. Superimposing BrSAH on a cofactor molecule co-crystallized with the small molecule methyltransferase NNMT highlights moderate steric clashes with A169 that may be accommodated by the enzyme at an energetic cost (Fig. 3F). We verified that BrSAH inhibits NNMT with a moderate IC50 of 1.5 μM (Fig. 4 and Table 1).
Finally, even though the design of BrSAH was derived from 5ITC, a potent inhibitor of the protein kinase haspin and adenosine kinase,14, 15 addition of the homocysteine tail is likely to preclude binding at ATP binding sites, and BrSAH is expected to be inactive against kinases. We confirmed that no binding to the haspin catalytic domain was observed with 10 μM BrSAH by differential scanning fluorimetry (see material and methods).
3. Discussion
We have established that BrSAH is a novel, potent and selective DOT1L inhibitor. We note here that we were unable to find cellular activity reported for any close SAH analog against any methyltransferase, even though biochemically active compounds were reported in the literature.9, 19 We attribute this liability - an apparently general phenomenon - to the high polarity of SAH analogs, which prevents crossing cell membranes. The discovery of EPZ004777 as a potent SAM competitor teaches us an important lesson: DOT1L tolerates significant chemical modifications at the amino-acid end of the cofactor (Fig. 1). Together, these observations outline a strategy towards the development of improved SAM-competitive inhibitors, where (1) addition of a bulky hydrophobic group such as a bromide or tri-fluoromethyl at the N7 position of the adenine ring enhances potency, increases selectivity, and reduces hydrophilicity of the adenosine moiety of SAH and (2) chemical modifications at the homocysteine, such as the one observed in EPZ004777, increase hydrophobicity at the amino-acid end to further improve cell penetrance.
Available structures clearly indicate the presence of a strong electrostatic potential at the amino-acid end of the native cofactor pocket that can only be satisfied with polar, and poorly bioavailable molecules. Overcoming this paradigm shared by SAM mimetics appears particularly challenging. Yet, EPZ004777, a hydrophobic SAM competitor that lacks the amino-acid moiety of the cofactor, contradicts this paradigm, suggesting that the cofactor pocket may adopt an altered conformation in the absence of the cofactor’s homocysteine moiety. The recent crystal structures of DOT1L in complex with 5-ITC and EPZ004777 confirm this hypothesis (PDB codes 3uwp and 4er3/4eki respectively): a large loop immediately N-terminal to the conserved methyltransferase fold closes like a lid on the cofactor (PDB code 1nw3), but adopts dramatically altered conformations in the 5-ITC and EPZ004777 complexes (Figure 5C)6,7. This loop motion significantly alters the size and electrostatics of the cofactor pocket, which may be sufficient to accommodate more hydrophobic and cell penetrant SAM analogs. Interestingly, this structural rearrangement observed in the DOT1L structure is reminiscent of another structural rearrangement reported for PRMTs: the alpha-X helix of CARM1, positioned at the N-terminus of the conserved methyltransferase fold, closes like a lid on the bound cofactor, but in the absence of SAM or SAH, this helix is partially disordered, which again alters the geometry and electrostatics of the cofactor pocket (Fig. 5A).18 Cell-penetrant SAM analogs that exploit this altered geometry may be able to inhibit CARM1 or other PRMTs.
A sequential mechanism of substrate binding has been proposed for SET-domain HKMTs, whereby cofactor binding is necessary for proper folding of the Post-SET domain and formation of the substrate binding groove.16 A striking parallel can be made for PRMTs and DOT1L, where cofactor binding stabilizes the catalytically competent conformation of the secondary element (alpha-helix in PRMTs, activation loop in DOT1L) immediately adjacent to the conserved methyltransferase fold. Whether this similarity in structural mechanism is driven by related evolutionary pressures remains an open question.
4. Conclusion
We have shown here that addition of a single halogen atom can dramatically increase the potency of DOT1L inhibitors, and improve their selectivity versus other lysine, arginine, DNA and small molecule methyltransferases. The structural mechanism underlying these improved properties was dissected. This discovery, combined with chemical modifications at the homocystein end, provide a framework for future developments of DOT1L inhibitors.
