Abstract
G-quadruplexes formed on DNA and RNA can be roadblocks to movement of polymerases and ribosome on template nucleotides. Although folding and unfolding processes of the G-quadruplexes are deliberately studied in vitro, how the mechanical and physical properties of the G-quadruplexes affect intracellular biological systems is still unclear. In this study, mRNAs with G-quadruplex forming sequences located either in the 5′ untranslated region (UTR) or in the open reading frame (ORF) were constructed to evaluate positional effects of the G-quadruplex on translation suppression in cells. Periodic fluctuation of translation suppression was observed at every three nucleotides within the ORF but not within the 5′ UTR. The results suggested that difference in motion of ribosome at the 5′ UTR and the ORF determined the ability of the G-quadruplex structure to act as a roadblock to translation in cells and provided mechanical insights into ribosomal progression to overcome the roadblock.
Guanine-rich (G-rich) sequences can form stable non-canonical structures called G-quadruplexes under physiological conditions1,2. Recent biological studies have revealed that both DNA and RNA G-quadruplexes form inside living cells3,4,5. Thus, this unique structure likely has a role in regulation of gene expression6,7.
DNA polymerase, RNA polymerase, reverse transcriptase, and ribosome, which show processive movement on template nucleotides, have fundamental functions for replication and expression of genomic information. Progression of all these proteins is known to be disturbed when G-quadruplex structure was formed on the template strand6,8,9,10,11. The proteins should unwind the G-quadruplex to continue their reactions. In vitro biophysical12,13,14 and mechanical15,16,17 analyses, and in silico simulations18,19,20 of folding and unfolding processes of the G-quadruplexes have suggested that the G-quadruplex unfolds through intermediate structures of partially folded quadruplex or G-triplex. Recent mechanical disruption studies done by Mao et al. demonstrated that the unfolding process of G-quadruplex differed depending on the positions, where the attracting force was applied, and resulted in changes in stability and unfolding kinetics of the G-quadruplex21,22. Based on the characteristic properties, progression of the polymerases and ribosome is expected to be differently affected by the G-quadruplex depending on their mechanisms to overcome the G-quadruplex region, although it is still challenging to correlate with intracellular biological reactions affected by the G-quadruplex.
G-quadruplexes located in both the 5′ untranslated region (UTR) and the open reading frame (ORF) of mRNAs reduce protein expression levels by acting as roadblocks to the ribosome6,23,24,25,26. From a mechanical viewpoint, only the small ribosomal subunit interacts with 5′ UTR and scans the region until a start codon is recognized, whereas the mature ribosome consisting of small and large subunits moves along the ORF by step-by-step translocation, which shows ratchet-like movement27,28,29,30. The mechanical driving forces of the small ribosomal sub unit and the mature ribosome are different31,32. In addition, since the mature ribosome progresses three nucleotides at an every step of translocation, there can be zero, one, or two nucleotides between the mRNA entry site of ribosome and the G-quadruplex when the mature ribosome approaches a G-quadruplex within the ORF. Thus, the ability of the G-quadruplex to block translation likely differs depending not only on whether it is located in the 5′ UTR or the ORF but also on its positions within ORF. However, little is known about positional effects of G-quadruplexes or of other RNA structures, such as pseudoknots and hairpins, that have suppressive effects on translation33,34.
In this study, we systematically evaluated the positional effect of a G-quadruplex forming sequence on the suppression of translation in cells. Reporter mRNAs, which form RNA G-quadruplex in the 5′ UTR or the ORF with stepwise displacement of their positions at single nucleotide levels, were designed. Based on a periodic fluctuation of translation suppression caused by RNA G-quadruplex within the ORF but not within the 5′ UTR, we propose mechanisms of single-step or two-step translocation to unwind the G-quadruplex during the translation elongation depending on the position of the G-quadruplex.
Results
Formation of G-quadruplex on reporter mRNA
Reporter mRNAs were constructed using a G-rich sequence derived from E. coli eutE gene; the reporter is based on a previously constructed reporter gene designed to express Renilla luciferase with a T7 tag at the N-terminus26. Mutant G-rich sequences with replacements of the guanine bases with adenine to prevent G-quadruplex formation were also constructed. Wild-type and mutant G-rich sequences were placed upstream or downstream of the start codon (Fig. 1a). Reporters containing wild-type G-rich sequences were created with stepwise displacement of the G-quadruplex forming sequence by a single nucleotide relative to the wt + 0 construct such that the G-quadruplex forming sequence was placed at seven different positions. This allowed us to evaluate the positional effect of the G-quadruplex with single-nucleotide resolution in both the 5′ UTR and the ORF (Fig. 1b).
Figure 1. Design of reporter mRNAs.