5 Materials and Methods
5.1. Protein expression and purification
A construct of human DOT1L covering residues 1-420 was subcloned into a modified pET28-MHL vector with an N-terminal His tag. The protein was overexpressed in E.coli BL21 (DE3) V2R-pRARE in Terrific Broth medium in the presence of 50 μg/ml of kanamycin and chloramphenicol. Cells were grown at 37°C to an OD600 of 1.5, induced by isopropyl-1-thio-D-galactopyranoside (IPTG, final concentration 1 mM) and incubated overnight at 15°C. The cell pellets were frozen in liquid nitrogen and stored at −80°C. For purification, the cell paste was thawed and resuspended in lysis buffer with 1mM phenylmethyl sulfonyl fluoride (PMSF). DOT1L (1-420) was purified by Ni-NTA column (Qiagen) and processed by TEV protease to remove the His tag. The protein was then incubated in 50 mM Tris-HCl pH 8.0, 0.1 mg/ml BSA, 1 mM MgCl2 with benzonase nuclease for 2 hours at room temperature. Filtered protein sample was diluted with 50 mMK2HPO4/ KH2PO4 pH 7.0, and further purified by HiTrap-SP (GE Healthcare). The protein was further purified by gel filtration (Superdex 200, GE Healthcare).
5.2. Virtual screening
Receptor preparation
The DOT1L-SAM complex structure (PDB code 1nw3) was used. The receptor was prepared with Maestro Protein Preparation Wizard (Schrodinger, NY) using default settings. One important structural water molecule W1025 was retained and included during docking. Protonation states were set at pH 7.4 using Epik. H-bond assignment was optimized by Protassign, including exhaustive sampling and minimization of hydrogens of altered species at neutral pH. Water orientation was also sampled for the conserved water molecule. The receptor was refined during Impref minimization with RMSD=0.3 Angstrom set for heavy atoms convergence under OPLS2005 forcefield. A receptor grid was centered on bound SAM. All hydroxyl groups accessible from the cofactor binding site were set rotatable.
Chemical library
the ZINC clean-drug-like set (downloaded on January 13, 2011) containing 3.7 million commercially available compounds was used as ligand library for docking. The library was prepared with LigPrep (Schrodinger, NY) during which protonation states of all ligands were set at pH=7.4 using Epik.
Docking
The virtual screening was implemented in two consecutive steps using Schrodinger Glide Virtual Screening Workflow. Initially, the whole 3.7-million-compound library was docked into the receptor using Glide HTVS mode. The resulting top 100,000 ligands were retained for docking with Glide SP where six hydrogen bond constraints were set based on previous analysis.13: N of F223, N of K187, OE2 of E186, O of G163, O of water W1025, and O of D161. The top 777 compounds satisfying at least 4 out of the six constraints were retained and clustered based on their chemical similarity. Twelve representatives of each cluster and 12 SAM analogs occupying a previously identified hydrophobic cleft at Val249, including 5-ITC (docking score: −9.54), were selected for experimental validation.
5.3. Differential scanning fluorimetry screening
Thermal denaturation experiments were carried out in a Mx3005p real-time PCR machine (Agilent) using a protein concentration of 2 μM and an inhibitor concentration of 10 μM. Samples were buffered in 10 mM HEPES, pH 7.5, 500 mM NaCl and a 1:1000 dilution of Sypro Orange (Invitrogen, CA). The assay and data evaluation were carried out as previously described.20
5.4. DOT1L methyltransferase assay
The assay conditions reported by Daigle et al were used with minor modifications.4 Nucleosome purified from chicken blood cells was used as substrate. Nucleosome (final concentration of 60 nM) was added into 20 μl assay mixture (20 mM Tris-HCl, 5 mM DTT, 10 mM MgCl2, 0.01% Triton X-100, 0.25 nM recombinant DOT1L (1-420) with inhibitors at different concentrations or DMSO as a control). The mixture was incubated at room temperature for 30 minutes before 3H-SAM (PerkinElmer, catalog number NET155001MC) was added to start the reaction (final concentration of 0.75 μM). Reaction mixture was incubated at room temperature for one hour and quenched by adding 160 μl of 10% trichloroacetic acid (TCA). The mixture was transferred into a glass fiber filter plate (Millipore, catalog number MSFBN6B) and washed twice with 10% TCA and once with ethanol. 100 μl Microscint Zero (PerkinElmer, catalog number 6013611) was finally added into filter plates and centrifuged to flow through filters. Tritium incorporation was monitored using TopCount (PerkinElmer).
5.5. Crystallization
Purified protein DOT1L (1-420) was concentrated to 16 mg/mL in a buffer containing 20 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, and 1 mM TCEP. To prepare the complex, DOT1L (1-420) was mixed with BrSAH by directly adding a 5 fold excess of compound to the protein solution and crystallized by using the sitting drop vapor diffusion method at 18 °C. Crystals of DOT1L with BrSAH were grown in conditions containing 1.5 M (NH4)2SO4, 100 mM sodium acetate, pH 4.6. Prior to flash-freezing the crystals in liquid nitrogen, the crystals were soaked in reservoir solution with added glycerol (20%) as a cryoprotectant.
5.6. Diffraction data collection and model refinement
Diffraction data were collected at GM/CA collaborative access team (GMCA-CAT) beamline 23ID-D of the Advanced Photon Source, Argonne National Laboratory, at a wavelength of 1.0332 Å. For the refinement data set, intensities from one hundred consecutive single-degree oscillation frames were integrated with DENZO and scaled with SCALEPACK.21 Amplitudes were derived with the program TRUNCATE.22 The unit cell was isomorphous to Protein Data Bank (PDB) entry 3QOW8 which provided initial coordinates for refinement, as well as flags for cross-validation.23 Geometry restraints for BrSAH were calculated with PRODRG.24 The current model was deposited in the PDB after iterative geometry validation on the MOLPROBITY server,25 re-building with COOT26 and refinement with REFMAC.27
5.7. Selectivity assays
Selectivity assays for histone methyltransferases (HMTs) were performed as described by Siarheyeva et al.28 Selectivity experiments for compounds using different HMTs (G9a, EHMT1, SUV39H2, PRMT3, SETD7, SETD8, and PRMT5-MEP50, PRC2 and MLL1 complexes) were performed by monitoring the incorporation of tritium-labeled methyl group to lysine or arginine residues of peptide substrates using scintillation proximity assay (SPA). The enzymatic reactions were conducted in triplicate at room temperature with 1 hour incubation of a 20 μl reaction mixture in 20 mM Tris-HCl, pH 8.0, 5 mM DTT, and 0.01% Triton X-100 containing 3H-SAM (Cat.# NET155V250UC; Perkin Elmer), around Km value for each enzyme (G9a, 8μM; EHMT1, 8μM; SUV39H2, 3 μM; PRMT3, 14 μM; SETD7, 2 μM; SETD8, 60 μM; PRMT5-MEP50, 2 μM; PRC2, 2 μM and MLL1, 2 μM). Protein concentration in the assay for each HMT was as follows: G9a (5 nM), EHMT1 (5 nM), SUV39H2 (20 nM), PRMT3 (100 nM), SETD7 (20 nM), SETD8 (50 nM), PRMT5-MEP50 (15 nM), PRC2 (20 nM), and MLL1 (500 nM). Corresponding biotinylated peptides (H3 (1-25) for G9a, EHMT1, SUV39H2, SETD7 and MLL1 complex; H3 (21-44) for PRC2 complex; H4 (1-24) for PRMT3 and SETD8) were used at about five times the Km value for each enzyme (G9a, 0.8 μM; EHMT1, 0.6 μM; SUV39H2, 0.5 μM; PRMT3, 0.6 μM; SETD7, 2μM; SETD8, 40 μM; PRMT5-MEP50, 120 nM; PRC2 complex, 1 μM; and MLL1 complex, 2 μM). IC50 values were determined using compound concentrations ranging from 100 nM to 100 μM. Since all assays were performed at cofactor concentration equal to Km values for SAM, Ki values were calculated as IC50/2 as described by Cer et al.29 To stop the enzymatic reactions, 20 μl of 7.5 M Guanidine hydrochloride was added, followed by 180 μl of buffer (20 mM Tris, pH 8.0), mixed and then transferred to a 96-well Flashplate (Cat.# SMP103; Perkin Elmer). After mixing, the reaction mixtures in Flashplate were incubated for 1 hour and the CPM counts were measured using Topcount plate reader (Perkin Elmer). The CPM counts in the absence of compound for each data set were defined as 100% activity. The CPM counts in the absence of the enzyme in each data set were defined as background (0%). IC50 values were calculated with the software SigmaPlot.