(a) A G-rich sequence derived from E. coli eutE gene or a variant was inserted into the 5′ UTR or the ORF of an mRNA encoding T7-tagged Renilla luciferase. (b) Sequences of G-rich sequence variants. Wild-type and mutant G-rich sequence regions are underlined. Positions of substitutions from guanine to adenine in the mutant sequence are in italics. Amino acid sequences encoded by the G-rich sequences within the ORF are shown under the nucleotide sequences.
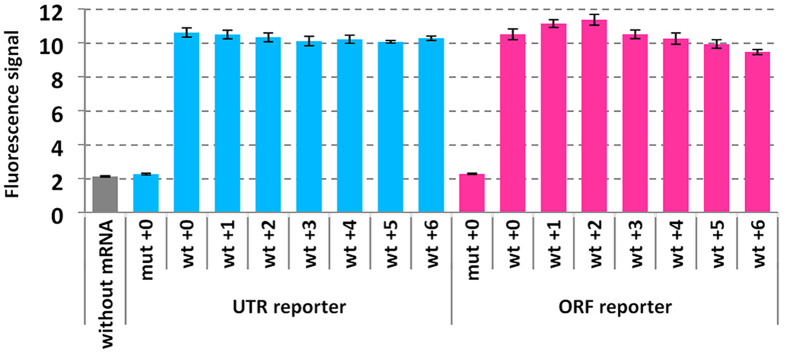
The reporter mRNAs were transcribed and formation of G-quadruplex on the mRNAs was evaluated by analysis of the fluorescence signal of N-methyl mesoporphyrin (NMM). NMM fluorescence is enhanced upon binding to a G-quadruplex35,36. Fluorescence signals of NMM with reporter mRNAs containing the wild-type G-rich sequence were higher than those without mRNA and with mRNAs containing mutant sequence irrespective of position of the G-rich sequence (Fig. 2). The signal intensities among the wild-type mRNAs were almost the same in the presence of 10-fold excess of NMM. In our previous study, a short RNA oligonucleotide corresponding to the wild-type G-rich sequence formed parallel G-quadruplex26. The flanking sequences of the wild-type G-rich sequence that may affect G-quadruplex stability and topology37,38 are designed to be all the same with 5′ GCC and 3′ AA. Thus, it was suggested that the wild-type G-rich sequences in the reporter mRNAs uniformly formed parallel G-quadruplex.
Figure 2. Analysis of NMM fluorescence in the presence of reporter mRNAs.
Reporter mRNAs (500 nM) were mixed with NMM (5 μM) in a buffer containing 30 mM HEPES-KOH (pH 6.8), 100 mM KCl, 0.1% DMSO, and 0.01% Tween 20. Florescence intensities were measured after incubation at 37 °C for 30 min using 400 nm excitation and 615 nm emission. mRNAs were purified and refolded by heating at 70 °C for 5 min and cooling to 25 °C at 1 °C min−1 before addition of NMM.
Intracellular translation suppression caused by G-quadruplex
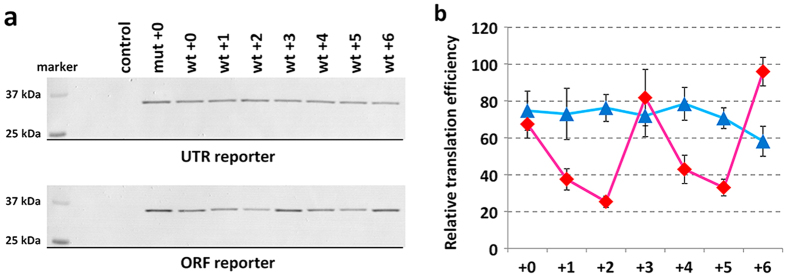
Effects of the G-quadruplexes on protein expression were evaluated in a cell-based assay. Plasmids encoding the reporter mRNAs under control of Cytomegalovirus (CMV) promoter were transfected into MCF7 human breast carcinoma cells. Plasmid vector (pSV40-FLuc), which expresses firefly luciferase under Simian vacuolating virus 40 (SV40) enhancer and early promoter, was co-transfected to normalize transfection efficiency of the experimental samples. Cells were lysed 24 hours after transfection and expression levels of firefly luciferase were evaluated by luminescence signals (see Fig. S1a, in the Supporting Information). Expression levels of Renilla luciferase in the same lysate were evaluated by western blotting using an anti-T7 tag antibody (Fig. 3a). The signal intensities of the western blotting were normalized by luminescence signals of firefly luciferase to obtain relative expression levels of Renilla luciferase (R/F protein ratio) (see Fig. S1b, in the Supporting Information). The R/F protein ratios in cells that expressed the reporter mRNAs with the wild-type G-rich sequence both in the 5′ UTR and in the ORF were lower than in cells that expressed the mRNAs with the mutant G-rich sequence at cognate locations. The R/F protein ratios were periodically changed depending on the position of wild-type G-rich sequence within the ORF while those were the same within experimental error irrespective of the position in the 5′ UTR.