For DNMT1, the IC50 determination was performed as described above by using dsDNA as a substrate. The dsDNA substrate was prepared by annealing two complementary strands (biotinylated forward strand: B-GAGCCCGTAAGCCCGTTCAGGTCG and reverse strand: CGACCTGAACGGGCTTACGGGCTC), synthesized by Eurofins MWG Operon. DNMT1 (100 nM) was incubated with dsDNA substrate (100 nM), 3H-SAM (2 μM, ~ 60 μCi/ml) in buffer (20 mM Tris-HCl, pH 8.0, 5 mM DTT, 0.01% Triton X-100) and the reactions were incubated for 1 hour at 37 °C before quenching.
Inhibition of methyltransferase activity of recombinant full-length human NNMT was measured using SAHH-coupled assay30. The reaction mixture (15 μL) consisted of 50 mM Tris-HCl, pH 7.5, 5 μM SAM, 150 μM nicotinamide, 5 μM SAH hydrolase, 15 μM ThioGlo 1 (Calbiochem), 0.5 μM NNMT and variable amount of BrSAH ranging between 0.1 and 100 μM. The methylation reaction was followed by measuring the increase in fluorescence using Synergy 4 microplate reader (BioTek) with 360/40 nm excitation filter and 528/20 nm emission filter for 30 min in 384-well plate format. Reaction rates were calculated from the initial linear slopes using Gen5 Software (BioTek). Data was converted to percent activity, assuming 100% activity in the absence of BrSAH. IC50 values were calculated using a four parameter logistic equation using Sigma Plot software (Systat Software). Standard deviations were calculated from three independent experiments.
5.8. Chemistry
All oxygen and/or moisture sensitive reactions were carried out under N2 atmosphere in glassware purged with N2 prior to use. All reagents and laboratory grade solvents were purchased from commercial vendors and used as received, without further purification. Nuclear Magnetic Resonance (NMR) spectra were recorded on a Bruker Avance-III 500 MHz spectrometer (500 MHz 1H, 125 MHz 13C). Proton chemical shifts are reported in ppm (δ) referenced to the NMR solvent; DMSO-d6 (2.50 ppm for 1H) or D2O (4.79 ppm for 1H). Data are reported as follows: chemical shifts (δ), multiplicity (br = broad, s = singlet, d = doublet, t = triplet, q = quartet, quint = quintuplet, m = multiplet); coupling constant(s) (J) in Hz; integration; assignment. Unless otherwise noted, NMR data were collected at 25 °C. Flash column chromatography was performed using a Biotage SP1 system fitted with a KP-SIL SNAP Silica Gel (60 Å mesh) Flash Cartridge (FSKO-1107). Purity determination was conducted by UV absorbance at 254 nm during Tandem Liquid Chromatography/Mass Spectrometry (LCMS) using a Waters Acquity separations module. Identity was determined via low-resolution mass spectra (LRMS) acquired in positive ion mode using a Waters Acquity SQD mass spectrometer (electrospray ionization source) fitted with a PDA detector. Mobile phase A consisted of 0.01% formic acid in water, while mobile phase B consisted of 0.01% formic acid in acetonitrile. The gradient ran from 5% to 95% mobile phase B over 5 minutes at 0.7 mL/min. An Acquity CSH C18, 1.7 μm, 2.1 × 50 mm column was used with column temperature maintained at 25 °C. The sample solution injection volume was 5 μL. Analytical thin-layer chromatography (TLC) was performed on aluminum sheets, silica gel 60 F254 (0.2 mm, VWR International, Darmstadt, Germany). Visualization was accomplished with UV light and aqueous potassium permanganate (KMnO4) stain followed by heating.