Figure 3. Translation suppression in MCF7 cells caused by RNA G-quadruplexes located in the 5′ UTR and ORF.
(a) Translated products from reporter mRNAs detected by western blotting. (b) Translation efficiencies of 5′ UTR reporter (blue) and ORF reporter (red) mRNAs. The values were calculated by dividing relative R/F protein ratio by relative R/F mRNA ratio (see Figs S2b and S3, in the Supporting Information).
Previous studies have suggested that the transcription of G-rich sequence induced formation of a DNA/RNA hybrid G-quadruplex that suppresses transcription levles39,40,41. To accurately investigate the positional effect of G-quadruplex on translation reaction, levels of mRNA transcripts should be considered. Transcription levels of Renilla luciferase mRNA relative to those of firefly luciferase mRNA (R/F mRNA ratio) were evaluated by real-time PCR (see Fig. S2, in the Supporting Information). Moderately reduced R/F mRNA ratios in the cells that transcribed mRNAs with wild-type G-rich sequences comparing to the cells that transcribed mRNA having mutant G-rich sequence suggest that the transcription of the wild-type G-rich sequence induced formation of the DNA/RNA hybrid G-quadruplex39,40,41 in cells. However, there was no positional effect of the wild-type G-rich sequence on the R/F mRNA ratio irrespective of the 5′ UTR or the ORF.
Intracellular translation efficiencies of the mRNAs with the wild-type G-rich sequence relative to the mRNA with the mutant G-rich sequence were evaluated from R/F protein ratio and R/F mRNA ratio (Fig. 3b). The translation efficiencies of the wild-type G-rich sequence in all positions in the 5′ UTR and the ORF were lower than that of the mutant sequence. The results clearly showed periodic fluctuation of translation suppression at every three nucleotides within the ORF but not within the 5′ UTR, although the translation suppression caused by the ORF wt + 6 was slight and probably within error of the mutant reporter.
General property of the translation suppression in various cell lines
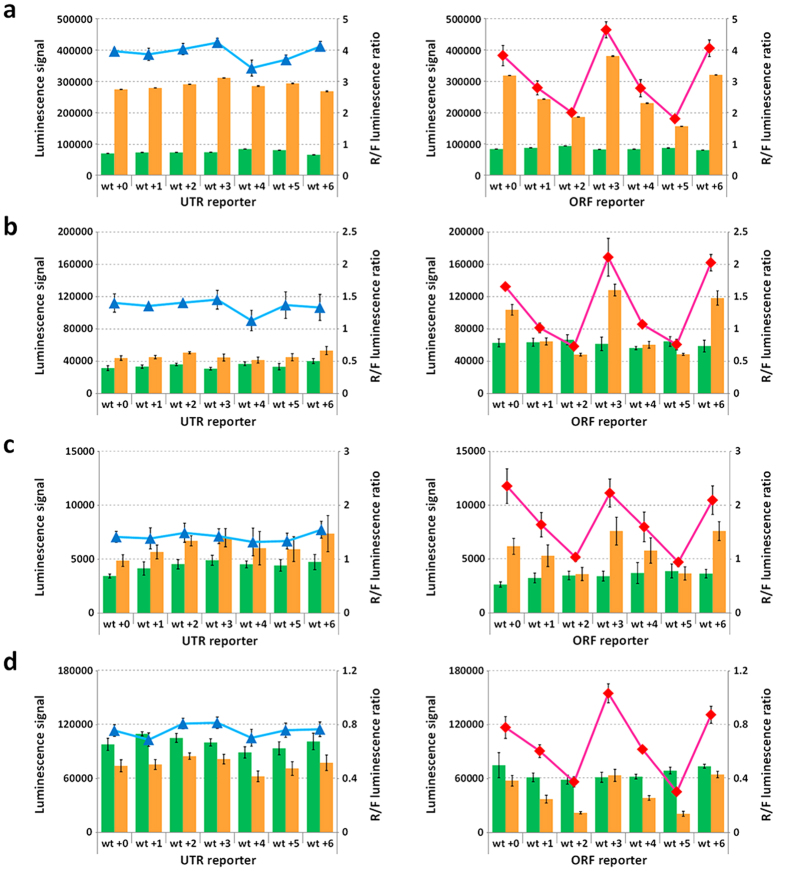
To confirm whether the periodic fluctuation of translation suppression can be observed in different cell lines, we quantitatively analyzed protein expression levels of Renilla luciferase by using dual luciferase assay. We assumed that the differences in amino acid sequences at the G-rich sequence region do not affect the relative activity of Renilla luciferase. This assumption was justified since the luminescence signals of Renilla luciferase relative to firefly luciferase (R/F luminescence ratio) in MCF7 cells (Fig. 4a) almost corresponded to the R/F protein ratio (Fig. S1b, in the Supporting Information). The periodic fluctuations of R/F luminescence ratios within the ORF but not within the 5′ UTR were also observed in human embryonic kidney (Flp-In 293, Invitrogen), cervix epithelioid carcinoma (HeLa), and hepatocellular carcinoma (HepG2) cells (Fig. 4b–d). In addition, the fluctuation of R/F luminescence ratios was also observed when the G-rich sequence variants were located at 792-nucleotides downstream of the start codon (Fig. S3, in the Supporting Information).