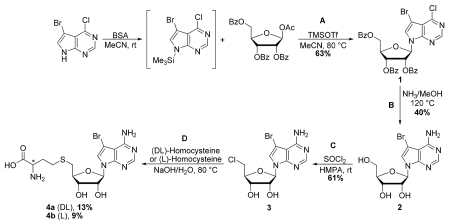
Steps A and B are adapted from Seela et al.31 Additional information relating to the characterization of reported compounds (1) and (2) can be found therein.
Steps C and D are adapted from Lim et al.32
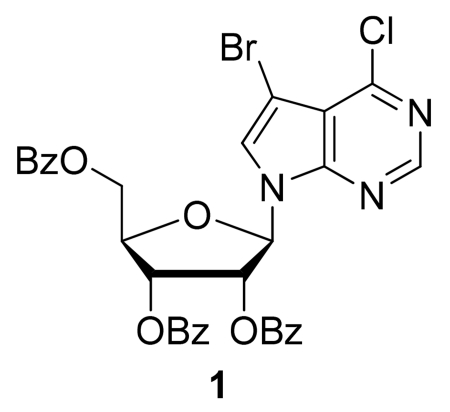
Step A: (2R,3R,4R,5R)-2-(Benzoyloxymethyl)-5-(5-bromo-4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)tetra-hydrofuran-3,4-diyl dibenzoate (1)
N,O-Bis(trimethylsilyl)acetamide (BSA, 0.56 mL, 2.27 mmol) was added to a stirred suspension of 5-bromo-4-chloro-7H-pyrrolo[2,3-d]pyrimidine (480 mg, 2.06 mmol) in dry acetonitrile (15 mL). After stirring at rt for 10 min, 1-O-acetyl-2,3,5-tri-O-benzoyl-β-D-ribofuranose (1.16 g, 2.31 mmol) was added, followed by the addition of trimethylsilyl trifluoromethanesulfonate (0.41 mL, 2.25 mmol). The reaction mixture was stirred at rt for 15 min, after which the flask was transferred to a preheated 80 °C oil bath. After stirring for 1 h at 80 °C, the reaction mixture was cooled to rt and diluted with EtOAc (75 mL). The organic phase was sequentially washed with aqueous saturated NaHCO3 solution and brine, dried (Na2SO4), filtered, and concentrated under reduced pressure to afford the crude nucleoside. Purification by flash column chromatography (silica gel, 20-100% EtOAc/hexanes gradient) provided (1) as a colorless foam (870 mg, 63%). TLC (silica gel, hexanes/ethyl acetate 3:1): Rf = 0.45. LRMS (ESI): 677.3, 675.2 and 673.2 [M+H]+.