Figure 4.
Dual luciferase assay in cell lysates from (a) MCF7, (b) Flp-Iin 293, (c) HeLa, and (d) HepG2 cells transfected with reporter and control plasmids. Luminescence signals of firefly (green) and Renilla (orange) luciferases (left axes) are plotted in bar graphs; R/F luminescence ratios of 5′ UTR reporter (blue) and ORF reporter (red) (right axes) are plotted as data points. Values are means ± S.D. obtained from 4 or 5 wells.
ORF reporter mRNAs having G-rich sequence derived from human E4F transcription factor 1 (E4F1) gene, which is encoding a key regulator protein of p53 tumor suppressor protein42,43, at different positions were also constructed (Fig. S4a, in the Supporting Information). The periodic fluctuations of R/F luminescence ratios at every three nucleotides were observed in all cells tested (Fig. S4b–e, in the Supporting Information). From these results, it was suggested that the positional effect of the G-quadruplex on translation suppression is general property caused by the structure that depends on mechanical and physical properties of the G-quadruplex and the translational machinery.
Discussion
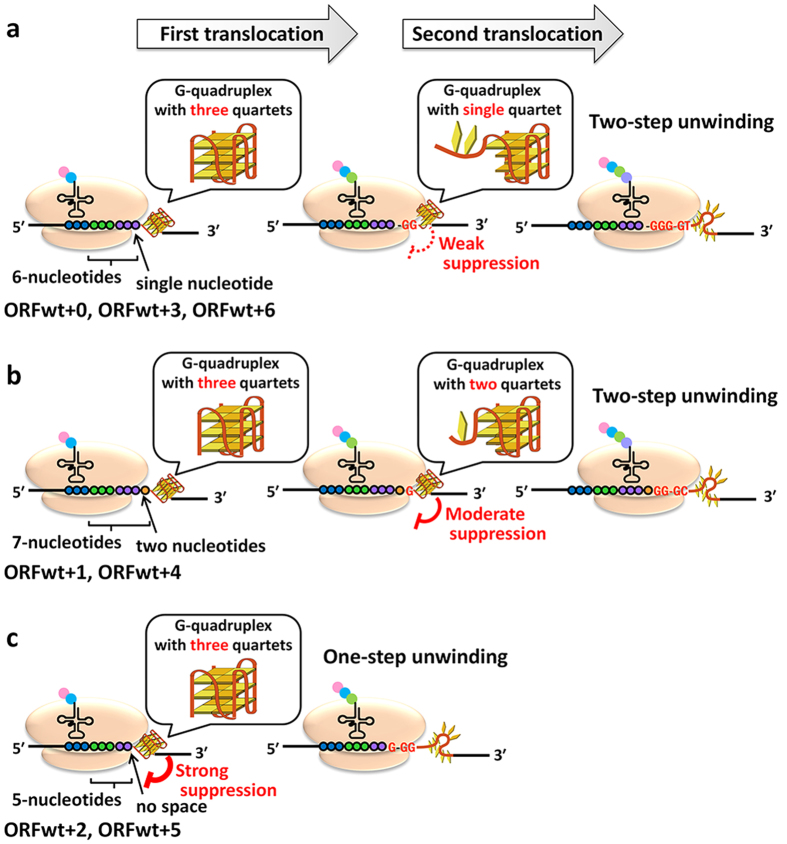
Previous studies demonstrated that, within the 5′ UTR of an mRNA, the extent of translational inhibition depended on stability of the G-quadruplex and its relative position to a site for ribosomal binding44,45. The G-quadruplex positions investigated in the previous studies were dispersed broadly across the 5′ UTR. In contrast to the studies, here, we evaluated effects of displacement of the G-quadruplex within a range of seven adjacent nucleotides with considering levels of mRNA transcripts. We observed no differences in translation efficiency from the UTR reporter mRNAs with the wild-type G-quadruplex sequences at positions 0 (UTR wt + 0) to 6 (UTR wt + 6). The results suggest that small displacement on 5′ UTR do not affect the translation efficiency. In contrast, the translation efficiency fluctuated significantly when the G-quadruplex forming sequences derived from both E.coli and human genes were altered in single-nucleotide steps in the ORF; translation efficiency oscillated with a three-nucleotide period when the G-quadruplex was located in the ORF. The oscillation of the translation suppression was irrespective of the G-rich sequences and their locations within ORF. In our previous in vitro study, translation elongation was halted when there were 5, 6 or 7 nucleotides between a codon in the A-site and the first guanine of the G-rich sequence35. A similar phenomenon was observed in mammalian cells46. Various in vitro mechanical studies and in silico simulations suggest that the G-quadruplex unfolds through intermediate of partially unfolded G-quadruplex or G-triplex, while there are possibilities to form multiple intermediate states15,16,17,18,19,20,47,48. Here, based on these previous reports and results in this study, we hypothesize that the ribosome unwinds the G-quaduplex in two-step translocation through the intermediate of partially unwound G-quadruplex or G-triplex, and propose the model shown in Fig. 5. When the translation elongation is halted before the G-quadruplex formed on ORF wt + 0, wt + 3, and wt + 6 mRNAs, there is a single-nucleotide space between the mRNA entry site of ribosome and the downstream G-quadruplex. In this case, the ribosome overcomes the G-quadruplex region through an intermediate, in which two guanine bases in the quartets are pulled into the ribosome and a single G-quartet is remained (Fig. 5a, middle part). For ORF wt + 1, and wt + 4 mRNAs, the two-step translocation goes through the intermediate, in which a single guanine is pulled into the ribosome and two G-quartets are present, because there is two-nucleotides spacer between the ribosome and the G-quadruplex before the first translocation (Fig. 5b, middle part). The intermediate should be more stable for the ORF wt + 1 and wt + 4 mRNAs than for the ORF wt + 0, wt + 3, and wt + 6 mRNAs, and this correlates with the lower translation efficiencies of ORF wt + 1 and wt + 4 mRNAs compared to those of ORF wt + 0, wt + 3 and wt + 6 mRNAs. The translation efficiency of the ORF wt + 2 and wt + 5 mRNAs were expected to be the lowest as the ribosome should unwind the G-quadruplex by one-step translocation since all quartets of the G-quadruplex are “in frame” with the first translocation (Fig. 5c). This should enforce ribosome the highest energy to overcome the G-quadruplex region.
Figure 5.
Schematic illustration of effects of RNA G-quadruplexes located at (a) wt + 0, wt + 3, and wt + 6, (b) wt + 1 and wt + 4, and (c) wt + 2 and wt + 5 positions of ORF on translation.
Recent studies have suggested that a stable RNA structure within an ORF causes arrhythmic translation by inducing a translational halt49,50,51,52,53. Stalling of a ribosome can result in internal translation initiation52, ribosomal frameshift53, no-go mRNA decay49, or altered co-translational folding of nascent protein51. The results detailed here reveal that the same G-quadruplex forming sequence suppressed translation in both the 5′ UTR or the ORF. More interestingly, the extent of translation suppression due to the G-quadruplex in the ORF depended on the relative position and distance between ribosome and the G-quadruplex at single nucleotide level. We expect that the thermodynamic stabilities of G-quadruplexes formed were the same independent of location because the G-quadruplex forming and flanking sequences were identical. Thus, the intracellular periodic fluctuation of the translation suppression observed when the G-quadruplex is formed in the ORF seems due to differences in motion of ribosome to unwind a G-quadruplex. Triplet periodicity within the ORF region is also observed in natural biological phenomena such as siRNA efficiency and mRNA stabilities54,55,56. Our study sheds light on these intriguing biological fluctuations from mechanics of step-by-step translocation and unfolding of RNA structures by the ribosome.
Methods
Construction of plasmid vectors
Plasmid vectors encoding Renilla luciferase with a G-quadruplex forming sequence in various positions in the 5′ UTR or the ORF under control of a cytomegalovirus promoter were constructed based on the previously described pCMV-TnT-T7-RL reporter vector26. Plasmid vectors for ORF wt + 0 and ORF mut + 0 were previously constructed26. DNA fragments having G-rich sequence derived from E. coli eutE and human E4F1 genes at different positions were prepared by annealing sense and antisense oligonucleotides shown in Tables S1 and S2, in the Supporting Information. The DNA fragments were cloned into NheI and XhoI sites or EcoRI and SalI sites of pCMV-TnT-T7-RL to construct 5′ UTR or ORF reporter vectors, respectively. All sequences inserted into the plasmid were confirmed. The coding sequence of AcGFP was excised from pAcGFP-C1 (Clontech) using NheI and XhoI, and was cloned into same restriction enzyme sites of ORF reporter vector to construct reporter genes having G-rich sequence at the middle of ORF.