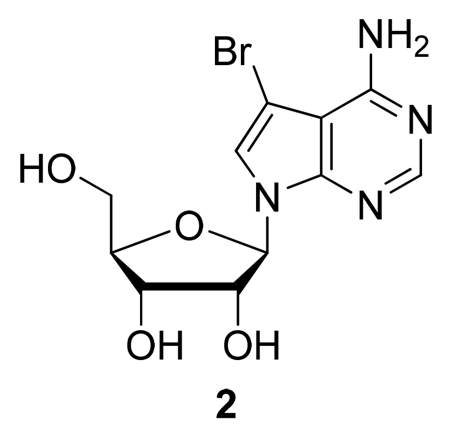
Step B: (2R,3R,4R,5R)-2-(4-Amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(hydroxymethyl)tetra-hydrofuran-3,4-diol (2)
A solution of (2R,3R,4R,5R)-2-(Benzoyloxymethyl)-5-(5-bromo-4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)tetrahydrofuran-3,4-diyl dibenzoate (1) (870 mg, 1.29 mmol) in methanolic ammonia (7 N, 55 mL) was heated at 120 °C in a screw-cap pressure flask for 2 h. The mixture was then cooled to rt, and the solvent evaporated under reduced pressure to afford the crude nucleoside. Purification by flash column chromatography (silica gel, 5-15% MeOH/CH2Cl2 gradient) provided (2) as an off-white solid (179 mg, 40%). TLC (silica gel, CH2Cl2/MeOH 6:1): Rf = 0.35. 1H NMR (500 MHz, DMSO-d6) δ: 8.10 (s, 1H, 6-H); 7.66 (s, 1H, 2-H); 6.81 (br s, 2H, 4-NH2); 6.05 (d, J = 6.0 Hz, 1H, 1′-H); 5.32 (d, J = 6.2 Hz, 1H, 2′-OH); 5.15 (d, J = 5.4 Hz, 1H, 3′-OH); 5.11 (d, J = 4.7 Hz, 1H, 5′-OH); 4.35 (m, 1H, 2′-H); 4.07 (m, 1H, 3′-H); 3.89 (m, 1H, 4′-H); 3.62 (m, 1H, 5′-H); 3.54 (m, 1H, 5′-H). LRMS (ESI): 346.2 and 344.4 [M+H]+.
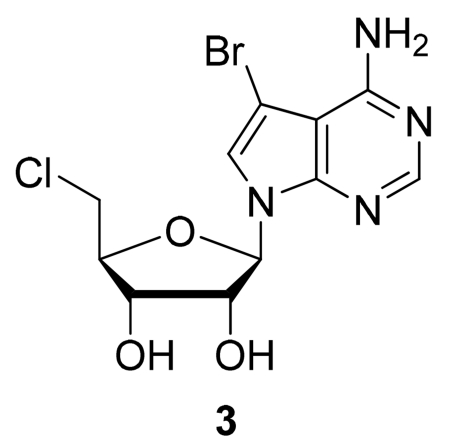
Step C: (2R,3R,4R,5R)-2-(4-Amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(chloromethyl)tetra-hydrofuran-3,4-diol (3)
A solution of (2R,3R,4R,5R)-2-(4-Amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (2) (160 mg, 0.463 mmol) in hexamethylphosphoramide (2 mL) was cooled to 0 °C. Thionyl chloride (0.202 mL, 2.78 mmol) was added dropwise with stirring, and after 10 min, the solution was warmed to rt and stirred a further 18 h. Following dilution with water (5 mL) and adjustment to pH = 8 with aqueous NaOH solution (1 M), the aqueous layer was extracted with EtOAc (3 × 5 mL). The combined organic extracts were dried (Na2SO4), filtered, and concentrated under reduced pressure to afford the crude nucleoside. This material was then dissolved in CH2Cl2 and triturated with hexanes to produce a precipitate. The precipitate was collected by filtration, providing pure (3) as an off-white solid (102 mg, 61%). TLC (silica gel, CH2Cl2/MeOH 6:1): Rf = 0.45. LRMS (ESI): 362.1, 364.0 and 365.9 [M+H]+ .
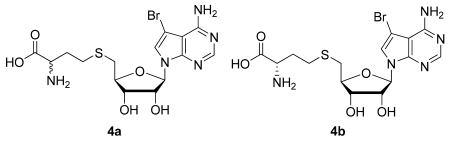
Step D: (DL)- and (L)-2-Amino-4-(((2S,3S,4R,5R)-5-(4-amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxy-tetrahydrofuran-2-yl)methylthio)butanoic acid (4a and 4b)
To a solution of DL- or L-homocysteine (26 mg, 0.19 mmol) in 1 N NaOH/H2O (0.60 mL) was added (2R,3R,4R,5R)-2-(4-amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-5-(chloromethyl)tetrahydrofuran-3,4-diol (3) (50 mg, 0.14 mmol) and KI (10 mg). The reaction mixture was stirred vigorously at 80 °C for 2 h, cooled to rt, and applied to a column (1 × 15 cm) of Amberlite XAD-4 ion exchange resin. The column was washed [0.2 N NH4OAc (30 mL); H2O (20 mL)] and eluted [H2O (20 mL); ethanol (30 mL)] to afford 4a or 4b as an off-white solid (8 mg, 13% for 4a and 6 mg, 9% for 4b). 1H NMR (500 MHz, D2O) δ; 7.89 (s, 1H, 6-H); 7.25 (s, 1H, 2-H); 6.01 (m, 1H, 1′-H); 4.75 (m, 1H, 2′-H); 4.45 (m, 1H, 3′-H); 4.22 (m, 1H, α-H); 4.19 (m, 1H, 4′-H); 3.70 (m, 2H, 5′-H); 2.60 (br t, 2H, γ-H); 2.11 (m, 2H, β-H). LRMS (ESI): 463.0 and 461.1 [M+H]+.