Preparation of mRNAs
DNA templates for in vitro transcription of reporter mRNAs were prepared by PCR amplification from the plasmid vectors using T7 promoter primer, 5’-CTTAATACGACTCACTATAGG-3’, and the antisense primer, 5′–CACTGCATTCTAGTTGTGGTTTG–3′. Reporter mRNAs were transcribed using ScriptMAX Thermo T7 Transcription Kit (Toyobo). Transcripts were purified using the RNeasy Mini Kit (Qiagen) and desalted using Centrisep Spin columns (Princeton).
Fluorescence measurements
mRNAs were refolded in a buffer containing 30 mM HEPES-KOH (pH 6.8) and 100 mM KCl by heating to 90 °C followed by cooling at a rate of 1 °C min–1. NMM (5 μM) was mixed with the refolded mRNAs (500 nM) in a buffer containing 30 mM HEPES-KOH (pH 6.8), 100 mM KCl, and 0.1% DMSO. Fluorescence intensity of NMM was measured at 37 °C using a microwell plate reader (Infinite 200 Pro; Tecan) with 400 nm excitation and 615 nm emission.
Transfection
MCF7, Flp-In 293, HeLa, and HepG2 cells were cultured in EMEM, DMEM, DMEM, and EMEM, respectively, supplemented with 10% fetal bovine serum and antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin), and maintained at 37 °C under 5% CO2. Cells were plated on collagen-coated 96- or 24-well plates and incubated overnight. Reporter plasmids (50 or 250 ng) and control plasmid, pSV40-FLuc (5 or 25 ng) were co-transfected into cells cultured on 96- or 24- well plates, respectively, with 0.3 or 1.5 μL of X-tremeGENE 9 (Roche), respectively, according to the manufacturer’s protocol.
Western blotting
Cell lysates in a Passive Lysis Buffer (Promega) were separated on a 12.5% e-PAGEL (Atto). Proteins in the gel were transferred to PVDF membrane using iBlot Dry Blotting System (Thermo Fisher Scientific), and Renilla luciferase was detected by an anti-T7 tag mouse monoclonal antibody (Novagen) and anti-mouse IgG antibody conjugated with alkaline phosphatase (Promega) using iBind Western System (Thermo Fisher Scientific). Signal intensities on the membrane were calculated using Multi Gauge software (Fuji Film) after the membrane was scanned as a tif image.
Real-time PCR
Total RNAs were obtained using the RNeasy Mini Kit (Qiagen) 24 h after transfection and treated with DNase. mRNAs were converted to cDNAs by PrimeScript RT-PCR Kit (Takara) using oligo-dT primer. Levels of firefly and Renilla luciferase mRNAs were evaluated by StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) using primers sets shown in Table S3, in the Supporting Information.
Dual luciferase assay
Cells were lysed in a Passive Lysis Buffer at 24 h after transfection. Aliquots of the lysates were transferred to wells of a 384-well white plate. Luminescence signals from firefly and Renilla luciferases in the lysate were measured using the Dual-Glo Luciferase Assay System (Promega) in a microwell plate reader (Varioskan Flash; Thermo Scientific). Luminescence signal of Renilla luciferase relative to firefly luciferase (R/F luminescence ratio) was calculated after subtraction of luminescence signals obtained from Passive Lysis Buffer only.
Additional Information
How to cite this article: Endoh, T. and Sugimoto, N. Mechanical insights into ribosomal progression overcoming RNA G-quadruplex from periodical translation suppression in cells. Sci. Rep. 6, 22719; doi: 10.1038/srep22719 (2016).
Supplementary Material
Acknowledgments
This work was supported in part by Grants-in-Aid for Scientific Research and MEXT (Japan)-Supported Program for the Strategic Research Foundation at Private Universities (2014–2019) and The Hirao Taro Foundation of KONAN GAKUEN for Academic Research. We thank Misa Kinoshita, Nobuaki Hattori and Kenichi Hase for their help with experiments.
Footnotes
Author Contributions T.E. and N.S. conceived the study; T.E. designed the constructs and performed experiments; and T.E. and N.S. wrote the paper.