Supplementary Material
Acknowledgements
We thank Dr. Amy K. Wernimont for her review of the crystal structure and Dr. Ekaterina Kuznetsova for providing chicken nucleosome and NNMT assay. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from Canadian Institutes of Health Research, Eli Lilly Canada, Genome Canada, GlaxoSmithKline, the Ontario Ministry of Economic Development and Innovation, the Novartis Research Foundation, Pfizer, AbbVie, Takeda and the Wellcome Trust. C.H.A. holds a Canada Research Chair in Structural Genomics. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357
Footnotes
Accession codes: The coordinates and structure factors for human DOT1L in complex with BrSAH have been deposited in the Protein Data Bank (PDB) with accession number 3sx0
Supplementary data associated with this article can be found in the online version
REFERENCES
- 1.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, Pollock RM, Richon VM, Kung AL, Armstrong SA. Cancer Cell. 2011;20:66. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muntean AG, Hess JL. Annu Rev Pathol. 2012;7:283. doi: 10.1146/annurev-pathol-011811-132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nguyen AT, Taranova O, He J, Zhang Y. Blood. 2011;117:6912. doi: 10.1182/blood-2011-02-334359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, Jin L, Kuntz KW, Chesworth R, Moyer MP, Bernt KM, Tseng JC, Kung AL, Armstrong SA, Copeland RA, Richon VM, Pollock RM. Cancer Cell. 2011;20:53. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anglin JL, Deng L, Yao Y, Cai G, Liu Z, Jiang H, Cheng G, Chen P, Dong S, Song Y. J Med Chem. 2102;55:8066. doi: 10.1021/jm300917h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basavapathruni A, Jin L, Daigle SR, Majer CR, Therkelsen CA, Wigle TJ, Kuntz KW, Chesworth R, Pollock RM, Scott MP, Moyer MP, Richon VM, Copeland RA, Olhava EJ. Chem Biol Drug Des. 2012;80:971. doi: 10.1111/cbdd.12050. [DOI] [PubMed] [Google Scholar]
- 7.Yu W, Chory EJ, Wernimont AK, Tempel W, Scopton A, Federation A, Marineau JJ, Qi J, Barsyte-Lovejoy D, Yi J, Marcellus R, Iacob RE, Engen JR, Griffin C, Aman A, Wienholds E, Li F, Pineda J, Estiu G, Shatseva T, Hajian T, Al-Awar R, Dick JE, Vedadi M, Brown PJ, Arrowsmith CH, Bradner JE, Schapira M. Nat Commun. 2012;3:1288. doi: 10.1038/ncomms2304. [DOI] [PubMed] [Google Scholar]
- 8.Richon VM, Johnston D, Sneeringer CJ, Jin L, Majer CR, Elliston K, Jerva LF, Scott MP, Copeland RA. Chemical Biology & Drug Design. 2011;78:199. doi: 10.1111/j.1747-0285.2011.01135.x. [DOI] [PubMed] [Google Scholar]
- 9.Yao Y, Chen P, Diao J, Cheng G, Deng L, Anglin JL, Prasad BV, Song Y. J Am Chem Soc. 2011;133:16746. doi: 10.1021/ja206312b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kernstock S, Davydova E, Jakobsson M, Moen A, Pettersen S, Maelandsmo GM, Egge-Jacobsen W, Falnes PO. Nat Commun. 2012;3:1038. doi: 10.1038/ncomms2041. [DOI] [PubMed] [Google Scholar]
- 11.Magnani R, Dirk LM, Trievel RC, Houtz RL. Nat Commun. 2010;1:43. doi: 10.1038/ncomms1044. [DOI] [PubMed] [Google Scholar]