References
- Collie G. W. & Parkinson G. N. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 40, 5867–5892 (2011). [DOI] [PubMed] [Google Scholar]
- Nakano S., Miyoshi D. & Sugimoto N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem. Rev. 114, 2733–2758 (2014). [DOI] [PubMed] [Google Scholar]
- Biffi G., Di Antonio M., Tannahill D. & Balasubramanian S. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat. Chem. 6, 75–80 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E. Y., Beraldi D., Tannahill D. & Balasubramanian S. G-quadruplex structures are stable and detectable in human genomic DNA. Nat. Commun. 4, 1796 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G., Tannahill D., McCafferty J. & Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 5, 182–186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaut A. & Balasubramanian S. 5′-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 40, 4727–4741 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S., Moine H. & Vagner S. G-quadruplexes in RNA biology. Wiley Interdiscip. Rev. RNA 3, 495–507 (2012). [DOI] [PubMed] [Google Scholar]
- Pontier D. B., Kruisselbrink E., Guryev V. & Tijsterman M. Isolation of deletion alleles by G4 DNA-induced mutagenesis. Nat. Methods 6, 655–657 (2009). [DOI] [PubMed] [Google Scholar]
- Broxson C., Beckett J. & Tornaletti S. Transcription arrest by a G quadruplex forming-trinucleotide repeat sequence from the human c-myb gene. Biochemistry 50, 4162–4172 (2011). [DOI] [PubMed] [Google Scholar]
- Tateishi-Karimata H., Isono N. & Sugimoto N. New insights into transcription fidelity: thermal stability of non-canonical structures in template DNA regulates transcriptional arrest, pause, and slippage. PLoS ONE 9, e90580 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagihara M., Yoneda K., Yabuuchi H., Okuno Y. & Nakatani K. A reverse transcriptase stop assay revealed diverse quadruplex formations in UTRs in mRNA. Bioorg. Med. Chem. Lett. 20, 2350–2353 (2010). [DOI] [PubMed] [Google Scholar]
- Gray R. D., Buscaglia R. & Chaires J. B. Populated intermediates in the thermal unfolding of the human telomeric quadruplex. J. Am. Chem. Soc. 134, 16834–16844 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaglia R., Gray R. D. & Chaires J. B. Thermodynamic characterization of human telomere quadruplex unfolding. Biopolymers 99, 1006–1018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncina M., Lah J., Prislan I. & Vesnaver G. Energetic basis of human telomeric DNA folding into G-quadruplex structures. J. Am. Chem. Soc. 134, 9657–9663 (2012). [DOI] [PubMed] [Google Scholar]
- de Messieres M., Chang J. C., Brawn-Cinani B. & La Porta A. Single-molecule study of G-quadruplex disruption using dynamic force spectroscopy. Phys. Rev. Lett. 109, 058101 (2012). [DOI] [PubMed] [Google Scholar]
- Yangyuoru P. M. et al. Mechanochemical properties of individual human telomeric RNA (TERRA) G-quadruplexes. Chem. Bio. Chem. 14, 1931–1935 (2013). [DOI] [PubMed] [Google Scholar]
- Li W., Hou X. M., Wang P. Y., Xi X. G. & Li M. Direct measurement of sequential folding pathway and energy landscape of human telomeric G-quadruplex structures. J. Am. Chem. Soc. 135, 6423–6426 (2013). [DOI] [PubMed] [Google Scholar]
- Stadlbauer P., Trantirek L., Cheatham T. E. 3rd, Koca J. & Sponer J. Triplex intermediates in folding of human telomeric quadruplexes probed by microsecond-scale molecular dynamics simulations. Biochimie 105, 22–35 (2014). [DOI] [PubMed] [Google Scholar]
- Stadlbauer P., Krepl M., Cheatham T. E. 3rd, Koca J. & Sponer J. Structural dynamics of possible late-stage intermediates in folding of quadruplex DNA studied by molecular simulations. Nucleic Acids Res. 41, 7128–7143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergues-Pupo A. E., Arias-Gonzalez J. R., Moron M. C., Fiasconaro A. & Falo F. Role of the central cations in the mechanical unfolding of DNA and RNA G-quadruplexes. Nucleic Acids Res. 43, 7638–7647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z. et al. Click chemistry assisted single-molecule fingerprinting reveals a 3D biomolecular folding funnel. J. Am. Chem. Soc. 134, 12338–12341 (2012). [DOI] [PubMed] [Google Scholar]
- Ghimire C. et al. Direct Quantification of Loop Interaction and pi-pi Stacking for G-Quadruplex Stability at the Submolecular Level. J. Am. Chem. Soc. 136, 15537–15544 (2014). [DOI] [PubMed] [Google Scholar]
- Gomez D. et al. A G-quadruplex structure within the 5′-UTR of TRF2 mRNA represses translation in human cells. Nucleic Acids Res. 38, 7187–7198 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder K., Wieland M. & Hartig J. S. Predictable suppression of gene expression by 5′-UTR-based RNA quadruplexes. Nucleic Acids Res. 37, 6811–6817 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin J. D. & Perreault J. P. 5′-UTR G-quadruplex structures acting as translational repressors. Nucleic Acids Res. 38, 7022–7036 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh T., Kawasaki Y. & Sugimoto N. Suppression of gene expression by G-quadruplexes in open reading frames depends on G-quadruplex stability. Angew. Chem. Int. Ed. 52, 5522–5526 (2013). [DOI] [PubMed] [Google Scholar]