- 12.Min J, Feng Q, Li Z, Zhang Y, Xu RM. Cell. 2003;112:711. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 13.Campagna-Slater V, Mok MW, Nguyen KT, Feher M, Najmanovich R, Schapira M. J Chem Inf Model. 2011;51:612. doi: 10.1021/ci100479z. [DOI] [PubMed] [Google Scholar]
- 14.Eswaran J, Patnaik D, Filippakopoulos P, Wang F, Stein RL, Murray JW, Higgins JM, Knapp S. Proc Natl Acad Sci U S A. 2009;106:20198. doi: 10.1073/pnas.0901989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ugarkar BG, DaRe JM, Kopcho JJ, Browne CE, 3rd, Schanzer JM, Wiesner JB, Erion MD. J Med Chem. 2000;43:2883. doi: 10.1021/jm000024g. [DOI] [PubMed] [Google Scholar]
- 16.Schapira M. Curr Chem Genomics. 2011;5:85. doi: 10.2174/1875397301005010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin JL, McMillan FM. Curr Opin Struct Biol. 2002;12:783. doi: 10.1016/s0959-440x(02)00391-3. [DOI] [PubMed] [Google Scholar]
- 18.Troffer-Charlier N, Cura V, Hassenboehler P, Moras D, Cavarelli J. EMBO J. 2007;26:4391. doi: 10.1038/sj.emboj.7601855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dowden J, Hong W, Parry RV, Pike RA, Ward SG. Bioorg Med Chem Lett. 2010;20:2103. doi: 10.1016/j.bmcl.2010.02.069. [DOI] [PubMed] [Google Scholar]
- 20.Niesen FH, Berglund H, Vedadi M. Nat Protoc. 2007;2:2212. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. Macromolecular Crystallography, Pt A. 1997;276:307. [Google Scholar]
- 22.French S, Wilson K. Acta Crystallographica Section A. 1978;34:517. [Google Scholar]
- 23.Brunger AT. Nature. 1992;355:472. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 24.Schuttelkopf AW, van Aalten DMF. Acta Crystallographica Section D-Biological Crystallography. 2004;60:1355. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 25.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. Acta Crystallographica Section D-Biological Crystallography. 2010;66:12. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta Crystallographica Section D-Biological Crystallography. 2010;66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. Acta Crystallographica Section D-Biological Crystallography. 2011;67:355. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siarheyeva A, Senisterra G, Allali-Hassani A, Dong A, Dobrovetsky E, Wasney GA, Chau I, Marcellus R, Hajian T, Liu F, Korboukh I, Smil D, Bolshan Y, Min J, Wu H, Zeng H, Loppnau P, Poda G, Griffin C, Aman A, Brown PJ, Jin J, Al-Awar R, Arrowsmith CH, Schapira M, Vedadi M. Structure. 2012;20:1425. doi: 10.1016/j.str.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Cer RZ, Mudunuri U, Stephens R, Lebeda FJ. Nucleic Acids Res. 2009;37:W441. doi: 10.1093/nar/gkp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allali-Hassani A, Wasney GA, Siarheyeva A, Hajian T, Arrowsmith CH, Vedadi M. J Biomol Screen. 2011;17:71. doi: 10.1177/1087057111422256. [DOI] [PubMed] [Google Scholar]
- 31.Seela F, Ming X. Tetrahedron. 2007;63:9850. [Google Scholar]
- 32.Lim J, Winkler WC, Nakamura S, Scott V, Breaker RR. Angew Chem Int Ed Engl. 2006;45:964. doi: 10.1002/anie.200503198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.