- Fischer N., Konevega A. L., Wintermeyer W., Rodnina M. V. & Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature 466, 329–333 (2010). [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75, 434–467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell 15, 1109–1123 (1978). [DOI] [PubMed] [Google Scholar]
- Shoji S., Walker S. E. & Fredrick K. Ribosomal translocation: one step closer to the molecular mechanism. ACS Chem. Biol. 4, 93–107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. et al. Direct measurement of the mechanical work during translocation by the ribosome. eLIFE 3, e03406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T. et al. Initiation factor 2, tRNA, and 50S subunits cooperatively stabilize mRNAs on the ribosome during initiation. Proc. Natl. Acad. Sci. USA 109, 4881–4885 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M. & Imanaka T. mRNA secondary structure in an open reading frame reduces translation efficiency in Bacillus subtilis. J. Bacteriol. 171, 4080–4082 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P., Jenner A. J., Brierley I. & Inglis S. C. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell Biol. 13, 6931–6940 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh T., Kawasaki Y. & Sugimoto N. Translational halt during elongation caused by G-quadruplex formed by mRNA. Methods 64, 73–78 (2013). [DOI] [PubMed] [Google Scholar]
- Endoh T., Kawasaki Y. & Sugimoto N. Stability of RNA quadruplex in open reading frame determines proteolysis of human estrogen receptor alpha. Nucleic Acids Res. 41, 6222–6231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. & Yang D. Sequence, stability, and structure of G-quadruplexes and their interactions with drugs. Curr. Protoc. Nucleic Acid Chem. Chapter 17, Unit17 15 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora A., Nair D. R. & Maiti S. Effect of flanking bases on quadruplex stability and Watson-Crick duplex competition. FEBS J. 276, 3628–3640 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang J. Y., Zheng K. W., Xiao S., Hao Y. H. & Tan Z. Mechanism and manipulation of DNA:RNA hybrid G-quadruplex formation in transcription of G-rich DNA. J. Am. Chem. Soc. 136, 1381–1390 (2014). [DOI] [PubMed] [Google Scholar]
- Zheng K. W. et al. Co-transcriptional formation of DNA:RNA hybrid G-quadruplex and potential function as constitutional cis element for transcription control. Nucleic Acids Res. 41, 5533–5541 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. Y., Zheng K. W., Zhang J. Y., Hao Y. H. & Tan Z. Formation of DNA:RNA Hybrid G-Quadruplex in Bacterial Cells and Its Dominance over the Intramolecular DNA G-Quadruplex in Mediating Transcription Termination. Angew. Chem. Int. Ed. 54, 2447–2451 (2015). [DOI] [PubMed] [Google Scholar]
- Sandy P. et al. p53 is involved in the p120E4F-mediated growth arrest. Oncogene 19, 188–199 (2000). [DOI] [PubMed] [Google Scholar]
- Le Cam L. et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell 127, 775–788 (2006). [DOI] [PubMed] [Google Scholar]
- Kumari S., Bugaut A. & Balasubramanian S. Position and stability are determining factors for translation repression by an RNA G-quadruplex-forming sequence within the 5′ UTR of the NRAS proto-oncogene. Biochemistry 47, 12664–12669 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holder I. T. & Hartig J. S. A Matter of Location: Influence of G-Quadruplexes on Escherichia coli Gene Expression. Chem. Biol. 21, 1511–1521 (2014). [DOI] [PubMed] [Google Scholar]
- Endoh T. & Sugimoto N. Unusual -1 ribosomal frameshift caused by stable RNA G-quadruplex in open reading frame. Anal. Chem. 85, 11435–11439 (2013). [DOI] [PubMed] [Google Scholar]
- Wu W. Q., Hou X. M., Li M., Dou S. X. & Xi X. G. BLM unfolds G-quadruplexes in different structural environments through different mechanisms. Nucleic Acids Res. 43, 4614–4626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvam S., Yu Z. & Mao H. Exploded view of higher order G-quadruplex structures through click-chemistry assisted single-molecule mechanical unfolding. Nucleic Acids Res. 44, 45–55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M. K. & Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J. R. & Stansfield I. Halting a cellular production line: responses to ribosomal pausing during translation. Biol. Cell 99, 475–487 (2007). [DOI] [PubMed] [Google Scholar]
- Watts J. M. et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460, 711–716 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J. et al. Ribosome stalling regulates IRES-mediated translation in eukaryotes, a parallel to prokaryotic attenuation. Mol. Cell 17, 405–416 (2005). [DOI] [PubMed] [Google Scholar]
- Kontos H., Napthine S. & Brierley I. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell Biol. 21, 8657–8670 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y. et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700 (2014). [DOI] [PubMed] [Google Scholar]
- Shabalina S. A., Ogurtsov A. Y. & Spiridonov N. A. A periodic pattern of mRNA secondary structure created by the genetic code. Nucleic Acids Res. 34, 2428–2437 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T. & Suzuki T. Specific residues at every third position of siRNA shape its efficient RNAi activity. Nucleic Acids Res. 35, e27 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.